Abstract
Spinal infections pose a significant clinical challenge due to the difficulty of early diagnosis. Traditional Tissue Culture Technique (TCT) has limitations, while Metagenomic Next-Generation Sequencing (mNGS) has emerged as a promising diagnostic tool. However, reports on the diagnostic performance of mNGS and TCT for spinal infections are inconsistent, and there has been a lack of systematic analysis of the evidence. This systematic review and meta-analysis included 10 studies involving a total of 770 patients to compare the diagnostic accuracy of mNGS and TCT. A comprehensive literature search was conducted using PubMed, Embase, Web of Science, Cochrane Library, and SinoMed to identify studies evaluating the diagnostic accuracy of mNGS and TCT for spinal infections. Data were analyzed using Review Manager 5.3 and Stata 16.0 to compute the sensitivity, specificity, and Area Under the Summary Receiver Operating Characteristic Curve (AUC). The meta-analysis revealed pooled estimates for mNGS with a sensitivity of 0.81 (95% CI, 0.74–0.87), specificity of 0.75 (95% CI, 0.48–0.91), and an AUC of 0.85 (95% CI, 0.82–0.88). In contrast, pooled estimates for TCT showed a sensitivity of 0.34 (95% CI, 0.27–0.43), specificity of 0.93 (95% CI, 0.79–0.98), and an AUC of 0.59 (95% CI, 0.55–0.63). While mNGS demonstrates higher sensitivity and overall diagnostic accuracy than TCT, its lower specificity suggests that it may be most effective when used alongside conventional methods to enhance diagnostic reliability in spinal infections.
Similar content being viewed by others
Introduction
Spinal infection, defined as an infectious process involving the vertebral body, intervertebral disc, paraspinal soft tissue, or epidural space and confirmed by clinical presentation, imaging findings, and microbiological or histopathological evidence, presents significant diagnostic challenges due to its insidious onset, non-specific symptoms, and often inconclusive laboratory findings1,2,3,4,5,6. Timely and accurate identification of pathogens is essential for guiding appropriate antimicrobial therapy. Traditionally, diagnosis has relied on isolating pathogens from cultured spinal tissue or fluid samples; however, the low detection rates of conventional microbiological cultures pose substantial challenges for accurate diagnosis and targeted antimicrobial therapy7,8.
Metagenomic next-generation sequencing (mNGS) has emerged as a promising diagnostic method for infectious diseases due to its ability to detect a broad range of pathogens, including rare and unculturable organisms9,10,11,12,13. Compared to TCT, mNGS typically offers faster turnaround and higher detection rates. However, its diagnostic accuracy in spinal infections remains controversial, with studies reporting considerable variability in sensitivity and specificity.
For instance, Zhang Yi et al.14, in a prospective study involving 38 patients with suspected spinal infections, found that mNGS had a sensitivity of 0.86 and specificity of 1.00, while TCT showed a sensitivity of 0.49 and the same specificity of 1.00. In contrast, Ma Chi-yuan et al.15 reported lower diagnostic performance, with mNGS yielding a sensitivity of 0.69 and specificity of 0.27, and TCT showing even poorer sensitivity at 0.15 but a similarly high specificity of 1.00. These inconsistencies may be attributed to differences in sample sizes, specimen acquisition methods (e.g., open surgery versus percutaneous biopsy), and sequencing platforms used across studies. These variations highlight the need for a systematic evaluation to synthesize current evidence and identify the true diagnostic performance of mNGS compared to TCT.
To date, no meta-analysis has consolidated the diagnostic efficacy of mNGS and TCT in spinal infections. This study aims to assess the diagnostic value of both mNGS and TCT for spinal infections by including analyses from studies that utilized both testing methods.
Materials and methods
Search strategy
This study was registered in the PROSPERO database (CRD42022383002) and adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The PubMed, Embase, web of science, Cochrane library and SinoMed databases were searched until December 2023, respectively. The literature on the comparative diagnostic accuracy of mNGS and TCT was searched using specific terms including “mNGS”, “metagenomic next-generation sequencing”, “culture”, “spinal”, “spine”, “infection”, “infective”, and “spinal infection”.
