Abstract
Brain Magnetic Resonance image diagnostics employs image processing, but aberrations such as Intensity Inhomogeneity (IIH) distort the image, making diagnosis difficult. Clinical diagnostic methods must address IIH discrepancies in brain MR scans, which occur often. Accurate brain MR image processing is difficult but required for clinical diagnosis. In this study, we introduced a more energy-efficient intensity inhomogeneity correction (IIC) method that makes use of the Modified Energy-based Generative Adversarial Network. This method uses reconstruction error in the discriminator architecture to save energy by altering the cost function. The generator’s performance is also improved by this reconstruction error. As the reconstruction error decreases, the discriminator collects latent information from real images to enhance output. To prevent mode collapse, the model has a drawing away term (PT). The generator design is improved by using skip connections and information modules that collect features at various scales. The suggested method beats state-of-the-art methods in metrics such as Peak Signal to Noise Ratio (PSNR), Structural Similarity Index (SSIM), Multi-Scale Structural Similarity Index (MSSSIM), Mean Squared Error (MSE), and Root Mean Square Error (RMSE).
Similar content being viewed by others
Introduction
The human brain is by far the most important organ in the human body. Any brain condition could have major consequences for an individual’s health because it controls important activities such as thinking, perceiving, and involuntary and voluntary muscular action. Brain Magnetic Resonance Imaging (MRI) is an important diagnostic tool in medicine because of its capacity to create precise images of the brain’s structure and function. It enables doctors and medical experts to see the brain’s internal architecture and discover anomalies including tumors, haemorrhages, and other diseases that may impair brain function. Brain MRI is a non-invasive procedure that does not use ionizing radiation, therefore it is a safe alternative for patients. It can also identify subtle changes in the brain that other imaging techniques would miss. This allows for early diagnosis of problems, which leads to earlier intervention and better outcomes for patients. As a result, brain MRI is a significant diagnostic tool in the medical sector since it may identify, diagnose, and monitor a variety of neurological illnesses, resulting in better patient treatment and results.
One of the most essential uses of brain MRI is tumor identification. This is achieved through the segmentation procedure. The MRI picture of the brain is initially preprocessed to remove any noise or artifacts that may interfere with segmentation. The MRI is then segmented to separate the various features in the image, including brain tissue, cerebrospinal fluid, and tumor region. This can be achieved using a variety of segmentation methods, including region-growing, thresholding, and clustering. When the segmentation is finished, the tumor region is recognized based on its location and characteristics, such as form, size, and intensity. The tumour can thus be categorized as benign or malignant. Aside from the diagnosis of brain tumors, segmentation has begun to be used in a variety of disciplines1. IIH has a significant impact on such diagnoses because it disrupts the segmentation process. It has been shown that images impacted by artifacts after segmentation result in a reduction in image details, thereby failing the diagnosis algorithms and resulting to inaccurate diagnosis. Medical picture segmentation is a critical step in a variety of medical applications, including post-surgical evaluation, surgical planning2, and abnormality identification3. As a result, the INU affects all of these applications that rely significantly on image segmentation and noise removal.
Mathematically, the IIH can be represented as follows.
where \(V(p,q)\) is IIH affected image, \(W(p ,q)\) is ground truth, \(B\left(p,q\right)\) is bias field, and \(N(p,q)\) stands for any additive noise.
Over the years, many methods have been introduced to reduce this artifact. Traditional approaches include filters such as Block Matching And 3D Filtering (BM3D), is one of the best conventional denoising methods available in the literature. It operates using the idea of doing collaborative filtering in a 3D transform domain and grouping comparable image blocks. The foundation of NLM (Non-Local Means Algorithm) is the notion that similar patterns appear throughout natural imagery. To cut down on noise, it averages pixels with like neighbourhoods. Weighted Nuclear Norm Minimization (WNNM) is a matrix-completion-based denoising method. It relies on the fact that groups of similar patches can be arranged into low-rank matrices.Although these approaches give a fair performance in terms of IIC techniques, it cannot be improved beyond a certain limit. Then, machines and deep learning methods began to appear.
The Generative Adversarial Network (GAN) deep learning method is adept at creating modified/corrected images. The original GAN had a generator block that would generate these images and a discriminator that would discriminate between generated and real images. The generator aims to fool the discriminator and the aim of the discriminator is to catch the generated images. It was thus a mini-max two-player game. Many IIC4 models have been developed using this very network.
Generative-Adversarial-Networks (GANs) can be classified based on various criteria such as architectural enhancements, training strategies, and specific applications. Deep Convolutional GANs (DCGANs) are GANs that employ convolutional neural networks (CNNs) in their architectures, known for generating high-quality images, StyleGAN introduces style-based generator architectures for controlling specific visual attributes of generated images, Self-Attention GANs (SAGANs) incorporate self-attention mechanisms to capture long-range dependencies in images, improving overall image quality, CycleGANs are specialized in unpaired image-to-image translation tasks, enforcing cycle consistency between the original and translated images, Pix2Pix are used for paired image-to-image translation tasks, learning mappings between input and output images, BigGAN are Scaled-up GAN architectures designed for generating high-quality images at high resolutions, and Conditional GANs (cGANs) are applying condition to both the generator and discriminator on additional information, such as class labels, enabling controlled generation. The above discussed GANs are comes under Architectural Enhancements based GANs.
