Abstract
The genera Cryptosporidium, Eimeria, and Cystoisospora cause gastrointestinal diseases in pigs that can lead to economic losses in the pig industry. Despite their importance, the molecular epidemiology and species diversity of these parasites remain poorly understood. Therefore, this study aimed to investigate the distribution and diversity of these genera Cryptosporidium, Eimeria, and Cystoisospora in pigs in Korea and to evaluate their potential influencing factors, including geographical location and season. A total of 700 fecal samples were collected from 103 pig farms between May 2020 and February 2023. PCR identified the genera Cryptosporidium, Eimeria, and Cystoisospora in 49 (7.0%), 24 (3.4%), and 6 (0.9%) samples, respectively. At the farm level, 43 (41.8%) out of 103 farms had at least one pig infected with these parasites. According to the region, Eimeria spp. showed the highest prevalence in Gyeongsangnam-do (8.5%; 17/200) with a statistically significant difference. Seasonal analysis revealed a statistically significant difference for Eimeria spp. with higher prevalence in summer (6.4%; 15/233) and winter (4.7%; 7/149). Phylogenetic analyses revealed Cryptosporidium (Cr.) scrofarum and Cr. suis, and confirmed the presence of Eimeria (E.) debliecki, E. perminuta, E. spinosa, and E. suis, as well as Eimeria sp. genotype 1–4. All Cystoisospora (Cy.) positive samples were confirmed as Cy. suis. This study examined the nationwide distribution of the genera Cryptosporidium, Eimeria, and Cystoisospora in pigs in Korea, providing molecular evidence of these parasites. The results improve our understanding of the distribution and diversity of apicomplexan protozoa in pigs in Korea. Notably, Cr. scrofarum and Cr. suis identified in this study are known to infect humans, indicating potential zoonotic risks. These findings highlight the importance of continued surveillance to mitigate economic losses on the pig industry and to address public health concerns.
Similar content being viewed by others
Introduction
Pigs are a major livestock species globally, playing a significant role in meat production. As reported by the Korean Statistical Information Service, the global pig population was estimated to exceed 900 million in 2022, with the annual per capita pork consumption in Korea being 28.5 kg. The high demand for pork underscores the economic significance of maintaining pig health. To this end, the control and prevention of diseases represent a paramount objective. A variety of gastrointestinal parasites infect pigs, including Entamoeba (Ent.) polecki, Ent. suis, Giardia (G.) duodenalis (synonyms G. intestinalis and G. lamblia), Ascaris suum, coccidia, Cryptosporidium spp., and strongyles. These infections result in a loss of productivity1,2,3,4,5.
The phylum Apicomplexa is a large group of parasitic protozoans that are distinguished by the presence of an apical complex, a specialized structure that is utilized for the penetration of host cells6. This phylum encompasses numerous significant pathogens in both human and veterinary medicine, including Plasmodium spp., Toxoplasma gondii, Babesia spp., Cryptosporidium spp., Eimeria spp., and Cystoisospora spp.7. These parasites are responsible for the development of severe clinical symptoms, particularly in hosts with compromised immune systems. Among these parasites, the genera Cryptosporidium, Eimeria, and Cystoisospora infect the small intestines of various hosts, including pigs, resulting in diarrhea, dehydration, weight loss, and, in some cases, fatality1,2. It can be reasonably deduced that these infections contribute to significant economic losses in the pig industry.
The genus Cryptosporidium is a protozoan parasite that commonly infects the gastrointestinal tract and respiratory epithelial cells of various vertebrate hosts2. The transmission of this parasite occurs via the fecal–oral route, as well as indirectly through the ingestion of Cryptosporidium-contaminated water or food8. The life cycle of this parasite includes both asexual and sexual reproduction, which can be completed within a single host8. Pigs are considered one of the major hosts susceptible to Cryptosporidium spp. Previous studies have identified a number of Cryptosporidium species in pigs, including Cryptosporidium (Cr.) parvum, Cr. felis, Cr. meleagridis, Cr. muris, Cr. tyzzeri, Cr. scrofarum, and Cr. suis. Of these, Cr. scrofarum and Cr. suis are the most prevalent species in pigs with a worldwide distribution9,10. While many infections are subclinical, clinical symptoms may manifest in neonatal and immunocompromised pigs, including diarrhea, weight loss, and even death11. Additionally, humans have been documented as infected with Cr. scrofarum and Cr. suis, indicating the potential for zoonotic transmission12,13,14,15.