Inclusion and exclusion criteria
Inclusion Criteria: (1) Studies involving subjects with suspected spinal infections; (2) Studies comparing the diagnostic performance of mNGS and TCT; (3) Studies containing necessary data for extraction, such as counts of False Positives (FP), True Positives (TP), False Negatives (FN), and True Negatives (TN); (4) Diagnostic criteria based on histopathological tests and the Infectious Diseases Society of America (IDSA) standards16. Exclusion Criteria: (1) Studies focusing solely on mNGS or TCT; (2) Studies lacking comparative analysis of mNGS and TCT; (3) Studies with incomplete data; (4) Case reports, animal studies, limited analyses, and review comments.
Data extraction
Two independent researchers(YZY, YCL) initially screened titles and abstracts according to the inclusion and exclusion criteria, followed by full-text reviews of eligible studies. Discrepancies were resolved through discussion. Common reference standards used for diagnosing spinal infections included histopathological tests and IDSA criteria. Extracted data comprised: (1) the first author’s name, year of publication, and study type; (2) number of cases and distribution in the diagnostic 4-fold table (FP, TP, FN, TN); (3) type of mNGS sequencing platform; (4) sampling method; (5) diagnostic reference standard for spinal infections.
Quality assessment
The Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool was used to evaluate the methodological quality of included studies across four domains: patient selection, index test, reference standard, and flow/timing. Two independent researchers performed the assessment, with discrepancies resolved through group consensus. Studies classified as high risk in any domain were retained in the primary analysis but subjected to sensitivity analysis by excluding them to assess their impact on pooled estimates. Additionally, meta-regression was performed to evaluate whether QUADAS-2 risk categories (low/unclear/high) explained heterogeneity in sensitivity or specificity. No studies were excluded solely based on QUADAS-2 scores unless critical methodological flaws (e.g., non-consecutive enrollment without justification, lack of blinding between index test and reference standard) rendered diagnostic accuracy data unreliable.
Statistical analysis
Review Manager 5.3 and Stata 16.0 were used to calculate pooled sensitivity, specificity, and 95% confidence intervals (CI) using a random-effects model to account for anticipated heterogeneity across studies. Summary Receiver Operating Characteristic (SROC) curves and the Area Under the Curve (AUC) were generated. Heterogeneity was quantified using the Q-test and I² statistic, with I² ≥ 50% indicating substantial heterogeneity. When significant heterogeneity (I² ≥ 50% or p < 0.1) was detected, threshold effect analysis, sensitivity analysis (e.g., excluding studies with high QUADAS-2 risk scores), and meta-regression were performed to identify sources of variability. All statistical tests used a significance threshold of p < 0.05.
Results
Search results and quality assessment
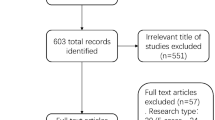
The article selection process followed the PRISMA guidelines and is depicted in Fig. 1. A comprehensive search across the specified databases yielded 1,406 articles. After duplicate removal, 623 articles remained. Initial screening of titles and abstracts narrowed the selection to 22 articles. Full-text review led to the exclusion of 12 articles for various reasons: six did not meet the inclusion criteria, two lacked necessary data for extraction, three were case reports, and one was a narrative review. Ultimately, 10 studies were included in this review, consisting of two prospective and eight retrospective studies, involving a total of 770 patients. The basic characteristics of the included studies are summarized in Table 1. The methodological quality of these studies was assessed using the QUADAS-2 scale, as illustrated in Figs. 2 and 3. Most studies exhibited a low risk of bias concerning reference standards, procedures, and timing. However, four studies presented an unclear risk of bias regarding consecutive inclusion.