There exists training strategy based GANs like Least Squares GANs (LSGANs) are used to minimize the least squares loss function instead of using the binary cross-entropy loss, Adversarial Autoencoders combine the concepts of GANs with autoencoders for unsupervised representation learning, and Energy-based GANs (EBGANs) formulate GAN training as an energy minimization problem.
GANs can be used for specific applications also. For example Text-to-Image GANs are used to generate images from textual descriptions, Video Generation GANs are extend GANs to generate realistic videos, Medical Image Synthesis GANs are used to generate synthetic medical images for tasks like data augmentation and cross-modality image translation, Super-Resolution GANs are preferred to generate high-resolution images from low-resolution inputs, Image-to-Image Translation GANs are used to translate images from one domain to another, such as converting satellite images to maps or day to night images, and Conditional Image Generation based models generate images conditioned on specific attributes or input images.
Residual Cycle Generative Adversarial Network (Res-Cycle GAN) proposed in 2020, this approach leverages a residual cycle generative adversarial network to correct intensity non-uniformity in MRI5. The network integrates residual blocks into a cycle-consistent GAN framework, facilitating an inverse transformation between uncorrected and corrected MRI images. This design enforces consistency by generating both corrected and synthetic corrected images. The generator, a fully convolutional neural network with residual blocks, enhances the transformation from raw to corrected MRI images. Evaluations on abdominal MRI data have shown that this method achieves higher accuracy and better tissue uniformity compared to traditional algorithms.
Integrated Coil Inhomogeneity Correction and Brain Extraction Using Deep Learning6 proposed a 3D generative adversarial network (GAN) to automate coil inhomogeneity correction and brain extraction in MRI data processing. This network was trained on both spin-echo and gradient-echo echo-planar imaging (EPI) data, demonstrating high accuracy and consistency compared to manual methods. The approach streamlines the data processing pipeline, reducing operator-dependent variations and biases.
Probabilistic Hadamard U-Net (PHU-Net)7, a model aimed at correcting bias fields in prostate MRI. This network combines the Hadamard transform with U-Net architecture to extract low-frequency scalar fields, which are then multiplied by the original input to obtain corrected images. In the frequency domain, high-frequency components are eliminated using a trainable filter, hard-thresholding layer, and sparsity penalty. A conditional variational autoencoder encodes possible bias field-corrected variants into a low-dimensional latent space, allowing the generation of multiple plausible corrected images. Experimental results have demonstrated PHU-Net’s effectiveness in correcting bias fields in prostate MRI, with a fast inference speed and improved segmentation accuracy.
Most of the existing deep learning techniques, however, do not consider the energy efficiency of the model. We propose a model that employs modified EBGAN to create an energy-efficient deep learning model to reduce IIH in brain MR images. Unlike the previous GAN methods that used the discriminator as a binary classifier, the proposed model also tries to increase the number of targets for the discriminator to fit the datasets better.
The primary outcome of our proposed work:
-
The modified EBGAN architecture requires fewer training images to generate the denoised images, thus consuming less energy.
-
The model consists of a discriminator that is supplemented with an autoencoder and a reconstruction error, which is also used by the generator to improve its performance. This helps in altering the discriminator cost function by changing its role as a mere classifier.
-
As the reconstruction error falls below a certain threshold, the discriminator gradually shifts its focus to extracting latent features from real images. This causes the system to return an even more precise output.
-
Furthermore, the generator architecture also uses information modules that capture multiple features at multiple scales.
-
The proposed model incorporates the precision achieved by skip connections as seen in previous works, by including these in the generator block.
In this paper, “Literature survey” section discussed over the literature survey, and “Proposed method” section presents the proposed model in greater detail. “Simulation results” section shows the outcomes obtained using the proposed model and its comparative analysis with other existed traditional methods, and “Discussion” section discusses the simulation results followed by a conclusion of the results obtained and states the possible shortcomings of the model.
Literature survey
Overall, IIC methods may be divided into two approaches, namely prospective, and retrospective methods. The prospective methods aim to calibrate and enhance the process of capturing the image, whereas the retrospective methods solely work on the acquired image data, and occasionally on prior knowledge. In this literature survey, we shall discuss a few retrospective methods.
Traditionally, filters have been the running solution for the issue of IIH in brain MR images. Overall, Linear, and non-linear filters are the two categories of filters. Kushwaha et al.8 conducted a study in 2020, reviewing the performance of various filters that fall under the two categories. They concluded that non-linear filters give the superior results in terms of edge and peak preservation in images. Thus, filters such as BM3D, NLM, and bilateral filters can be seen to give a fair performance in brain MR-image denoising and IIC9.
In 2017, Saladi et al.10 analyzed the performance of various non-linear filters, bilateral filters, Non-Local Means (NLM), principal component-analysis non-local means, and spatially adaptive NLM filters for brain MR-image denoising. This study concluded that spatially adaptive NLM gave the best performance.
In 2018, Ali et al11. also performed a similar analysis to test the performance of a different set of filters namely, their proposed median filter, adaptive median filter, and adaptive Weiner filter for brain MR-image denoising. They concluded that the median filter achieved the best performance. Wavelet domain filters have also been proven to be fair contenders in the field of IIC in brain MR images. In 2010, Anand et al12. proposed a wavelet domain-based bilateral filtering technique that was shown to give admirable results in reducing Rician noise in brain MR images.
