Abstract
The species of Aquilaria and Gyrinops are renowned for being the primary sources of Agarwood, a valued resin, produced by these trees in response to fungal infections or physical damage. The major derivatives of Agarwood are wood chips and oil which are used in perfumes, incenses, cosmetics, and medicines. Due to high international trade and exploitation in the wild, the members of both genera are listed under Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). Aquilaria khasiana, endemic to the Indian state of Meghalaya, is a less-studied CITES-listed species. The present study reveals its occurrence in an additional location i.e., in Jeypore Reserve Forest, Assam. The species identity was confirmed using morphological and barcode markers (ITS2, rbcl, and matK). The chloroplast genome was sequenced for the first time to explore plastome characteristics and reconstruct well-resolved phylogeny. Additionally, its conservation status assessment was carried out considering its new distributional records. Phylogenetic analyses, based on chloroplast genome data and the three barcode markers showed that both Aquilaria and Gyrinops are paraphyletic, with some species of Aquilaria nested within the Gyrinops clade. Despite possessing distinct morphological traits of Aquilaria, A. khasiana cluster with the Gyrinops along with the only Indian Gyrinops species, viz. G. walla as in the case of all previous phylogenetic analyses. Based on the present and previous molecular studies, it is concluded that Gyrinops is paraphyletic to Aquilaria. The chloroplast genome contained 90 protein-coding genes, 38 tRNAs, and 8 rRNAs. The present conservation status assessment supports retaining the species as Critically Endangered but with modified IUCN criteria and subcriteria.
Similar content being viewed by others
Introduction
Agarwood, also known as ‘Gaharu’, ‘Oud’, ‘Aloewood’, ‘Eaglewood’, is the trade name of a resinous substance obtained from infected trees, particularly belonging to the genus Aquilaria Lam. and Gyrinops Gaertn. (Thymelaeaceae). The major products derived from agarwood are wood chips and agar oil. The chips are burned for their unique aroma, while oil, often called “liquid gold”, is used in perfumes, incenses, air fresheners, cosmetics, medicine, and aromatherapy1. Additionally, the bark and wood of certain species are used to make ropes, cloths, and fuel for fumigation2,3. The dried leaves of A. malaccensis are used for herbal tea, and Boya oil, derived from non-infected wood is used in smokeless flavoured tobacco products3,4. Agarwood also holds significant cultural and religious importance and is utilized by different communities1,2. Currently, Agarwood has become one of the world’s most valuable wildlife commodities5,6. However, due to an increase in global demand and the slow formation of the resinous wood, the supply of agarwood has decreased7 and wild populations are threatened with extinction8.
The genus Aquilaria is native to Assam, Bangladesh, Borneo, Cambodia, China South-Central, China Southeast, East Himalaya, Hainan, Laos, Malaya, Maluku, Myanmar, New Guinea, Philippines, Sumatera, Thailand, Vietnam, and represented by 21 species9. Gyrinops is reported in India, Laos, Lesser Sunda Island, Maluku, New Guinea, Sri Lanka, Sulawesi, and Thailand and is represented by 9 species9. In India 2 species of Aquilaria, viz. Aquilaria khasiana Hallier f. and A. malaccensis Lam. are known to occur in the wild. Whereas, Gyrinops is represented by 1 species in India (Kerala and Tamil Nadu), i.e., Gyrinops walla Gaertn.
Aquilaria malaccensis is the first species of Agarwood listed in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). The species was listed under CITES Appendix II (w.e.f. 16 February 1995) based on India’s proposal to the 9th Conference of Parties of CITES (CoP9) held in Fort Lauderdale in the USA in 1994. Subsequently, all species under the genera Aquilaria and Gyrinops were listed in Appendix II of CITES based on Indonesia’s proposal at CoP13 in 2004 (effective from 12 January 2005).
Aquilaria khasiana has been known to be endemic to the Indian state of Meghalaya10. Recently, during field surveys in connection with the study on some CITES-listed species, the authors located A. khasiana in Mawkasain, Mawsynram subdivision of East Khasi Hills district in Meghalaya, which is the same location reported by Mir et al.10. Interestingly, the species was also found in a new location, i.e., in Joypur Reserve Forest in Dibrugarh district of Assam. It was tentatively identified as A. khasiana or a species of Gyrinops by Bhattacharjee et al.4 due to the absence of flowers/ fruits. However, its identity is now confirmed as A. khasiana based on specimens collected in flowering conditions during May–June 2024 from both locations.