The genus Eimeria comprises numerous species, each of which is highly host-specific16. The transmission of the parasite occurs via the fecal–oral route, and Eimeria infections (coccidiosis) are prevalent in a wide range of livestock, including ruminants and pigs, as well as in poultry17. To date, at least eight Eimeria species are known to infect pigs. The following Eimeria species have been identified in pigs: Eimeria (E.) debliecki, E. neodebliecki, E. perminuta, E. polita, E. porci, E. scabra, E. spinosa, and E. suis. Among these, E. neodebliecki, E. scabra, and E. spinosa have been documented to induce clinical symptoms in pigs on occasion, including diarrhea, anorexia, and weight loss18,19. Despite the widespread distribution of Eimeria spp. in pig populations, there is a paucity of research on clinical studies, pathogenesis, and molecular epidemiology.
The genus Cystoisospora was previously classified within the family Eimeriidae as the genus Isospora, based on its homoxenous life cycle and oocyst morphology. Following a comprehensive examination of both morphology and molecular analysis, a number of Isospora species infecting mammals were reclassified within the family Sarcocystidae, genus Cystoisospora20. Cystoisospora spp. is renowned for its strict host specificity and global distribution21. To date, Cystoisospora (Cy.) suis is the only species of the genus that has been identified in pigs, and it is of particular importance due to its pathogenicity in piglets22,23. Following a prepatent period of 3–5 days, Cy. suis infection typically manifests as watery or pasty yellow diarrhea, which usually emerges during the second week of life24. In severe cases, infection with Cy. suis can result in growth retardation and weight loss, leading to economic losses for the pig industry. Furthermore, co-infection with other enteropathogens, including rotavirus, transmissible gastroenteritis virus, Clostridia, or Escherichia coli, can exacerbate clinical signs and increase mortality rates25.
Despite the wide variety of parasites that can infect pigs and their economic significance, research on intestinal protozoan parasites in pigs is limited, especially using molecular techniques rather than traditional microscopy. This results in difficulties in accurate species identification and analysis of molecular characteristics of parasites. Therefore, this study investigated the distribution and diversity of the genera Cryptosporidium, Eimeria, and Cystoisospora in pigs in Korea using molecular techniques. In addition, this study evaluated their potential influencing factors, including geographical location and season.
Material and methods
Sample collection
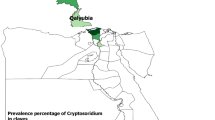
From May 2020 to February 2023, 700 pig fecal samples from 103 farms were collected from slaughterhouses in Korea (Fig. 1). The mean number of samples per farm was 6.8 with a standard deviation of 3.5. All pigs sampled were raised for meat production and at the time of sampling, they were approximately six months old and weighed approximately 115 kg. Clinical symptoms were not available. The regions where the pigs were raised (Gangwon-do, Gyeonggi-do, Chungcheongbuk-do, Chungcheongnam-do, Jeollabuk-do, Jeollanam-do, Gyeongsangbuk-do, Gyeongsangnam-do, and Jeju Island) and the date of sampling were recorded. Fecal samples were collected directly from the intestine by dissection to avoid environmental contamination and to ensure that no pig was sampled more than once. Samples were then transferred to the College of Veterinary Medicine, Chungbuk National University, Korea, and stored at 4 °C until processing.
Map of Korea showing the sampling region of pig fecal samples. The prevalence of Cryptosporidium, Eimeria, and Cystoisospora were determined by PCR. GW, Gangwon-do; GG, Gyeonggi-do; CB, Chungcheongbuk-do; CN, Chungcheongnam-do; JB, Jeollabuk-do; JN, Jeollanam-do; GB, Gyeongsangbuk-do; GN, Gyeongsangnam-do. No positive cases were identified in Gangwon-do and Chungcheongbuk-do. The map was originally created by Ksiom and adapted by Kwj2772 (Public domain, via Wikimedia Commons), and was subsequently modified using Microsoft Paint (Microsoft Corporation, Redmond, WA, USA).
DNA extraction, PCR, cloning, and sequencing
Genomic DNA was extracted from fecal samples using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA concentration was confirmed using a spectrophotometer (DeNovix, Wilmington, DE, USA). The extracted DNA was stored at -20 °C until further use.