Furthermore, techniques such as Fast Fourier Transform (FFT)13 denoising have also proven to be quite efficient in this domain. Gopinathan et al.14 proposed an FFT image denoising technique and concluded it to be giving a significant performance in terms of PSNR.
In addition to filtering methods, IIH artifacts can also be targeted directly for elimination. In 2015, Adhikari and colleagues15 introduced a nonparametric method for directly correcting IIH in brain MR images. Their approach involved merging several Gaussian surfaces to model the IIH as slowly varying multiplicative noise alongside the real tissue signals. The method was found to give admirable results with T2-weighted brain MR images. In 2016, Osadebey and colleagues16 used anatomical structural maps and regions of interest to correct the bias field in MRI scans. More recently, in 2020, Li and colleagues17 proposed an effective method for correcting the bias field and segmenting MRI data using multiplicative intrinsic component optimization (MICO). This approach has been well-received and has yielded promising results.
As the efficiency of MRI denoising methods increased, better segmentation methods could also be introduced. In 2016, Ivanovska18 devised a method for simultaneously correcting IIH and segmenting MR images. Their approach featured an energy function that facilitated direct reduction of the bias field term, which made it more flexible than previous models. To evaluate their method, they utilized both synthetic examples and actual MR scans of various organs and conducted subjective and quantitative assessments. Furthermore, various techniques using convolutional neural networks may be used for brain tumor segmentation19.
In 2018, saladi et al.20 also proposed a novel fuzzy factor for brain MR image segmentation. Liu and colleagues21 proposed a method for edge preserved IIC, and segmented liver using a level set approach. Meanwhile, in 2019, Kumar and Sridevi22 introduced a novel N3T-spline strategy, combined with a 3D convolutional neural network, to automatically segment brain tumors by correcting intensity fluctuations. Deep learning has recently emerged as a powerful tool in medical image processing23, offering solutions for image denoising, blurring, and semantic segmentation etc. Tian et al.24 studied numerous deep learning methods used for image denoising and demonstrated PSNR results that showed the great concert of deep learning methods over filtering techniques.
The Goodfellow et al.25 proposed Encoder-Decoder based Generative Adversarial Network (GAN) structure is a well-liked deep learning architecture for this use, which performs convolutions with separate receptive fields in parallel to gather multiscale contextual information, improving feature extraction and producing excellent results in recent studies, some also outperforming the standard filter methods. In26,27 used GAN model to generate synthetic brain positron emission tomography (PET) images exploiting Generative Adversarial Networks for three stages of Alzheimer’s disease—normal control (NC), mild cognitive impairment (MCI), and Alzheimer’s disease (AD).
Zhao et al.28 proposed StyleSwin transformer-based GAN for high resolution image generation. Style Swin is scalable at high resolutions, leveraging transformers’ strong expressivity to enhance both fine structures and coarse geometry. Huang et al.6 also proposed Enhanced balancing GAN which uses supervised autoencoder with an intermediate embedding model to disperse the labelled latent vectors which helps to encode the similar but different-class images dispersedly. A novel Generative Adversarial Network architecture is proposed for brain magnetic resonance imaging segmentation and brain tumor detection with improved resolution.
In29 suggested modified super resolution GAN model which reduces the IIH and improves the intensity of the brain MR images. The generator design of the improved SRGAN is based on the deep residual network (ResNet) architecture. Using a sequence of convolutional and deconvolutional layers, the generator network in the improved SRGAN aims to learn the mapping between low-resolution and high-resolution images.
However, information loss can occur during upsampling and downsampling, and to minimize this loss, skip connections have been introduced between encoder, and decoder blocks. To efficiently convey information from the encoder to the decoder and be useful for biological image analysis, a variety of skip connection blocks have been designed.
Attention is also a crucial component in achieving high semantic and spatial localization accuracy, and feature maps are given channel and spatial attention to facilitate proper feature selection. This idea has been demonstrated to be useful in various studies in the arena of medical image processing.
The energy-based GAN (EBGAN) is a comparatively novel type of GAN was proposed by Zhao et al.28. It has repeatedly shown its ability to generate more realistic and high-quality images. The use of EBGAN to develop a surface electromyography generator (SEMG), and the results obtained were shown to generate sufficient real EMG features. EBGAN-based model, IDS-EBGAN, to develop an anomaly intrusion detection system. It was shown that after training the given discriminator was able to achieve a perfect reconstruction of normal network traffic.
It can be seen from the literature survey that although deep learning may have indeed led to better results in brain MRI IIC, most of the previous works do not take into consideration the energy-efficiency of the model. To address this issue, a modified version of the EBGAN approach has been proposed in this study30,31,32,33.
Lu et al. introduced RanCom-ViT model uses a pre-trained Vision Transformer (ViT) as its backbone, selected on the basis of it outperforming convolutional neural networks (CNNs) on larfer datasets. The RanCom-ViT model utilizes transfer learning through the employment of a pre-trained vision transformer (ViT) as its base architecture. In this strategy, the ViT model34 that has been initially trained on a large dataset to perform any general task is fine-tuned on the particular task of classifying Alzheimer’s disease from brain MRI images. This simply serves to bridge the gap of limited data because the model begins with weights that are nearer to the ideal solution.
