Abstract
Prohibitin2 (PHB2) is a mitochondrial endosomal protein that is closely linked to tumors; however, its specific molecular role remains unclear. Therefore, this study investigates the role of PHB2 in cancer. Multiple databases, including cBioPortal, PhosphoNET, and AlphaFold, were accessed to investigate this issue. The findings indicated that PHB2 expression in tumor tissues exceeded that in normal tissues, with PHB2 localized in mitochondria. Patients diagnosed with bladder and lung adenocarcinomas exhibiting elevated PHB2 expression demonstrated an increased survival rate compared to those with reduced expression levels. Conversely, the opposite was observed in patients with renal clear cell carcinoma and breast cancer. In pan-cancer, the mutation rate of PHB2 was elevated, with several overlapping amino acid sites exhibiting tumor mutations and post-translational modifications, including phosphorylation at tyrosine 121, acetylation at threonine 263, and ubiquitination at lysine 296. Multiple overlapping sites of amino acid phosphorylation modifications and mutations were identified in PHB2, specifically at threonine 42, tyrosine 121, and threonine 263. The three-dimensional predicted structure of PHB2 was analyzed utilizing the AlphaFold database. The pathogenicity mutation heatmap indicated that the overlapping sites of amino acid mutations and post-translational modifications, including phosphorylation (threonine 42, tyrosine 121, and threonine 263), acetylation, and ubiquitination (lysine 296), were significantly pathogenic. In conclusion, PHB2 exhibits cancer-type-specific prognostic associations. Its overlapping mutation and modification sites suggest potential functional importance, supporting its role as a candidate biomarker for further validation in pan-cancer contexts.
Similar content being viewed by others
Malignant tumors represent a significant risk to human health and survival1,2. The absence of effective early diagnostic methods, coupled with the nonspecific symptoms in the initial stages of the disease, leads to the majority of patients being diagnosed at advanced stages, which adversely affects prognosis3,4. “Early detection, diagnosis, and treatment” are essential for the survival and prognosis of patients with malignant tumors5,6. Given the critical need for early detection, identifying reliable candidate biomarkers such as prohibitin2 (PHB2) could significantly enhance outcomes for patients with malignant tumors7.
PHB2 is a crucial gene for the self-renewal of human embryonic stem cells (hESCs)8. This protein encodes a highly conserved membrane scaffolding component recently recognized as a novel receptor in the mitochondrial inner membrane9. It exhibits multiple functions that are mediated in a cell-compartment- and tissue-specific manner10including roles in plasma membrane-associated cell signaling, mitochondrial chaperoning, and acting as a transcriptional co-regulator for transcription factors and sex steroid hormones within the nucleus11,12. Evidence indicates that PHB2 is significant in bladder cancer (BLCA), primary liver carcinoma, and colorectal cancer, as it facilitates cancer cell proliferation and tumorigenesis via oxidative phosphorylation13,14. Further research is necessary to investigate additional tumorigenic associations of PHB2 and their effects on the survival prognosis of cancer patients15.
This study aims to elucidate the expression patterns of PHB2 in pan-cancer and assess its potential as a diagnostic candidate biomarker, thereby contributing to improved early detection strategies.
Methods
Gene expression profiling interactive analysis (GEPIA) data download and analysis
Upon accessing the GEPIA database (http://gepia.cancer-pku.cn/) and entering “PHB2” in the search function, we conducted an analysis of the pan-cancer expression of PHB2 in both normal and tumor tissues. Subsequently, we examined the relationship between PHB2 expression levels and patient survival rates.
Tumor immune estimation resource (TIMER) data download and analysis
Access the TIMER database at http://timer.cistrome.org/, select “Gene_DE”, search for “PHB2”, and evaluate the pan-cancer expression of PHB2 in both normal and tumor tissues.
Kaplan-Meier plotter (KM-Plotter) data download and analysis
Access the KM-Plotter database at https://kmplot.com/analysis/ and select “Start KM Plotter for breast cancer (BRCA)” within the “mRNA RNA-seq” section to examine the relationship between PHB2 expression levels and the survival of BRCA patients.
Human protein atlas (HPA) data download and analysis
Access the HPA database at https://www.proteinatlas.org/. Investigate “PHB2”. Access the “SUBCELL” module. Choose the U2OS cell line. Disable the color rendering for “Microtubules”. Acquire images of nuclear and PHB2 immunofluorescence staining. Cell regions were selected for the analysis of the subcellular localization of PHB2. Access the “CANCER” module. Images of immunohistochemical staining of PHB2 in ovarian cancer (OV) tissues were chosen for download and analysis of localization.
cBioPortal data download and analysis
Access the cBioPortal database at https://www.cbioportal.org/, search for “PHB2”, and navigate to the “Cancer Types Summary” module to examine the mutation rate of PHB2 in tumor tissue. Additionally, utilize the “Mutations” module to investigate the tumor mutation hotspot of PHB2.