For molecular identification, PCR was performed targeting the 18S rRNA gene of Cryptosporidium spp. and Eimeria spp. and the ITS-1 region of Cystoisospora spp. (Table 1). PCR was performed using AccuPower HotStart PCR Premix Kit (Bioneer, Daejeon, Korea). All positive samples were sent to Macrogen (Daejeon, Korea) for bidirectional sequencing. In the case of Eimeria spp., some positive PCR products failed direct sequencing and were cloned into E. coli DH5α using the pTOP V2 vector (Enzynomics, Daejeon, Korea). At least three colonies per sample were selected and sequenced using a universal primer set (M13F and M13R) by Macrogen (Daejeon, Korea). The resulting sequences were aligned using MEGA 11, and the species were determined using the Basic Local Alignment Search Tool program and phylogenetic analysis.
Statistical analysis
For statistical analysis, the data of the samples were categorized as follows: regions as Gangwon-do, Gyeonggi-do, Chungcheongbuk-do, Chungcheongnam-do, Jeollabuk-do, Jeollanam-do, Gyeongsangbuk-do, Gyeongsangnam-do, and Jeju Island; and seasons as spring (March–May), summer (June–August), fall (September–November), and winter (December-February).
To analyze the relationships among region, season, and parasite infection, data were statistically analyzed with Chi-square test or Fisher’s exact test using R software. The p-value less than 0.05 was considered significant. In addition, 95% confidence intervals and odds ratio were calculated.
Phylogenetic analysis
To analyze the molecular characteristics of the identified parasites, phylogenetic trees were constructed using MEGA 11 by the neighbor joining method. The bootstrap method with 1000 replicates was used to assess the reliability of the trees. Sequences of other Cryptosporidium, Eimeria, and Cystoisospora species obtained from the GenBank database were also included in the analysis, considering the country and species. The sequences of E. meleagrimitis (KC305197), Cr. parvum (L25642), and Cr. baileyi (AF222997) were used as outgroup for the genera Cryptosporidium, Eimeria, and Cystoisospora phylogenetic trees, respectively.
Results
Molecular identification and diversity of the genera Cryptosporidium, Eimeria, and Cystoisospora by PCR
At the individual level, of the 700 pig fecal samples, PCR identified the genera Cryptosporidium, Eimeria, and Cystoisospora in 49 (7.0%), 24 (3.4%), and 6 (0.9%) samples, respectively (Table 2). Of the samples tested, 75 (10.7%) were positive for at least one of the parasites and 625 were negative. At the farm level, 43 (41.8%) of 103 farms had at least one pig infected with the parasites. In addition, 33 (32.0%), 11 (10.7%) and 3 (2.9%) farms had at least one pig infected with the genera Cryptosporidium, Eimeria and Cystoisospora, respectively.
Of the 49 Cryptosporidium-positive samples, Cr. scrofarum was identified in 47 (95.9%) samples and Cr. suis in two (4.1%) samples. For the genus Cystoisospora, all six positive samples were identified as Cy. suis. Of the 24 Eimeria-positive samples, at least seven Eimeria species were identified, including three known species and four unspecified species: single infection as seven (29.2%) E. suis, one (4.2%) E. spinosa, one (8.3%) E. debliecki, seven (29.2%) Eimeria sp. genotype 1, one (4.2%) Eimeria sp. genotype 2, two (8.3%) Eimeria sp. genotype 3, and one (4.2%) Eimeria sp. genotype 4; double infection as one (4.2%) sample with E. suis and Eimeria sp. genotype 2, one (4.2%) with E. suis and E. debliecki; triple infection as one (4.2%) with E. suis, Eimeria sp. genotype 3, and Eimeria sp. genotype 4 and one (4.2%) with E. suis, E. perminuta, and Eimeria sp. genotype 3 (Table 3).
At the genus level, four cases showed co-infection: two (8.3%) cases of Cryptosporidium and Eimeria co-infection and two (8.3%) cases of Cryptosporidium and Cystoisospora co-infection. There were no cases of co-infection with all three parasites (Table 4).
Prevalence according to the province and season
According to region, Cryptosporidium spp. prevalence was highest in Gyeongsangbuk-do (12.1%; 12/99), followed by Gyeonggi-do (8.7%; 11/127), but no statistical difference was observed (p > 0.05). Eimeria spp. prevalence was highest in samples from Gyeongsangnam-do (8.5%; 17/200), followed by Jeju Island (4.0%; 1/25), Jeollabuk-do (2.7%; 2/74), and Gyeonggi-do (1.6%; 2/127) with statistical significance (p < 0.05). In addition, Cy. suis showed zero to two positive cases depending on the region, and it was not statistically significant (p > 0.05) (Table 2).
Regarding the season, Eimeria spp. prevalence was highest in summer (6.4%; 15/233), followed by winter (4.7%; 7/149), spring (0.7%; 1/134) and fall (0.5%; 1/184) with a statistically significant difference (p < 0.05). Cryptosporidium spp. and Cy. suis showed no statistically significant difference according to season (p > 0.05) (Table 2).