Lu et al. suggested CTBViT, a Vision Transformer (ViT)35 variant model designed for tuberculosis (TB) classification using X-ray images. The model utilizes an efficient block structure to minimize computational complexity while ensuring high classification accuracy. Moreover, CTBViT uses a randomized classifier to improve its generalizability across various datasets. The authors claim that CTBViT excels in its performance over state-of-the-art models, making it a prospective tool for autonomous TB diagnosis. The paper further emphasizes the usefulness of transfer learning to enhance the efficiency of computing.
Proposed method
The proposed model relies on deep learning to rectify IIH in brain MR images, and utilizes energy-based generative adversarial networks. It consists of three stages: the generator block, the discriminator block, and the skip connections block. These stages will be thoroughly explained in this method. Figure 1 depicts the basic architecture of GAN.
Generator network
To create images without IIH, the generator block is employed. It is composed of three components: encoders skip connections, and decoders.
Encoders
Fixing pixel intensity variation can be achieved by identifying and eliminating faulty pixel intensity values. To obtain a more detailed understanding of pixel values, one approach is to use large kernels during the convolution process. However, this approach can lead to increased complexity and execution time. Alternatively, dilated convolution can be used to reduce the computational complexity, although this may result in the loss of information about pixel intensities. To overcome this problem, horizontal and vertical convolutions are used in addition to the dilated convolution, which provides more local pixel intensity information.
The model uses four 3 × 3 kernels for dilated convolution, each with a different dilation value of 1, 2, 4, and 6, horizontal, and vertical kernels are applied to the resulting matrices for each dilation value. The system contains four encoder blocks, and each block reduces the feature map by a factor of two. The encoder blocks aim to extract features and generate a deep representation of the data.
Attention-driven skip connections
When downsampling occurs in each encoder block, we lose spatial information. Passing on this information directly to decoder blocks would result in an inaccurate output. To address this issue, attention-driven skip connections are employed. These skip connections use two feature maps: the main feature map and the enhancing feature map. The amount of local information increases with the next encoder block, while spatial information decreases, and vice versa.
The outcome of the corresponding encoder block is the primary feature map, while the enhancing feature map is the output of the encoder block with more spatial/local information. Each attention block provides channel, and spatial attention to the enhancing feature map, resulting in two attention maps that are used to scale the main feature map and recover missing information. For example, the first attention block uses the output of the last encoder block as its enhancing feature map, and so on for the remaining blocks.
Spatial information is compressed and some local pixel intensity details may be lost when the encoder down samples. Repetitive or unnecessary information transfer may result from simply passing feature maps from the encoder to the decoder (as in conventional U-Net architectures). This is addressed by the model’s introduction of attention-driven skip connections, which balance spatial and local pixel intensity information, prioritize important features while disregarding less important ones, and selectively improve feature maps to improve reconstruction quality.
Two essential elements make up each attention-driven skip connection: The output of the current encoder block is the subject of the first one, the Main Feature Map. The second is Enhancing Feature Map, which uses more local or spatial information from the output of an earlier encoder block. In particular, this can be further developed as flows.
-
1.
Extracting Features from Encoder Blocks Each encoder block in the model reduces the size of the feature map by a factor of two. Deep representations of the input image are extracted by each encoder block.
-
2.
2. Attention Module Processing Data from a previous encoder layer is passed to the matching decoder layer via each skip connection. The skip connection refines features using an attention mechanism rather than passing raw features directly.
The attention module generates two attention maps namely channel attention Map and Spatial attention Map. The channel attention map identifies important channels for feature refinement on the other hand Spatial Attention Map identifies crucial spatial regions for information transfer.
-
3.
3. Feature Enhancement through Attention Maps The attention maps are used to refine the enhancing feature map.This eliminates extraneous noise while preserving crucial details.The primary feature map is combined with the scaled feature maps.
-
4.
4. Fusion in the Decoder The matching decoder block receives the output of the attention-driven skip connection. The decoder reconstructs the corrected image with improved intensity correction after up sampling the feature maps.
Decoder
The inputs to all the decoders are the outputs of the attention blocks and the previous decoder’s output. The outcome of the last encoder is fed into the first decoder. The decoders use the pixel shuffle technique to upsample the input feature maps and then concatenate them with the feature maps enriched with attention from the attention-driven skip connection modules. The decoder blocks produce two outputs, one is passed on as input to the next decoder, and the other is the final output, which is an IIC image. The dimensions of the output image are adjusted to match those of the ground truth image by performing convolution and pixel shuffle operations. Finally, before producing the final corrected image, a 3 by 3 kernel convolution operation is applied.
Discriminator network
Without any clear probabilistic meaning, the discriminator might be considered an energy function (or contrast function). It is possible to think of the energy function, the discriminator computes as a trainable cost function for the generator. The discriminator is taught to provide areas with high data densities the lower energy values and provide higher energy values for those outside of these areas.
Furthermore, the generator may be thought of as a trainable parametric function that generates samples in parts of the space where the discriminator has assigned low energy. In the context of this study, the discriminator has an autoencoder architecture, and the energy is the reconstruction error, i.e., the MSE.
To direct the energy function, the discriminator’s output is subjected to an objective function, which assigns lower energy to genuine data samples and higher energy to artificially created ones. To provide higher-quality gradients in the case when the generator is far from convergence, two distinct losses are utilized, one to train D and the other to train G, in a manner like that used with the probabilistic GAN.