PhosphoNET data download and analysis
Access the PhosphoNET database at http://www.phosphonet.ca/, search for “PHB2”, and examine the phosphorylation modification sites associated with PHB2.
AlphaFold data download and analysis
Access the AlphaFold database (https://alphafold.ebi.ac.uk/), conduct a search for “PHB2”, and examine the three-dimensional predicted structure of PHB2 along with the pathogenic implications of amino acid mutation sites.
Results
The expression level of PHB2 in most tumor tissues was higher than that in normal tissues
GEPIA was employed to analyze the expression levels of PHB2 across various cancers in both normal and tumor samples. The findings indicated that PHB expression varied across 30 tumors, exhibiting elevated levels in 27, including BRCA, colon adenocarcinoma (CHOL), lung squamous carcinoma (LUSC), and OV. Simultaneously, it exhibited reduced levels in three tumors: cervical squamous carcinoma (CESC), acute myeloid leukemia-like tumor (LAML), and pheochromocytoma and paraganglioma (PCPG) (Figs. 1A, B).
An analysis of PHB2 expression across various cancers utilizing TMIER indicated that PHB2 was significantly expressed in ten tumor types, including glioblastoma (GBM), renal chromophobe cell carcinoma (KICH), and renal clear cell carcinoma (KIRC) (Fig. 2).
The expression level of PHB2 in most tumor tissues was higher than that in normal tissues. A Scatter plot illustrating the differential expression levels of PHB2 in tumor versus normal tissues, derived from GEPIA data. B Histogram illustrating the differential expression levels of PHB2 in tumor versus normal tissues based on data from GEPIA. Red bars indicate tumor-related tissues, while black bars denote normal tissues.
PHB2 was found to be localized in the mitochondria of both normal and tumor cells
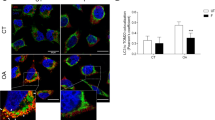
The subcellular localization of PHB2 was examined utilizing the “SUBCELL” module of the Human Protein Atlas database. Immunofluorescence staining revealed that green-stained PHB2 exhibited a dotted filamentous distribution surrounding the blue-stained nucleus on the mitochondria in U2OS cells (Fig. 3A).
The “CANCER” module facilitated the analysis of PHB2 subcellular localization within tumor tissues. Immunohistochemistry revealed that the brown PHB2 staining did not co-localize with the blue nucleus; rather, the protein was found surrounding the nucleus and localized within the mitochondria (Fig. 3B).
PHB2 was localized in the mitochondria of both tumor and normal cells. A Immunofluorescence staining verified the PHB2 localization in the mitochondria in normal cells from the HPA database. B Immunohistochemistry confirmed the PHB2 localization in the mitochondria in tumor cells from the HPA database.
The relationship between PHB2 expression levels and survival rates varied across different tumors
GEPIA was utilized to examine the relationship between PHB2 expression levels and patient survival rates. The findings indicated that in BLCA, patients exhibiting high PHB2 expression had a lower survival rate compared to those with low expression (Logrank P < 0.05). In KIRC, patients exhibiting high expression of PHB2 demonstrated a higher survival rate compared to those with low expression (Logrank P < 0.05). In lung adenocarcinoma (LUAD), patients exhibiting high expression of PHB2 demonstrated a lower survival rate compared to those with low expression (Logrank P < 0.05). KM-plotter analysis indicated that patients exhibiting high PHB2 expression had a higher survival rate compared to those with low expression in BRCA (Logrank P < 0.05) (Fig. 4A, B, C, D). The dual prognostic role of PHB2 (pro-tumor vs. anti-tumor) may reflect tissue-specific biological contexts, necessitating further mechanistic studies.
PHB2 exhibited a significant mutation rate across pan-cancer tissues, with overlapping amino acid positions for various tumor mutations and post-translational changes identified in these tissues
The analysis of the “Cancer Type Summary” module within the cBioPortal database indicated a high mutation rate of PHB2 in pan-cancer tissues, approximately 6% in OV tissues (Fig. 5A). The findings from the “Mutation” module indicated the presence of 34 tumor mutation hotspots on PHB2, with the 95th glycine to cysteine hotspot potentially serving as a significant mutation site in tumor cells. Furthermore, PHB2 undergoes various post-translational modifications, including phosphorylation, acetylation, and ubiquitination. Notably, tyrosine 121 and threonine 263 are identified as both tumor mutation hotspots and sites of post-translational phosphorylation, while lysine 296 serves as a tumor mutation hotspot and a potential site for post-translational acetylation and ubiquitination modifications (Fig. 5B). The overlapping sites of amino acid mutations and modifications may be significantly associated with the development of malignant tumor cells and could serve as potential screening sites for early diagnosis of malignant tumors in the future.