Sequencing and phylogenetic analysis
The 49 Cryptosporidium-positive samples (773–774 bp) were successfully sequenced and showed 99.9–100% identity within the same species. The sequences of Cr. scrofarum and Cr. suis obtained in this study showed 99.7–100% and 100% identity, respectively, with Cr. scrofarum (MT071828) and Cr. suis (MT071826) deposited in GenBank database. Of the 24 Eimeria-positive samples, 16 were successfully sequenced by direct sequencing, and 24 colonies were selected from the other eight positive samples, all with sequences (409–511 bp) showing 97.1–100% interspecies identity. Six Cystoisospora-positive samples (396–423 bp) were also successfully sequenced, with 98.6–99.8% intraspecies identity. Sequences of Cy. suis obtained in this study showed 98.4–99.3%, 98.8–100%, and 98.8–99.8% identity with GenBank sequences from China (KR139985), Myanmar (MW959806), and Japan (LC085519), respectively. All sequences obtained in this study were deposited in the GenBank database (Cryptosporidium spp., PQ615448-PQ615458; Eimeria spp., PQ615640-PQ615679; Cystoisospora suis, PQ621784-PQ621789).
Phylogenetic analysis showed a clear separation of Cr. scrofarum and Cr. suis obtained in this study (Fig. 2). Furthermore, phylogenetic analysis confirmed the presence of E. debliecki, E. perminuta, E. spinosa, and E. suis. In addition, sequences that differed from those of Eimeria deposited in the GenBank database were designated as Eimeria sp. genotype 1–4 (Fig. 3). The Cy. suis sequences obtained in this study were found to be identical to GenBank sequences from China (KR139985), Myanmar (MW959806), and Japan (LC085519) (Fig. 4).
Phylogenetic tree of 18S rRNA gene of Cryptosporidium spp. To construct the phylogenetic tree, the neighbor joining method was used with 1000 bootstrap replications. The bootstrap values less than 50 were omitted. The sequences analyzed in this tree were obtained from the GenBank databases. The sequences identified in this study are indicated with bold and arrows.
Phylogenetic tree of 18S rRNA gene of Eimeria spp. To construct the phylogenetic tree, the neighbor joining method was used with 1000 bootstrap replications. The bootstrap values less than 50 were omitted. The sequences analyzed in this tree were obtained from the GenBank databases. The sequences identified in this study are indicated with bold and arrows.
Phylogenetic tree of internal transcribed spacer 1 of Cystoisospora spp. To construct the phylogenetic tree, the neighbor joining method was used with 1000 bootstrap replications. The bootstrap values less than 50 were omitted. The sequences analyzed in this tree were obtained from the GenBank databases. The sequences identified in this study are indicated with bold and arrows.
Discussion
Previous studies have reported the presence of E. perminuta, E. debliecki, E. polita, E. scabra, and Isospora suis in pigs in Korea28, as well as Cryptosporidium spp.29,30. However, these studies were conducted several decades ago and relied on microscopic examination for species identification without molecular information. Therefore, there is a lack of molecular and epidemiologic data on the genera Cryptosporidium, Eimeria, and Cystoisospora infections in domestic pigs in Korea.
Cryptosporidium spp. is distributed worldwide, and previous studies have reported its prevalence in pigs in different countries2,9,10,30,31,32,33,34,35,36,37. For example, the reported prevalence included 14.4% in Switzerland31, 9.0% in Argentina34, 25.8% in Canada10, 14.5% in Vietnam35, 44.2% in Zambia36, 21.8% in Czech Republic2, 27.7% in Poland32, 32.6% in Japan33, 3.3% in China11, 1.6% in Texas37, and 20.8% in Thailand38. To the best of our knowledge, only two studies have reported the prevalence of Cryptosporidium spp. in pigs using microscopic methods, with 19.6% (98/500) and 10.5% (62/589) in Jeollabuk-do and Chungcheong-do, respectively29,30. In the present study, the overall prevalence of Cryptosporidium spp. was found to be 7.0% (49/700), which is comparatively lower than reported rates in other countries.