The discriminator’s loss function is designed to assign lower energy (reconstruction error) to real images and higher energy to generated ones. Considering a positive margin m, a data sample x (ground truth or IIH free image, in the context of the study) and a generated sample G(z) from pseudo noise z, the discriminator loss \({L}_{D },\) and the generator loss \({L}_{G}\) are formally described as
In energy-efficient GANs with autoencoders, energy typically refers to the reconstruction error from an autoencoder. The discriminator in this case is an autoencoder that tries to reconstruct the input image. The autoencoder i.e., discriminator is trained to reconstruct real images well. So, when a real image is input, the reconstruction error (energy) is low.
Energy is defined as:
where: D(z) = Output of the autoencoder discriminator, x = Input image (real or generated), E(z) = Energy (reconstruction loss).
Figure 2 depicts the autoencoder architecture where X corresponds to the ground truth. Autoencoder consists of encoder and decoder blocks that measure the MSE. The encoder used within the autoencoder system of the discriminator is a set of four convolutional layers with 64, 128, 256, and 512 filters which is shown in Fig. 3. It performs convolution on the input to extract a detailed feature map that is compressed in size compared to the input. The decoder has two transpose convolutional layers, which in turn perform transpose convolution on the output of the encoder, thus reverting the enhanced feature map to its original size, so that the inputs to the MSE block are of the same size.
The reconstruction-based output provides a variety of targets for the discriminator rather than training the model with only one bit of target information, as in a regular GAN where the discriminator acts as a binary classifier. Only two objectives are conceivable with the binary logistic loss; therefore, inside a mini batch, the gradients that correspond to different samples are probably far from orthogonal. Because of this, training becomes ineffective, and in current technology, lowering the mini batch sizes is frequently not a possibility. The reconstruction loss, on the other hand, is likely to result in highly varied gradient directions inside the mini batch, enabling a higher mini batch size without sacrificing efficiency.
Autoencoders have long been used to depict organically arising energy-based models. Autoencoders can learn an energy manifold with no guidance or negative examples when trained with a few regularization factors. This suggests that the discriminator helps to uncover the data manifold on its own even when an autoencoding model is trained to recreate an actual sample. In contrast, a discriminator trained using binary logistic loss loses all utility in the absence of negative samples from the generator.
Loss functions
The loss functions used for the optimization of the system are given below.
Guidance L1 loss
The given equation can represent it
The symbols yp and yg represent the pixel intensity and ground truth values, respectively. The equation aims to reduce the sum of the absolute differences between the predicted and ground truth values. The predicted image is obtained from the output of the decoder, which helps the model generate a more precise representation.
Adversarial loss
The given equation can represent it.
The primary significance of this loss function lies in its fundamental role in the system. It is based on the principle that the generator block intends to curtail the ability of the discriminator block to differentiate between an actual image and a generated image, while the discriminator block tries to maximize its ability to do so.
Histogram correlation loss
As we are aware, a histogram is a graphical representation of the pixel intensities in an image. Since the histograms of the IIH images and ground truth images differ, this loss function aims to increase the correlation between them. The correlation equation and loss function equation are as follows: –
3D loss
The format used to represent each 2D image in the system is a 3D matrix with layers ranging from 0 to 255. Each layer is a 2D binary layer where pixels with the corresponding layer value are marked as 1 and the rest are marked as 0. Both the predicted and ground truth images are represented in this format, and the average absolute difference between the two is calculated to determine the loss. The following is an expression for the loss equation.
The height, width, and depth of an image are represented by H, W, and D, respectively. The 3D matrix of the predicted image is expressed by Pp, and that of the ground truth image is represented by Pg.
Pulling-away term
To prevent the autoencoder model from creating samples that are concentrated in only or a small number of modes of the data samples, a “repelling regularizer” that integrates well into the model is utilized. A representation-level running pulling-away term (PT) is used to implement the repelling regularizer. Let \(S \in {R}^{s\bullet N}\) be a collection of sample representations obtained from the encoder output layer. Thus, PT is defined as
Complete loss function
The equation can be represented as given.
where α, β, and γ are the weighted terms that contribute to controlling the efficiency of the model.
Figure 4 shows the generator training stage after discriminator training with the ground truth (X), and Y refers to the IIH corrupted image.
To make the generated data statistically identical to real data is the primary goal of incorporating these loss functions into the suggested model. Table 1 illustrates the types of losses and their purpose to calculate and how effectively boosting the output of the model.
Among all the losses, Histogram losses are more important than remaining because it will help to improve the contrast of the image.
Simulation results
The training dataset used consists of simulated along with real brain MR images. The simulated dataset, which consisted of T1-weighted MR images, was attained from BrainWeb. All the ground truth images used were obtained from this database. To increase the ground truth image dataset, data augmentation techniques including rotation of images, and flipping of images were applied. Furthermore, the IIH affected images were obtained from BrainWeb (40%, 20% and 10% IIH). Additional IIH affected images were also obtained from the Kaggle brain MRI datasets, which consisted of real brain MRI data.
Parametric metrics
-
1.
The computation of the mean squared error (MSE) involves determining the mean of the squared differences between the pixels of the original and reconstructed image. This measure provides information on how far the reconstructed image deviates from the original.
$$MSE = \left(\frac{1}{N}\right)* \sum \left({\left(X\left[m,n\right]- Y\left[m,n\right]\right)}^{2}\right)$$(12)In the original image, X[m,n] represents the pixel’s intensity at location (m,n), while Y[m,n] represents the intensity of the corresponding pixel at location (m,n) in the reconstructed image. N is the total number of pixels in the image.