PHB2 had multiple amino acid phosphorylation modification sites
The PhosphoNET analysis revealed several phosphorylation sites on amino acids in PHB2 (Fig. 6).
The analysis of PHB2 identified a total of 299 amino acids, with three specific sites (threonine 42, tyrosine 121, and threonine 263) designated as potential tumor mutation hotspots. A post-translational phosphorylation modification occurred, specifically the simultaneous mutation of functional sites, indicating that these three amino acid sites and their corresponding gene loci were associated with the development of tumors (Figs. 5B and 6).
AlphaFold predicted the three-dimensional structure of PHB2 and analyzed the amino acid mutation sites as highly pathogenic
Through analysis of the AlphaFold database, we derived a three-dimensional predicted structure of PHB2. In this structure, dark blue peptides represent regions with very high confidence (pLDDT > 90), blue peptides indicate areas with relatively high confidence (70 < pLDDT < 90), and yellow peptides denote regions with low confidence (50 < pLDDT < 70) (Fig. 7A). The heat map of pathological mutations indicated that the proximity of the amino acid mutation site score to 1.0 is significant. A closer proximity of the color to red indicates a higher pathogenicity of the mutation. Amino acid mutations at overlapping sites for post-translational modifications, including threonine 42, tyrosine 121, threonine 263, and lysine 296, exhibit varying pathogenicity when altered to different amino acids. Pathogenicity may be classified as low, indeterminate, or high. Some mutation sites with scores near 1.0 consistently exhibit high pathogenicity (Fig. 7B). Consequently, these amino acid mutation sites may play a significant role in tumorigenesis.
Discussion
PHB2 is a gene characterized by a high mutation rate across pan-cancer tissues, encoding a protein that is localized in mitochondria. The three-dimensional structure of this protein was predicted using the AlphaFold database. The protein comprises 299 amino acids, featuring four specific amino acid sites: threonine at position 42, tyrosine at position 121, threonine at position 263, and lysine at position 296. These sites function as tumor mutation hotspots and are subject to post-translational modifications, including phosphorylation, acetylation, and ubiquitylation, which are influenced by the epigenetic regulation of the gene16,17. Further analysis of the heatmap of pathogenicity mutations revealed that the overlapping sites of amino acid mutations and post-translational modifications exhibited varying pathogenicity when mutated to different amino acids, categorized as low pathogenicity, indeterminate cases, and high pathogenicity. All of them exhibited mutations with scores approaching 1.0, indicating high pathogenicity. Data analysis from GEPIA and TIMER indicated that PHB2 expression levels were elevated in the majority of tumor tissues compared to normal tissues, aligning with prior studies18,19,20,21. This study posits that amino acid mutation sites and their associated gene loci play a critical role in the initiation and progression of tumor cells.
We examined the relationship between PHB2 and tumorigenesis, as well as the impact of PHB2 expression levels on the survival prognosis of cancer patients, utilizing the GEPIA and KM-plotter databases. The findings indicated variability in effects depending on the tumor type, with prior studies predominantly demonstrating that elevated PHB2 expression correlated with negative prognostic outcomes in patients18,19,20,22highlighting an additional innovative aspect. In BRCA and KIRC, patients exhibiting high PHB2 expression demonstrated a superior survival rate compared to those with low expression. Conversely, in LUAD and BLCA, patients exhibiting high PHB2 expression demonstrated a lower survival rate compared to those with low expression. We hypothesize that varying tumorigenesis may account for the differing effects of PHB2 on tumor cell genesis, metastasis, and apoptosis, which will inform future research on the regulation of cancer by PHB2.
While PHB2 demonstrates pan-cancer overexpression and mutation hotspots, its inconsistent survival associations (e.g., detrimental in LUAD/BLCA vs. beneficial in KIRC/BRCA) suggest it may not serve as a standalone universal biomarker. Future studies should explore whether combining PHB2 with tissue-specific co-factors improves prognostic stratification. Moreover, Our findings are derived solely from bioinformatic analyses. Experimental validation is critical to confirm PHB2’s functional roles and biomarker utility, particularly given its context-dependent prognostic effects.
PHB2 exhibits elevated expression and mutation rates across multiple cancers, with specific amino acid sites (threonine 42, tyrosine 121, threonine 263, and lysine 296) harboring both mutations and post-translational modifications. However, its context-dependent survival associations highlight the complexity of applying PHB2 into clinical practice. Further experimental studies are warranted to elucidate the tissue-specific functions of PHB2 and evaluate its utility as part of combinatorial biomarker panels.