Cr. scrofarum and Cr. suis are the most dominant species infecting pigs worldwide, both of which possess zoonotic potential as they have been reported in human31,39. This study used genus Cryptosporidium-targeted primers that have been demonstrated to detect a broad range of Cryptosporidium species, thereby providing molecular evidence of both species that had not been previously identified in Korea26. Furthermore, electrograms obtained from direct sequencing showed no other Cryptosporidium spp. infection (data not shown). Phylogenetic analysis revealed that the sequences of Cr. scrofarum and Cr. suis in this study were identical to those previously reported in China, suggesting the possibility of international transmission or common genetic lineages across East Asia. Furthermore, the observed higher prevalence of Cr. scrofarum compared to Cr. suis in this study may be due to the age of the pigs, which averaged about six months. Previous studies have shown age-related differences in the prevalence of these two species, with Cr. suis being more prevalent in younger pigs and Cr. scrofarum being more prevalent in older pigs33,40.
Previous studies reported that the prevalence of Eimeria spp. in pigs was 2.6% in Poland (including E. polita, E. debliecki, E. suis, E. scabra, and E. perminuta)41, 25.3% in Zimbabwe (including E. polita, E. debliecki, E. suis, E. scabra, E. porci, E. spinosa, E. neobliecki and E. perminuta)42, 78% in Indonesia43, 33% in Kenya (E. polita, E. deliecki, E. suis, E. scabra, and E. porci)44, and 8.6% in Germany (E. polita, E. debliecki, E. suis, E. scabra, E. porci, E. spinosa, and E. perminuta)45. However, only one epidemiologic study of coccidial infection in pigs has been conducted in Korea without identification of the species, reporting a prevalence of 22.3% (88/395) in samples from Chungju City28. In the current study, the prevalence of Eimeria spp. was found to be 3.4% (24/700), which is comparatively lower than the rates reported in other countries.
The Eimeria species identified in this study were E. debliecki, E. perminuta, E. spinosa, E. suis, and Eimeria sp. genotype 1–4. Although these species have been reported in other countries such as Germany, India, Poland, and China17,22,41,45, however, these findings represent the initial molecular identification of these species in Korea. In addition, phylogenetic analysis revealed different sequences that diverged from the reference Eimeria sequences in GenBank, suggesting novel Eimeria species infecting pigs. Given the limited molecular and pathogenetic studies on Eimeria spp. infecting pigs, further molecular research is needed.
Cystoisospora suis is recognized as the most pathogenic coccidian species in pigs, particularly affecting suckling piglets, which often present with symptoms of pasty to watery diarrhea22,46. Previous studies reported a high prevalence of Cy. suis in pigs, including 27.8% in Poland41, 32.8% in Brazil47, 52.1% in Venezuela48, 21.9% in China22, 8.8% in Switzerland31. In contrast, the prevalence of Cy. suis in this study was 0.9% (6/700). As the genus Cystoisospora has not been investigated in pigs in Korea, it is difficult to make a direct comparison. However, considering that the previous prevalence of coccidial parasites was 22.3% (88/395), it can be assumed that overall environmental improvements in the pig industry have contributed to a reduction in prevalence28. In addition, the age of the pigs sampled was approximately 6 months, which may be another reason for the low prevalence46. Because piglets are most susceptible to Cy. suis, often showing clinical manifestations, while older pigs are less susceptible46.
All of the Cystoisospora identified in this study were identified as Cy. suis and showed 98.4%-100% identity when compared to the sequences in GenBank by BLAST. In some cases, the identity was not high enough, which we believe is due to the relatively rapid evolution of the target gene, ITS-1. However, phylogenetic analysis showed that the sequences of Cy. suis identified in this study clustered with those previously reported from China (KR139985), Myanmar (MW959806), and Japan (LC085519).
According to the region, interestingly, none of the positive cases were observed in Jeju Island except for one Eimeria positive case. This may be due to the Jeju Island Public Notice prohibiting the importation and exportation of pigs. This regulation likely reduces the risk of parasite transmission, making it unlikely to spread within the island. Additionally, only the genus Eimeria demonstrated a statistically significant difference. Variations in regional prevalence may be attributed to factors such as hygiene practices and veterinary management. However, given the limited availability of detailed data specific to each region and the potential for sample size bias, the exact causes of these differences remain unknown.
Given that parasites generally thrive under moderate to high temperatures, a higher prevalence would be expected during the summer. However, this study revealed a lower prevalence in spring and fall with similar prevalence, while the prevalence was higher in both summer and winter. This pattern may be attributable to the intensive, industrialized breeding systems, which mitigate the impact of external seasonal factors such as temperature and humidity. Additionally, the observed results may have been influenced by the high concentration of Eimeria-positive cases reported in certain farms in Gyeongsangnam-do, particularly during summer and winter (data not shown).