-
2.
The root mean square error (RMSE) is calculated as the square root of the average of the squared differences between the pixels of the original and reconstructed image. This measure provides information on the degree of deviation of the reconstructed image from the original. It can be given as
$$RMSE = \left(\frac{1}{M}\right)* \sqrt{\sum {\left(X\left[m,n\right]- Y\left[m,n\right]\right)}^{2}}$$(13) -
3.
The difference between the original and reconstructed images is measured using a decibel (dB) metric called peak signal-to-noise ratio, or PSNR. It computes the peak signal power to noise power ratio in the image, providing information on the degree of dissimilarity between the two images. It is calculated as such:
$$PSNR = 10 * {log}_{10}(({MAX}^{2}) / MSE)$$(14)where MAX is the maximum possible pixel value in the image (e.g., 255 for an 8-bit image).
-
4.
The structural similarity index (SSIM) is a gauge of the likeness between two images that considers the way the human visual system perceives images.
$$SSIM = \frac{ (2*{u}_{1}*{u}_{2}+{c}_{1}^{2}) * (2*{\sigma }_{1}^{2}+{c}_{2}^{2}) }{(({u}_{1}^{2}+{u}_{2}^{2}+{c}_{1}^{2}) * ({\sigma }_{1}^{2} + {\sigma }_{2}^{2} + {c}_{2}^{2}))}$$(15) -
5.
The multi scale structural similarity index (MSSSIM) determines the similarity between two images by analyzing and contrasting their luminance, contrast, and structure.
$$\text{MSSSIM }(\text{P},\text{ Q}) = \left(\frac{1}{M}\right)* \sqrt{\sum \left(SSIM(P,Q)\right)}$$(16)
A comparative study is also performed using the SSIM, MSSSIM, PSNR, MSE and RMSE metrics as comparison parameters. In the study, staple methods such as BM3D, NLM, WNNM36, image denoising methods such as FFT, Bayes37, and VisuShrink30 are also used in the comparison. Furthermore, other deep learning methods such as InhomoNET, Pix2Pix and CycleGAN are also compared31.
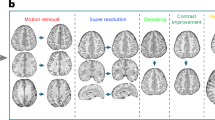
The metrics are applied to the resulting, corrected images from each method and their corresponding ground truths. Table 2 shows the results obtained using the proposed method, the IIH affected the image, and the ground truth. It is clear from the images that the proposed method gives a fair reduction in IIH levels.
The parameters SSIM, PSNR, MSE, and RMSE averages for ten images using each standard method are shown in Table 3. The SSIM, PSNR, MSSSIM, and MSE averages for ten images using each deep learning method are shown in Table 4. The results of comparative analysis with standard image denoising techniques are discussed in these tables. The comparative analysis with contemporary deep learning techniques, along with a comparison of the number of layers employed in network architecture is shown below.
Figure 5 depicts SSIM parameter for various traditional methods, and the proposed model shows a better value than the existing one. From Fig. 6, we can conclude that PSNR and MSE for proposed model have good compared with traditional methods. Figure 7 shows SSIM and MSSSIM metrics for proposed model with existing deep learning models. Figure 8 shows PSNR, and MSE of different deep learning models with proposed one. These findings show that for the objective of IIC correction on simulated brain MRI data, the suggested technique outperforms traditional and deep learning methods.
The generator and discriminator blocks are trained, and the losses occurring at individual blocks are shown in Fig. 9 depicts the training loss of generator with a final loss value of 0.01506. Figure 10 shows that the Discriminator loss variation throughout model training, with a final loss value of 0.002195. Figure 11 depicts total loss variation throughout model training, with a final loss value of 0.9424. From this we can conclude that the loss is very less while increasing the epochs.
The proposed model is compared with baseline model with respect to different metrices like FLOP (Floating Point Operations), Inference time, Memory Footprint and power consumption which is discussed in Table 5.
The proposed model performs better than existing model in terms of various parameters like Architecture, model mtrics, skip connections wise, usage of various activation functions, batch size etc., is described in Table 6. From this we can see how the proposed model gives better results.
Discussion
Input data is mapped by autoencoders to a lower-dimensional latent space representation. This latent space model, when used with autoencoders in modified EBGAN, captures the salient characteristics of the input pictures. The generator in modified EBGAN can create pictures that are more consistent with the data distribution by utilizing this learnt latent space, perhaps lowering noise.
Modified EBGAN formulates the training of GANs as an energy minimization problem. As an energy function, the discriminator in modified EBGAN assigns greater energy to created data and lower energies to genuine data. Cleaner, noise-free pictures are produced by the generator because of learning to create samples that are closer to the actual data distribution through the minimization of the energy function.
Besides the adversarial loss, modified EBGAN also includes an adaptive reconstruction loss. The generator is motivated by this loss to provide samples that the autoencoder can accurately recover. EBGAN with autoencoders can produce pictures that not only mislead the discriminator but also closely match the input data, resulting in less noise, by maximizing both the adversarial and reconstruction losses.
Based on the findings mentioned above, it can be said that the suggested technique provides a reliable solution for IIC in MR images. The technique not only outperforms conventional IIC and image denoising methods, but it also performs better than the stated deep learning methods.