Meta-analysis results
Efficacy of the two testing methods
mNGS demonstrated a pooled sensitivity of 0.81 (95% CI, 0.74–0.87) and specificity of 0.75 (95% CI, 0.48–0.91) (Fig. 4). In contrast, TCT showed a pooled sensitivity of 0.34 (95% CI, 0.27–0.43) and specificity of 0.93 (95% CI, 0.79–0.98) (Fig. 5). The positive likelihood ratio (PLR) for mNGS was 3.30 (95% CI, 1.30–7.90) and for TCT was 5.00 (95% CI, 1.70–14.80), while the negative likelihood ratio (NLR) was 0.25 (95% CI, 0.15–0.41) for mNGS and 0.70 (95% CI, 0.63–0.79) for TCT. In clinical diagnostic interpretation, a PLR > 10 or NLR < 0.1 is generally considered to provide strong diagnostic evidence. Neither mNGS (PLR = 3.30, NLR = 0.25) nor TCT (PLR = 5.00, NLR = 0.70) met these criteria, demonstrating that neither method alone can definitively confirm or exclude spinal infection. The wide confidence intervals for mNGS specificity (0.48–0.91) and TCT sensitivity (0.27–0.43) further emphasize substantial variability across studies. These findings underscore the necessity of a combined diagnostic strategy integrating clinical evaluation, imaging, and microbiological testing to optimize diagnostic accuracy. To further evaluate diagnostic accuracy, SROC curves were plotted, revealing an AUC of 0.85 (95% CI, 0.82–0.88) for mNGS (Fig. 6) and 0.59 (95% CI, 0.55–0.63) for TCT (Fig. 7).
Heterogeneity analysis
Sensitivity analysis was conducted on the 10 studies included, assessing Goodness-of-fit, Bivariate normality, Influence analysis, and Outlier detection for the TCT group, as shown in Fig. 8. However, Influence analysis and Outlier detection identified studies by Yuan Li 202221, Guanzhong Wang 202322, and Shi Shiyuan 202223 as potential sources of heterogeneity in the mNGS group (Fig. 9 and Supplementary Fig.S1). The Spearman correlation coefficient for sensitivity and specificity was 0.043 (p = 0.907) in the mNGS group and 0.411 (p = 0.238) in the TCT group, indicating that heterogeneity was not due to the threshold effect in either group. Further univariable meta-regression analysis was performed to explore the sources of heterogeneity. It was found that in the mNGS group, sensitivity was affected by sample size, and specificity was influenced by the sampling method. In the TCT group, sensitivity was influenced by the sampling method, while specificity was affected by both the sample size and the diagnostic reference standard (Tables 2 and 3, Supplementary Fig.S2 and S3).
Additionally, the Deeks funnel plot asymmetry test showed no significant publication bias in either the mNGS group (p = 0.59) or the TCT group (p = 0.96) (Supplementary Fig.S4 and S5).
In addition, meta-regression was performed to assess whether sequencing platform influenced diagnostic performance. No significant difference was observed in sensitivity (p = 0.06) or specificity (p = 0.95) between the Illumina and MGISEQ platforms.
Discussion
In recent years, mNGS has been increasingly utilized for diagnosing spinal infections, achieving promising clinical outcomes. In contrast, TCT, a classical method in the diagnosis of spinal infections, often suffers from low culture-positive rates. Consensus on the relative efficacy of these two diagnostic methods is still lacking. Consequently, this study reviewed the existing literature with the hope of filling this gap and providing a foundation for evidence-based clinical decision-making.
Most studies have indicated that mNGS offers significant clinical value in diagnosing spinal infections14,15,17. Our findings demonstrate that mNGS is more sensitive than TCT (0.81 vs. 0.34), albeit with a lower specificity (0.75 vs. 0.93). The PLR for mNGS and TCT was 3.30 and 5.00, respectively, while the NLR was 0.25 for mNGS and 0.70 for TCT. It is widely recognized that a PLR > 10 or an NLR < 0.1 can decisively confirm or exclude a diagnosis. Our findings suggest that neither mNGS nor TCT alone can definitively confirm or rule out spinal infections. However, mNGS exhibited not only higher sensitivity but also a more favorable diagnostic efficacy and a higher AUC compared to TCT, underscoring its potential as a more effective diagnostic tool in clinical settings.