The diagnosis and species-level differentiation of protozoan parasites has traditionally relied on morphological analysis of oocysts using microscopy; however, this method has significant limitations, including morphological similarity among species that makes accurate species differentiation difficult, low sensitivity, and the inability to assess molecular characteristics. In contrast, PCR-based molecular techniques have demonstrated high efficacy for the detection and differentiation of the genera Cryptosporidium, Eimeria, and Cystoisospora, regardless of parasite developmental stage or morphology24. These methods offer higher sensitivity and specificity, allowing accurate species identification at the molecular level.
In fact, in our previous study, only one coccidian oocyst was identified by microscopy in 364 porcine fecal samples without species identification3. Although this study tested more samples than the previous study, the PCR-based assay identified a significantly higher number of coccidia. Notably, Cryptosporidium spp. were not detected in our previous work, whereas this study identified two Cryptosporidium species at the molecular level. The results of this study demonstrated the advantages of PCR over microscopic examination, highlighting its improved detection rates and ability to confirm parasites at the species level.
This study assessed the nationwide distribution of the genera Cryptosporidium, Eimeria, and Cystoisospora in pigs in Korea between 2020 and 2023. As fecal samples were collected from approximately six-month-old pigs at slaughterhouses, data on clinical signs were unavailable, preventing direct assessment of the growth impact. Nevertheless, infections with the genera Cryptosporidium, Cystoisospora, and Eimeria are known to cause gastrointestinal disturbances such as diarrhea and dehydration, which may negatively affect growth performance1,2. To our knowledge, this is the first molecular evidence of the genera Cryptosporidium, Eimeria, and Cystoisospora infection in domestic pigs in Korea. This study improves our understanding of the distribution and diversity of apicomplexan protozoa in pigs in Korea.
Conclusion
This study presents prevalence and molecular characteristics of apicomplexan protozoa, such as the genera Cryptosporidium, Eimeria, and Cystoisospora, in Korean pigs. The results of this study revealed the nationwide distribution and species diversity of these parasites. Although the prevalence observed in this study was relatively lower compared to other countries, it is important to note that the study was limited to pigs about six months of age. Given that Cr. scrofarum and Cr. suis identified in this study can infect humans, their presence also raises public health concerns. Therefore, continuous monitoring of these parasites in pigs is warranted to prevent economic loss on the pig industry and potential zoonotic transmission.
Data availability
The raw sequence data were deposited in the NCBI GenBank database (18S rRNA of Cryptosporidium spp., PQ615448-PQ615458; 18S rRNA of Eimeria spp., PQ615640-PQ615679; ITS-1 region of Cystoisospora suis, PQ621784-PQ621789).
References
Worliczek, H. L., Gerner, W., Joachim, A., Mundt, H. & Saalmüller, A. Porcine coccidiosis - Investigations on the cellular immune response against Isospora suis. Parasitol. Res. 105, 151–156 (2009).
Němejc, K. et al. Occurrence of Cryptosporidium suis and Cryptosporidium scrofarum on commercial swine farms in the Czech Republic and its associations with age and husbandry practices. Parasitol. Res. 112, 1143–1154 (2013).
Lee, S. et al. Distribution of gastrointestinal parasitic infection in domestic pigs in the Republic of Korea: Nationwide survey from 2020–2021. Korean J. Parasitol. 60, 207–211 (2022).
Byun, J. et al. Identification of zoonotic Balantioides coli in pigs by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and its distribution in Korea. Animals 11, 2659 (2021).
Matsubayashi, M. et al. Genetic identification of Entamoeba polecki subtype 3 from pigs in Japan and characterisation of its pathogenic role in ulcerative colitis. Infect. Genet. Evol. 36, 8–14 (2015).
Morrison, D. A. Evolution of the Apicomplexa: Where are we now?. Trends Parasitol. 25, 375–382 (2009).
Striepen, B., Jordan, C. N., Reiff, S. & van Dooren, G. G. Building the perfect parasite: Cell division in Apicomplexa. PLoS Pathog. 3, e78–e78 (2007).
Pettersson, E., Ahola, H., Frössling, J., Wallgren, P. & Troell, K. Detection and molecular characterisation of Cryptosporidium spp. in Swedish pigs. Acta Vet. Scand. 62, 1–40 (2020).
Zintl, A. et al. Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitology 134, 1575–1582 (2007).
Budu-Amoako, E. et al. Occurrence of Giardia and Cryptosporidium in pigs on Prince Edward Island, Canada. Vet. Parasitol. 184, 18–24 (2012).