The limitations of the proposed model are, the autoencoders require careful tuning of the reconstruction loss to balance feature extraction and generation quality. The combined framework requires significant computational power due to dual adversarial training and reconstruction loss optimization. Hyperparameter tuning is complex and resource intensive. The autoenocders are Overfitting to specific noise or artifact patterns. The major challenges of the proposed model are the training instability in GANs leads to longer convergence time. Real-time performance is hard to achieve with large encoder-decoder and GAN models. By replacing the standard convolutions with depthwise separable convolutions and use of loss-aware energy optimization during training may help to avoid the above challenges.
The ADAM optimizer was implemented with a batch size of 1 image and an initial learning rate of 0.0001 for the initial 50 epochs, and it was subsequently lowered linearly to 0 over the following 50 epochs. The alpha, beta, and gamma values were assigned values of 10, 1, and 100, correspondingly, based on trial and error to achieve the best performance of the model. Each epoch is having 120 iterations. The implementation was performed using the TensorFlow network in Python.
The proposed model has effectively produced IIC free images. The performance parameters like PSNR is above 30, MSE is around 41, and SSIM is 0.9665 which is closest to 1.
The Proposed model combines Generative Adversarial Netwrok (GAN) with Auto Encoders. The Autoencoders helps in Save Energy because it has Fewer skip connections, no deep residual blocks, Reduces activations & memory usage mid-network, Requires less computation during reconstruction and lower power draw on GPU. In the proposed model the discriminator role is replaced with Autoencoders, which are effectively minimizes the noise involved in the images.
Conclusion
This paper proposes a modified energy-efficient deep learning model to correct IIH in brain MR-images, incorporating an energy-based discriminator, enhanced generator, and attention-driven skip connections, enhancing system performance by recovering latent features. Furthermore, the discriminator also includes a reconstruction error (MSE) which acts as ‘energy’. Thus, higher energy is attributed to generated images and lower energy to real images. This error is also used by the generator to improvise its own performance. The generator consists of information modules that apply dilated as well as horizontal and vertical convolutions to images to obtain maximum data on pixel intensities. Additionally, the attention-driven skip connections help the system to obtain a balance of both spatial and local pixel intensity information. Using effective feature extraction, robust architecture with autoencoders and GANs for improved performance and image denoising, and GAN-based data generation to improve generalization without conventional augmentation, the suggested method outperforms previously used approaches. The modified EBGAN has better values in performance evaluation parameters such as PSNR is 32.73, MSE is 41.24, RMSE and SSIM are 6.421 and 0.9665 respectively. The proposed system is found to give superior performance in terms of the quality of images obtained and IIH reduced in brain MR-images for further segmentation and classification to save the life of a human. Hence, the Coefficient of Joint Variation (CJV) metric can be used to judge the level of IIC achieved. Similarly, segmentation can also be performed on the IIC results to assess their quality.
Data availability
The data used to support the findings of this study are available from publicly available datasets. https://brainweb.bic.mni.mcgill.ca/. https://www.kaggle.com/datasets/mhantor/mri-based-brain-tumor-images
References
Balafar, M. A. et al. Review of brain MRI image segmentation methods. Artif. Intell. Rev. 33, 261–274 (2010).
Lin, Q. et al. A novel approach of surface texture mapping for cone-beam computed tomography in image-guided surgical navigation. IEEE J. Biomed. Health Inform. https://doi.org/10.1109/JBHI.2023.3298708 (2023).
Vovk, U., Pernus, F. & Likar, B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans. Med. Imaging 26(3), 405–421 (2007).
Venkatesh, V., Sharma, N. & Singh, M. Intensity inhomogeneity correction of MRI images using InhomoNet. Comput. Med. Imaging Graph. 84, 101748 (2020).
Dai, X. et al. Intensity non-uniformity correction in MR imaging using residual cycle generative adversarial network. Phys. Med. Biol. 65(21), 215025 (2020).
Chuang, K. H., Wu, P. H., Li, Z., Fan, K. H. & Weng, J. C. Deep learning network for integrated coil inhomogeneity correction and brain extraction of mixed MRI data. Sci. Rep. 12(1), 8578 (2022).
Zhu, X., Pan, H., Gundogdu, B., Jha, D., Velichko, Y., Murphy, A. B., & Bagci, U. A probabilistic hadamard U-Net for MRI bias field correction. In International Workshop on Machine Learning in Medical Imaging 208–217 (Springer, 2024).
Desai, B., Kushwaha, U. & Jha, S. Image filtering -techniques, algorithm and applications. Appl. GIS. 7, 101 (2020).
Thakur, P., Syamala, N., Karuna, Y., & Saritha, S. Performance analysis of IIC techniques for Brain MR-images. In 2023 10th International Conference on Signal Processing and Integrated Networks (SPIN) 351–357 (IEEE, 2023).
Saladi, S. & Amutha Prabha, N. Analysis of denoising filters on MRI brain images. Int. J. Imaging Syst. Technol. 27(3), 201–208 (2017).
Ali, H. M. MRI medical image denoising by fundamental filters. High-Resolut. Neuroimaging-Basic Phys. Princ. Clin. Appl. 14, 111–124 (2018).
Anand, C. S. & Sahambi, J. S. Wavelet domain non-linear filtering for MRI denoising. Magn. Reson. Imaging 28(6), 842–861 (2010).