The studies included in this analysis exhibited considerable heterogeneity. Meta-regression results indicated that small sample sizes (n < 50) and sampling methods contributed to the heterogeneity observed in the mNGS groups, while for the TCT group, the same factors plus reference standards were influential. Specifically, five studies14,15,19,22,24 had sample sizes under 50, potentially affecting the pooled results for both groups. In terms of sampling techniques, percutaneous puncture was used exclusively in three studies, while a combination of open surgery and percutaneous puncture was employed in seven. Percutaneous puncture biopsy is a critical method for obtaining specimens in spinal infections25 and is noted for minimizing skin contamination at the puncture site26. Jakrapun et al.27 conducted a meta-analysis on the efficacy of image-guided percutaneous puncture vertebral biopsy in spinal infections, demonstrating that sampling methods significantly impact diagnostic outcomes. Auid-Orcid et al.28 investigated sampling methods from 31 patients with suspected spinal tuberculosis. They found that the sensitivity for diagnosing tuberculosis was significantly higher with open surgical biopsy compared to percutaneous Xpert testing. Similarly, the specificity also showed a significant difference, being higher in open surgical biopsy versus percutaneous Xpert testing. Jin et al.29 reported similar findings, where the sensitivity of Xpert for diagnosing tuberculosis varied between open surgery (0.84) and percutaneous puncture (0.77). Our findings indicate that the specificity of mNGS and the sensitivity of TCT could be influenced by the sampling methods used. Additionally, this study’s results suggest that the specificity of TCT is affected by the reference standards employed, which may contribute to TCT’s lower sensitivity and relatively poor detection capability.
Different detection platforms have varying criteria for microbial positivity, which can impact test outcomes30,31. Zhang et al.32 utilized both Illumina and Nanopore mNGS platforms to assess their ability to detect microorganisms in alveolar lavage fluid. Their findings indicated that the Nanopore platform was superior in detecting Mycobacterium tuberculosis and fungi. Li Yulian et al.33 conducted a meta-analysis on the use of mNGS to detect Mycobacterium tuberculosis in the lungs, highlighting that variations in detection platforms could contribute to the heterogeneity in sensitivity observed. The mNGS platforms discussed in this paper include Illumina NextSeq and MGISEQ. The Illumina NextSeq platform is noted for its higher gene coverage, while the MGISEQ platform boasts greater sensitivity34. Nevertheless, our analysis found that the choice of detection platform did not account for the heterogeneity observed within the mNGS group. Meta-regression analysis revealed no statistically significant difference in diagnostic performance between the Illumina and MGISEQ platforms (sensitivity: p = 0.06; specificity: p = 0.95). However, the small number of Illumina-based studies (n = 4) may have limited the statistical power to detect subtle differences. These findings are consistent with a previous meta-analysis by Sike et al.35, which evaluated mNGS for diagnosing cerebrospinal fluid infections in pediatric patients and found that diagnostic performance did not significantly vary across Illumina and BGISEQ platforms.
Although meta-regression was employed to investigate the sources of heterogeneity in each group, it could not fully account for the observed variability. This study has several limitations potentially linked to various factors: (1) Antibiotic exposure prior to sampling could influence outcomes, as pre-sampling antibiotic use or brief discontinuation might eradicate microorganisms in tissues, thus affecting detection rates in both groups7,36,37. Husseini et al.38 recommend suspending antibiotics two weeks before a puncture biopsy to optimize microbial yield; (2) The results might also be influenced by the location and timing of the sample collection. Anderson et al.39 found that pus from punctures yields a higher positivity rate than bone or disc tissues, and biopsies from the upper thoracic spine show higher positivity rates. Alessandro et al.40 investigated factors affecting tissue cultures and found no significant differences related to the volume of tissue extracted or the biopsy area (bone tissue vs. intervertebral discs); however, sampling during the acute phase notably increased microbial detection rates; (3) mNGS can theoretically detect all potential pathogens in a sample by extracting nucleic acids41,42,43, but the sequencing platform’s dataset length might impact the accuracy of detection and the estimation of abundance44. Interpretation of mNGS results could be skewed by the presence of underlying microbiota, sample contamination, and host nucleic acids45. Four studies in our analysis reported possible contamination-related false positives, accounting for approximately 5.6% of mNGS-positive samples (15 out of 266), primarily involving low-abundance or environmental organisms. Regrettably, most of the 10 studies included did not fully report such data, preventing a thorough analysis of the relevant heterogeneity. This reinforces the importance of interpreting sequencing results in conjunction with clinical findings, conventional microbiology, and imaging; (4) Due to the high sensitivity of mNGS, there is an inherent risk of false-positive results stemming from sample contamination, environmental microorganisms, or colonizing species, particularly in specimens with low microbial burden. This limitation underscores the need for cautious interpretation of mNGS results and highlights the importance of integrating molecular findings with clinical judgment, imaging studies, and conventional microbiology to enhance diagnostic accuracy; (5) In addition to methodological concerns, practical barriers such as cost and accessibility limit the clinical applicability of mNGS. The test is expensive, requires advanced sequencing platforms, and depends on trained personnel and bioinformatics infrastructure—resources that may not be readily available in many healthcare settings. These economic and logistical constraints should be considered when evaluating the feasibility of widespread implementation; (6) Although the study included dual-test data from the same patient cohorts, we did not apply a contrast-based or arm-based model due to limitations in cross-tabulated reporting and the complementary nature of our research objective. Future studies may explore these models in head-to-head comparisons with standardized paired data; (7) Due to inconsistent or missing pathogen-specific data across the included studies, we were unable to assess whether diagnostic performance varied by microorganism type. Future studies incorporating standardized microbiological classifications may help clarify test-specific strengths for different spinal pathogens.