Zhang, Q., Wang, X. Y., Chen, J. W., Ling, D. & Zhao, G. H. Cryptosporidium suis infection in post-weaned and adult pigs in Shaanxi Province, Northwestern China. Korean J. Parasitol. 53, 113–117 (2015).
Sannella, A. R., Suputtamongkol, Y., Wongsawat, E. & Cacciò, S. M. A retrospective molecular study of Cryptosporidium species and genotypes in HIV-infected patients from Thailand. Parasit. Vectors. 12, 91 (2019).
Cama, V. A. et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 196, 684–691 (2007).
Kváč, M., Květoňová, D., Sak, B. & Ditrich, O. Cryptosporidium pig genotype II in immunocompetent man. Emerg. Infect. Dis. 15, 982–983 (2009).
Moore, C. E. et al. Molecular characterization of Cryptosporidium species and Giardia duodenalis from symptomatic Cambodian children. PLOS Negl. Trop. Dis. 10, e0004822 (2016).
Bangoura, B., Bhuiya, M. A. I. & Kilpatrick, M. Eimeria infections in domestic and wild ruminants with reference to control options in domestic ruminants. Parasitol. Res. 121, 2207–2232 (2022).
Sharma, D. et al. Discrimination, molecular characterisation and phylogenetic comparison of porcine Eimeria spp. in india. Vet. Parasitol. 255, 43–48 (2018).
Hill, J. E., Lomax, L. G., Lindsay, D. S. & Lynn, B. S. Coccidosis caused by Eimeria scabra in a finishing hog. J. Am. Vet. Med. Assoc. 186, 981–983 (1985).
Koudela, B. & Vı́tovec, J. Biology and pathogenicity of Eimeria neodebliecki Vetterlingin, 1965 experimentally infected pigs. Parasitol. Int. 47, 249–256 (1998).
Barta, J. R., Schrenzel, M. D., Carreno, R. & Rideout, B. A. The genus Atoxoplasma (Garnham 1950) as a junior objective synonym of the genus Isospora (Schneider 1881) species infecting birds and resurrection of Cystoisospora (Frenkel 1977) as the correct genus for Isospora species infecting mammals. J. Parasitol. 91, 726–727 (2005).
Scorza, A. V., Tyrrell, P., Wennogle, S., Chandrashekar, R. & Lappin, M. R. Experimental infection of cats with Cystoisospora felis. J. Vet. Intern. Med. 35, 269–272 (2021).
Gong, Q. et al. Prevalence of coccidia in domestic pigs in China between 1980 and 2019: A systematic review and meta-analysis. Parasit. Vectors 14, 248 (2021).
Joachim, A. & Schwarz, L. Coccidia of swine: Eimeria species, Cystoisospora (syn. Isospora) suis. In Encyclopedia of Parasitology (ed. Mehlhorn, S.) 537–541 (Springer, 2016).
Ruttkowski, B., Joachim, A. & Daugschies, A. PCR-based differentiation of three porcine Eimeria species and Isospora suis. Vet. Parasitol. 95, 17–23 (2001).
Joachim, A. & Shrestha, A. Coccidiosis of pigs. In Coccidiosis in livestock, poultry, companion animals, and humans (ed. Dubey, J. P.) 125–145 (CRC Press, 2020).
Xiao, L. et al. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67, 1097–1101 (2001).
Samarasinghe, B., Johnson, J. & Ryan, U. Phylogenetic analysis of Cystoisospora species at the rRNA ITS1 locus and development of a PCR-RFLP assay. Exp. Parasitol. 118, 592–595 (2008).
Jang, D. H. Survey for internal parasites of swine in Korea. Korean J. Vet. Res. 15, 309–314 (1975).
Yu, J. & Seo, M. Infection status of pigs with Cryptosporidium parvum. Korean J. Parasitol. 42, 45–47 (2004).
Rhee, J. K., Seu, Y. S. & Park, B. K. Isolation and identification of Cryptosporidium from various animals in Korea. I. Prevalence of Cryptosporidium in various animals. Korean J. Parasitol. 29, 139 (1991).
Schubnell, F. et al. Occurrence, clinical involvement and zoonotic potential of endoparasites infecting Swiss pigs. Parasitol. Int. 65, 618–624 (2016).
Rzeżutka, A., Kaupke, A., Kozyra, I. & Pejsak, Z. Molecular studies on pig cryptosporidiosis in Poland. Pol. J. Vet. Sci. 17, 577–582 (2014).