Heckbert, P. Fourier transforms and the fast Fourier transform (FFT) algorithm. Comput. Graph. 2, 15–463 (1995).
Gopinathan, S., Kokila, R. & Thangavel., P. Wavelet and FFT based image denoising using non-linear filters. Int. J. Electr. Comput. Eng. (2088-8708) 5 (5), (2015).
Adhikari, S. K. et al. A nonparametric method for intensity inhomogeneity correction in MRI brain images by fusion of Gaussian surfaces. Signal Image Video Process. 9(8), 1945–1954 (2015).
Osadebey, M., Nizar, B. & Arnold, D. Brain MRI intensity inhomogeneity correction using region of interest, anatomic structural map, and outlier detection. In Applied Computing in Medicine and Health 79–98 (Morgan Kaufmann, 2016).
Feng, C., Li, W., Hu, J., Yu, K. & Zhao, D. BCEFCM_S: Bias correction embedded fuzzy c-means with spatial constraint to segment multiple spectral images with intensity inhomogeneities and noises. Signal Process. 168, 107347 (2020).
Ivanovska, T. et al. An efficient level set method for simultaneous intensity inhomogeneity correction and segmentation of MR images. Comput. Med. Imaging Graph. 48, 9–20 (2016).
Yepuganti, K., Saladi, S. & Narasimhulu, C. V. Segmentation of tumor using PCA based modified fuzzy C means algorithms on MR brain images. Int. J. Imaging Syst. Technol. 30(4), 1337–1345 (2020).
Saladi, S. & Amutha Prabha, N. MRI brain segmentation in combination of clustering methods with Markov random field. Int. J. Imaging Syst. Technol. 28(3), 207–216 (2018).
Liu, H. et al. Liver MRI segmentation with edge-preserved intensity inhomogeneity correction. Signal Image Video Process. 12(4), 791–798 (2018).
Anand Kumar, G. & Sridevi, P. V. Intensity inhomogeneity correction for magnetic resonance imaging of automatic brain tumor segmentation. In Microelectronics, Electromagnetics and Telecommunications 703–711 (Springer, 2019).
Yao, Q. Y. et al. Image-based visualization of stents in mechanical thrombectomy for acute ischemic stroke: Preliminary findings from a series of cases. World J. Clin. Cases 11(21), 5047–5055. https://doi.org/10.12998/wjcc.v11.i21.5047 (2023).
Chunwei, T. et al. Deep learning on image denoising: An overview. Neural Netw. 131, 251–275 (2020).
Goodfellow, I. et al. Generative adversarial networks. Commun. ACM 63 (11), 139–144. https://doi.org/10.1145/3422622 (2020).
Zhu, Y. et al. Deep learning-based predictive identification of neural stem cell differentiation. Nat. Commun. 12(1), 2614. https://doi.org/10.1038/s41467-021-22758-0 (2021).
Liu, H., Yuan, H., Hou, J., Hamzaoui, R. & Gao, W. PUFA-GAN: A frequency-aware generative adversarial network for 3D point cloud upsampling. IEEE Trans. Image Process. 31, 7389–7402. https://doi.org/10.1109/TIP.2022.3222918 (2022).
Zhao, J., Mathieu, M., & LeCun, Y. Energy-based generative adversarial network. Preprint at arXiv:1609.03126. (2016).
Huo, R. et al. Associations between carotid atherosclerotic plaque characteristics determined by magnetic resonance imaging and improvement of cognition in patients undergoing carotid endarterectomy. Quant. Imaging Med. Surg. 12(5), 2891–2903. https://doi.org/10.21037/qims-21-981 (2022).
Zhu, J.-Y. et al. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE international conference on computer vision. (2017).
Isola, P. et al. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE conference on computer vision and pattern recognition. (2017).
Karthiga, R. et al. A novel exploratory hybrid deep neural network to predict breast cancer for mammography based on wavelet features. Multimed. Tools Appl. https://doi.org/10.1007/s11042-023-18012-y (2024).
Renugadevi, M. et al. Machine learning empowered brain tumor segmentation and grading model for lifetime prediction. IEEE Access 11, 120868–120880. https://doi.org/10.1109/ACCESS.2023.3326841 (2023).
Lu, S. Y., Zhang, Y. D. & Yao, Y. D. An efficient vision transformer for Alzheimer’s disease classification using magnetic resonance images. Biomed. Signal Process. Control 101, 107263 (2025).
Lu, S. Y., Zhu, Z., Tang, Y., Zhang, X. & Liu, X. CTBViT: A novel ViT for tuberculosis classification with efficient block and randomized classifier. Biomed. Signal Process. Control 100, 106981 (2025).
Ruikar, S. & Doye, D. D. Image denoising using wavelet transform. In 2010 International Conference on Mechanical and Electrical Technology. (IEEE, 2010).
Lebrun, M., Buades, A. & Morel, J.-M. A nonlocal Bayesian image denoising algorithm. SIAM J. Imag. Sci. 6(3), 1665–1688 (2013).
Author information
Authors and Affiliations
Contributions
All authors have contributed equally and agreed for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Karuna, Y., Syamala, N., Ravikumar, C.V. et al. Modified energy-based GAN for intensity in homogeneity correction in brain MR images. Sci Rep 15, 26409 (2025). https://doi.org/10.1038/s41598-025-08552-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08552-8