A key strength of this study is that, to the best of our knowledge, it represents the first meta-analysis to systematically and directly compare the diagnostic performance of mNGS and TCT specifically in spinal infections. This study offers a distinct contribution to the current body of literature. By synthesizing data from ten studies across varied patient populations, sampling methods, and sequencing platforms, this analysis provides robust evidence to guide clinical practice.
Conclusions
This study represents the first comparative meta-analysis of the diagnostic performance of mNGS versus TCT in spinal infections. The results demonstrate that mNGS provides higher sensitivity and overall diagnostic accuracy compared to TCT. However, its lower specificity indicates a risk of false positives, suggesting that mNGS may be more effective when used in conjunction with traditional diagnostic methods such as TCT to improve diagnostic confidence.
Data availability
The datasets are available from the corresponding author upon reasonable request.
Abbreviations
- mNGS:
-
Metagenomic next generation sequencing
- TCT:
-
Tissue culture technique
- AUC:
-
Area under the summary receiver operating characteristic curve
- PRISMA:
-
Preferred reporting items for systematic reviews and meta-analyses
- SROC:
-
Summary receiver operating characteristic
- CI:
-
Confidence interval
- FP:
-
False positives
- TP:
-
True positives
- FN:
-
False negatives
- TN:
-
True negatives
- IDSA:
-
Infectious Diseases Society of America
- PLR:
-
The positive likelihood ratio
- TN:
-
The negative likelihood ratio
References
Galhotra, R. D. et al. Utility of magnetic resonance imaging in the differential diagnosis of tubercular and pyogenic spondylodiscitis. J. Nat. Sci. Biol. Med. 6(2), 388–393 (2015).
Loke, T. K., Ma, H. T. & Chan, C. S. Magnetic resonance imaging of tuberculous spinal infection. Australas Radiol. 41(1), 7–12 (1997).
Lee, C. S. et al. Deep spinal infection after outpatient epidural injections for pain: A retrospective sample cohort study using a claims database in South Korea. Anesthesiology 134(6), 925–936 (2021).
Street, K. J., White, S. G. & Vandal, A. C. Clinical prevalence and population incidence of serious pathologies among patients undergoing magnetic resonance imaging for low back pain. Spine J. 20(1), 101–111 (2020).
Dayer, R. et al. Laboratory diagnostics for primary spinal infections in pediatric and adult populations: a narrative review. N Am. Spine Soc. J. 16, 100270 (2023).
Zhang, G. et al. Clinical application value of metagenomic next-generation sequencing in the diagnosis of spinal infections and its impact on clinical outcomes. Front. Cell. Infect. Microbiol. 13, 1076525 (2023).
Kim, C. J. et al. Microbiologically and clinically diagnosed vertebral osteomyelitis: impact of prior antibiotic exposure. Antimicrob. Agents Chemother. 56(4), 2122–2124 (2012).
Lee, Y. et al. Changes of biomarkers before and after antibiotic treatment in spinal infection. Korean J. Neurotrauma. 15(2), 143–149 (2019).
Xie, S. et al. Clinical features of varicella-zoster virus caused neurological diseases detected by metagenomic next-generation sequencing. Open. Med. (wars). 18(1), 20230744 (2023).