Yui, T. et al. Age-related detection and molecular characterization of Cryptosporidium suis and Cryptosporidium scrofarum in pre- and post-weaned piglets and adult pigs in Japan. Parasitol. Res. 113, 359–365 (2014).
De Felice, L. A., Moré, G., Cappuccio, J., Venturini, M. C. & Unzaga, J. M. Molecular characterization of Cryptosporidium spp. from domestic pigs in Argentina. Vet. Parasitol. Reg. Stud. Rep. 22, 100473 (2020).
Nguyen, S. T. et al. Molecular characterization of Cryptosporidium in pigs in central Vietnam. Parasitol. Res. 112, 187–192 (2013).
Siwila, J. & Mwape, K. E. Prevalence of Cryptosporidium spp. and Giardia duodenalis in pigs in Lusaka, Zambia. Onderstepoort. J. Vet. Res. 79, E1–E5 (2012).
Rodriguez-Rivera, L. D. et al. Prevalence and diversity of Cryptosporidium and Giardia identified among feral pigs in Texas. Vector Borne Zoonotic Dis. 16, 765–768 (2016).
Thathaisong, U., Siripattanapipong, S., Inpankaew, T., Leelayoova, S. & Mungthin, M. High prevalence of Cryptosporidium infection caused by Cryptosporidium scrofarum and Cryptosporidium suis among pigs in Thailand. Parasitol. Int. 77, 102122 (2020).
Bodager, J. R. et al. Complex epidemiology and zoonotic potential for Cryptosporidium suis in rural Madagascar. Vet. Parasitol. 207, 140–143 (2015).
Wang, R. et al. Prevalence and molecular identification of Cryptosporidium spp. in pigs in Henan, China. Parasitol. Res. 107, 1489–1494 (2010).
Karamon, J., Ziomko, I. & Cencek, T. Prevalence of Isospora suis and Eimeria spp. in suckling piglets and sows in Poland. Vet. Parasitol. 147, 171–175 (2007).
Chhabra, R. C. & Mafukidze, R. T. Prevalence of coccidia in pigs in Zimbabwe. Vet. Parasitol. 41, 1–5 (1992).
Widisuputri, N. K. A., Suwanti, L. T. & Plumeriastuti, H. A Survey for zoonotic and other gastrointestinal parasites in pig in Bali Province, Indonesia. IJTID 8, 54–65 (2020).
Kagira, J. M. et al. Prevalence of gastrointestinal protozoa and association with risk factors in free-range pigs in Kenya. J. Protozool. Res. 20, 1–9 (2010).
Daugschies, A., Imarom, S., Ganter, M. & Bollwahn, W. Prevalence of Eimeria spp. in sows at piglet-producing farms in Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 51, 135–139 (2004).
Shrestha, A. et al. Cystoisospora suis – a model of mammalian cystoisosporosis. Front. Vet. Sci. 2, 68 (2015).
Sperling, D., Calveyra, J., Karembe, H. & de Freitas Costa, E. Cystoisospora suis infection in suckling piglets in Brazil: Prevalence and associated factors. Vet. Parasitol. Reg. Stud. Rep. 36, 100796 (2022).
Pinilla León, J. C. & Silva Borges, N. D. Infection dynamics of Cystoisospora suis (Isospora suis) on a pilot swine farm in Carabobo State, Venezuela. Rev. Mex Cienc. Pecu. 10, 149–160 (2019).
Funding
This study was supported by the research fund for academic research projects of the Hyonsong Educational Cultural Foundation and by the Animal and Plant Quarantine Agency (APQA), Korea (No. B-1543069-2025-27-02).
Author information
Authors and Affiliations
Contributions
Conceptualization, J.W.B., S.H.L.; Sampling, S.H.L., D.H.K., M.H.H.; Data curation, S.L., B.A.; Formal analysis, S.L., K.D.M.; Methodology, S.L., B.K.K., J.W.B., S.H.L; Funding acquisition, S.H.L.; Project administration, B.K.K., J.W.B., S.H.L.; Software, S.L., K.D.M.; Supervision, S.H.L.; Writing—original draft, S.L.; Writing—review & editing, S.H.L., B.A., K.D.M., D.H.K., M.H.H., K.T.J., B.K.K., J.W.B. All authors have read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethical approval and consent to participate
Ethical review and approval were waived for this study, due to the study focused on post-slaughter samples.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lee, S., Lee, SH., Alkathiri, B. et al. Molecular identification and diversity of gastrointestinal apicomplexan protozoa in pigs in the Republic of Korea. Sci Rep 15, 20369 (2025). https://doi.org/10.1038/s41598-025-08200-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08200-1