Data availability
Publicly available datasets were analyzed in this study. The data can be found here: http://gepia.cancer-pku.cn/, http://timer.cistrome.org/, https://kmplot.com/analysis/, https://www.proteinatlas.org/, https://www.cbioportal.org/, http://www.phosphonet.ca/, https://alphafold.ebi.ac.uk/.
References
Hyuna, S. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71(3), (2021).
Seema, B., Sanober, A. & Naeem Ahmed, L. A heartrending burden of gynaecological cancers in advance stage at nuclear institute of medicine and radiotherapy Jamshoro Sindh. Pak J. Med. Sci. 32(1), (2016).
Matthew, J. R. et al. A multi-omic single-cell landscape of human gynecologic malignancies. Mol. Cell. 81(23), (2021).
Minghai, Z. et al. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim. Biophys. Acta Rev. Cancer 1875(2), (2021).
Liu, X. et al. Does the primary treatment sequence affect post-relapse survival in recurrent epithelial ovarian cancer? A real-world multicentre retrospective study. BJOG. Int. J. Obstet. Gynecol. 70–78 (2022).
Seebacher, V. et al. The impact of the duration of adjuvant chemotherapy on survival in patients with epithelial ovarian cancer - a retrospective study. PloS One 12(1), e0169272 (2017).
Książek, K. Molecular biology of ovarian cancer: from mechanisms of intraperitoneal metastasis to therapeutic opportunities. Cancers 13(7), 1661 (2021).
Roy, M. et al. Efficient PHB2 (prohibitin 2) exposure during mitophagy depends on VDAC1 (voltage dependent anion channel 1). Autophagy. 1–13 (2024).
Li, J. et al. Aldehyde dehydrogenase 2 lactylation aggravates mitochondrial dysfunction by disrupting PHB2 mediated mitophagy in acute kidney injury. Adv. Sci. e2411943 (2024).
Leytens, A. et al. Targeted proteomics addresses selectivity and complexity of protein degradation by autophagy. Autophagy 21 (2), 460–475 (2025).
Wilson, E. et al. Loss of prohibitin 2 in Schwann cells dysregulates key transcription factors controlling developmental myelination. Glia. 72(12), 2247–2267 (2024).
García-Chávez, D. et al. Prohibitins, Phb1 and Phb2, function as Atg8 receptors to support yeast mitophagy and also play a negative regulatory role in Atg32 processing. Autophagy 20(11), 2478–2489 (2024).
Ding, T. et al. Prohibitin 2 orchestrates long noncoding RNA and gene transcription to accelerate tumorigenesis. Nat. Commun. 15(1), 8385 (2024).
Liu, P. et al. HKDC1 functions as a glucose sensor and promotes metabolic adaptation and cancer growth via interaction with PHB2. Cell Death Differ. 31(12), 1595–1610 (2024).
Qiao, Y. et al. Selection and identification of a prohibitin 2-binding DNA aptamer for tumor tissue imaging and targeted chemotherapy. Int. J. Biol. Macromol. 259, 129002 (2024).
Akihide, Y. et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature. 574(7777), 273–277 (2019).
Baoxiang, C. et al. Metabolic Recoding of NSUN2‐mediated m5C modification promotes the progression of colorectal cancer via the NSUN2/YBX1/m5C‐ENO1 positive feedback loop. Adv. Sci. 11(28), 2309840 (2024).
Liang, X. et al. Prohibitin 2 confers NADPH oxidase 1-mediated cytosolic oxidative signaling to promote gastric cancer progression by ERK activation. Free Radic. Biol. Med. 224, 130–143 (2024).
Xu-Wei, C. et al. Tissue-based quantitative proteomics to screen and identify the potential biomarkers for early recurrence/metastasis of esophageal squamous cell carcinoma. Cancer Med. 7(6), 2504–2517 (2018).
Jie, Y. et al. Decreasing expression of Prohibitin-2 lowers the oncogenicity of renal cell carcinoma cells by suppressing eIF4E-mediated oncogene translation via MNK inhibition. Toxicol. Appl. Pharmacol. 466, 116458 (2023).
Jörg, M. et al. Differential immunization identifies PHB1/PHB2 as blood-borne tumor antigens. Oncogene. 23(44), 7430–7435 (2004).
Young Eun, L. et al. Overexpression of prohibitin 2 protein is associated with adverse prognosis in cytogenetically normal acute myeloid leukemia. Ann. Lab. Med. 42(5), 585–589 (2022).
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, J. Role of PHB2 as a potential biomarker in pan-cancer: a multi-database analysis. Sci Rep 15, 23933 (2025). https://doi.org/10.1038/s41598-025-07818-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-07818-5