Shanmuganathan, R. et al. Comparative metagenomic analysis of human intervertebral disc nucleus pulposus and cartilaginous end plates. Front. Cardiovasc. Med. 9, 927652 (2022).
Yang, S. et al. Spinal infection caused by Coxiella burnetii. BMC Infect. Dis. 23(1), 6 (2023).
Chen, S., Kang, Y., Li, D. & Li, Z. Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in Bronchoalveolar lavage fluid in patients with pulmonary infections: systematic review and meta-analysis. Int. J. Infect. Dis. 122, 867–873 (2022).
Tan, J. et al. The effectiveness of metagenomic next-generation sequencing in the diagnosis of prosthetic joint infection: A systematic review and meta-analysis. Front. Cell. Infect. Microbiol. 12, 875822 (2022).
Zhang, Y. et al. Evaluation of the metagenomic next-generation sequencing performance in pathogenic detection in patients with spinal infection. Front. Cell. Infect. Microbiol. 12, 967584 (2022).
Ma, C. et al. The potential of metagenomic next-generation sequencing in diagnosis of spinal infection: a retrospective study. Eur. Spine J. 31(2), 442–447 (2022).
Berbari, E. F. et al. 2015 infectious diseases society of America (IDSA) clinical practice guidelines for the diagnosis and treatment of native vertebral osteomyelitis in adults. CLIN. INFECT. DIS. 61(6), e26–46 (2015).
Huang, H. et al. Pathogen detection in suspected spinal infection: metagenomic next-generation sequencing versus culture. Eur. Spine J. 32(12), 4220–4228 (2023).
Xu, L., Zhou, Z., Wang, Y., Song, C. & Tan, H. Improved accuracy of etiological diagnosis of spinal infection by metagenomic next-generation sequencing. Front. Cell. Infect. Microbiol. 12, 929701 (2022).
Lin, W. et al. Diagnostic performance of metagenomic next-generation sequencing and conventional microbial culture for spinal infection: a retrospective comparative study. Eur. Spine J. 32(12), 4238–4245 (2023).
Cheng, H. et al. Diagnostic efficacy of metagenomic next-generation sequencing in patients with spinal infections: A retrospective study. Infect. Drug Resist. 16, 7613–7620 (2023).
Li, Y. et al. Diagnostic efficiency of metagenomic next-generation sequencing for suspected spinal tuberculosis in china: A multicenter prospective study [published correction appears in front microbiol. 2023;14:1198931]. Front. Microbiol. 13, 1018938 (2022).
Wang, G. et al. Application of metagenomic next-generation sequencing in the detection of pathogens in spinal infections. Spine J. 23(6), 859–867 (2023).
Shiyuan, S. Application value of metagenomic next generation sequencing in the diagnosis of non-tuberculous spinal infection. Chin. J. Orthop. 42(15), 961–967 (2022).
Zhang, D. et al. Etiological diagnosis value of metagenomic next generation sequencing in the patients with spinal infection. Chin. J. Infect. Dis. 41(3), 208–213 (2023).
Watt, J. P. & Davis, J. H. Percutaneous core needle biopsies: the yield in spinal tuberculosis. S Afr. Med. J. 104(1), 29–32 (2013).
Salzberg, S. L. et al. Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol. Neuroimmunol. Neuroinflamm. 3(4), e251 (2016).
Pupaibool, J., Vasoo, S., Erwin, P. J., Murad, M. H. & Berbari, E. F. The utility of image-guided percutaneous needle aspiration biopsy for the diagnosis of spontaneous vertebral osteomyelitis: a systematic review and meta-analysis. Spine J. 15(1), 122–131 (2015).
Waters, R. et al. Higher sensitivity of Xpert MTB/RIF ultra over tuberculosis culture for the diagnosis of spinal tuberculosis with open or computed Tomography-Guided biopsies. Open. Forum Infect. Dis. 11(1), ofad621 (2023).
Jin, Y. H. et al. Application value of Xpert MTB/RIF in diagnosis of spinal tuberculosis and detection of Rifampin resistance. Zhongguo Gu Shang. 30(9), 787–791 (2017).
Xing, X. W. et al. Metagenomic Next-Generation sequencing for diagnosis of infectious encephalitis and meningitis: A large, prospective case series of 213 patients. Front. Cell. Infect. Microbiol. 10, 88 (2020).
Miller, S. et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 29(5), 831–842 (2019).
Zhang, J. et al. Clinical value of metagenomic next-generation sequencing by illumina and nanopore for the detection of pathogens in Bronchoalveolar lavage fluid in suspected community-acquired pneumonia patients. Front. Cell. Infect. Microbiol. 12, 1021320 (2022).
Li, Y., Bian, W., Wu, S., Zhang, J. & Li, D. Metagenomic next-generation sequencing for Mycobacterium tuberculosis complex detection: a meta-analysis. Front. Public. Health. 11, 1224993 (2023).
Kanaujia, R., Biswal, M., Angrup, A. & Ray, P. Diagnostic accuracy of the metagenomic next-generation sequencing (mNGS) for detection of bacterial meningoencephalitis: a systematic review and meta-analysis. Eur. J. Clin. Microbiol. Infect. Dis. 41(6), 881–891 (2022).
He, S. et al. Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in cerebrospinal fluid in pediatric patients with central nervous system infection: a systematic review and meta-analysis. BMC Infect. Dis. 24(1), 103 (2024).
Ness, T. E. et al. Optimizing DNA extraction from pediatric stool for diagnosis of tuberculosis and use in Next-Generation sequencing applications. Microbiol. Spectr. 11(1), e0226922 (2023).
Zhao, M. et al. Metagenomic Next-Generation sequencing improves diagnosis of osteoarticular infections from abscess specimens: A multicenter retrospective study. Front. Microbiol. 11, 2034 (2020).
Husseini, J. S. & Huang, A. J. Discitis-osteomyelitis: optimizing results of percutaneous sampling. Skeletal Radiol. 52(10), 1815–1823 (2023).
Kuo, A. H., Cho, C. H., Huang, R. Y., Kim, C. J. & Lee, T. C. Target-specific yield rate and clinical utility of percutaneous tissue sampling in spinal infection. Clin. Imaging. 68, 257–262 (2020).
Cannavale, A. et al. Percutaneous spine biopsy under cone beam computed tomography guidance for spondylodiscitis: time is diagnosis. Neuroradiol. J. 34(6), 607–614 (2021).
Gu, W., Miller, S. & Chiu, C. Y. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338 (2019).
Chien, J. Y., Yu, C. J. & Hsueh, P. R. Utility of metagenomic next-generation sequencing for etiological diagnosis of patients with sepsis in intensive care units. Microbiol. Spectr. 10(4), e0074622 (2022).
Yu, Y., Wang, S., Dong, G. & Niu, Y. Diagnostic performance of metagenomic next⁃generation sequencing in the diagnosis of prosthetic joint infection using tissue specimens. Infect. Drug Resist. 16, 1193–1201 (2023).
Portik, D. M., Brown, C. T. & Pierce-Ward, N. T. Evaluation of taxonomic classification and profiling methods for long-read shotgun metagenomic sequencing datasets. BMC Bioinform. 23(1), 541 (2022).
Thoendel, M. et al. Impact of contaminating DNA in whole-genome amplification kits used for metagenomic shotgun sequencing for infection diagnosis. J. Clin. Microbiol. 55(6), 1789–1801 (2017).
Funding
This research was funded by Quzhou Science and Technology Tackling Project (2022K57).
Author information
Authors and Affiliations
Contributions
Yan-Cheng Li and Zhi-Yuan Yao conceptualized the study. Yan-Xiao Liu, Lei Li and Hua Wang helped in literature search. Zhi-Yuan Yao and Yan-Cheng Li extracted the data and assessed the quality. Yan-Cheng Li and Lei Li developed the software. Zhi-Yuan Yao and Hua Wang helped in formal analysis. Zhi-Yuan Yao and Hua Wang validated the study. Yan-Cheng Li wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.




Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, YC., Liu, YX., Li, L. et al. Metagenomic next-generation sequencing (mNGS) versus tissue culture technique (TCT) in diagnosis of spinal infection: a systematic review and meta-analysis. Sci Rep 15, 20926 (2025). https://doi.org/10.1038/s41598-025-06759-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-06759-3