The generic delimitation of Aquilaria and Gyrinops is controversial due to overlapping morphological and molecular characters. Hallier11 treated Gyrinops as a synonym of Aquilaria as he did not observe any significant morphological difference between them, whereas Hou12 treated these two as distinct genera based on number of stamens (10 in Aquilaria vs. 5 in Gyrinops). Hou12 also mentioned, “Aquilaria and Gyrinops seem to be very closely allied, the first being diplostemonous, the second haplostemonous, which is the only constant character”. However, variation in number of stamens (8–12) has been observed in Aquilaria, though all Gyrinops species have 5 stamens. The controversy was amplified when a molecular study by Eurlings and Gravendeel13 reported the paraphyletic placement of Aquilaria and Gyrinops. Farah et al.14 also doubted their recognition as two distinct genera because their study did not reveal a clear pattern of the grouping. Nevertheless, A. khasiana is not included in most of the phylogenetic analyses except the study by Eurlings and Gravendeel13 using only one marker (trnL-F).
The chloroplast/ plastid is an important organelle in plants. It plays a major role in photosynthesis and carbon fixation. It contains a circular genome that encodes ribosomal proteins associated with photosynthesis15,16. The size of the chloroplast genome (cp. genome) ranges from 120 to 170 kilo base pairs and is composed of four regions namely a large single copy (LSC) region, a small single copy (SSC) region, and two inverted repeats (IR) regions which separates both LSC and SSC regions17. The whole cp. genome sequence-based phylogenetic study is an effective tool in plant systematics due to its high resolution. Being highly conserved, the cp. genome allows resolving relationships even among closely related species18,19,20,21. Chloroplast genome structure comparison and phylogenetic analysis have been carried out in recent times in Aquilaria and Gyrinops due to the poor phylogenetic resolution in DNA marker-based phylogenetic analysis22,23. However, in genera exhibiting intergenus hybridization, the efficiency of cp. genome-based phylogenies is compromised due to processes like chloroplast capture and introgression which can mislead the phylogenetic outputs24,25.
The present study reports the existence of A. khasiana, a Critically Endangered species, at a new location (Jeypore Reserve Forest, Assam). To confirm the molecular identity of specimens collected from Joypur, Assam and to find the phylogenetic position of A. khasiana, phylogenetic analysis was carried out using nuclear and chloroplast markers. The study also aimed the sequencing and characterization of the whole chloroplast genome of A. khasiana collected from its type locality and a chloroplast genome-based phylogenetic analysis for the first time. An updated assessment of the species by following the IUCN’s guidelines26 is also provided.
Results
Aquilaria khasiana significantly differs from A. malaccensis morphologically. Aquilaria khasiana is up to 5 m high shrub or small tree, even sometimes almost looking like a climber (young plants); whereas A. malaccensis is up to 40 m tall tree. The inflorescence of A. khasiana is subsessile terminal or extra-axillary fascicle with up to 10 flowers, each having stamens with sessile anthers; whereas in A. malaccensis, the inflorescence is usually branched into 2–3 terminal umbellate subsessile to shortly peduncled cymes, each with 8–13 flowers, and the stamens are with subsessile anthers. The detailed taxonomic treatment of the species is provided here.
Taxonomic treatment
Aquilaria khasiana Hallier f., Meded. Rijks-Herb. 44: 18. 1922; Kanjilal, P.C. Kanjilal, R.N. De & Das, Fl. Assam 4: 113. 1940; J. Joseph, Fl. Nongpoh: 228. 1982; Mir, D.K. Roy & K. Upadhaya, Rheedea 27(2): 86. 2017. Type: INDIA. Meghalaya, ‘Khasia’, ‘3000 ped’ [3000 foot/ 914.4 m], J.D. Hooker & T. Thomson s.n. [Holotype L, barcode 0010149, digital image!; isotype BM, barcode BM014125546, digital image!; L, barcode 0010150, digital image!; GH, barcode 00443697, digital image! ] (Figs. 1 and 2).
Description
Shrubs or small trees up to 5 m high. Stem with whitish to greyish, thin, smooth bark, branchlets pale brown, slender, terete. Leaves ramal, alternate, spirally arranged, exstipulate, petiolate, petiole terete, sericeous, 2–3 mm long, lamina simple, oblong to elliptic or obovate to oblanceolate, sometimes elliptic, 8–15 × 2.5–5 cm, acute at base, acuminate at apex (acumen 10 to 15 mm), margins revolute, midrib pubescent abaxially, unicostate, reticulate, pinnately veined; basal sericeous scale-like appendages covering axillary vegetative buds, scales lanceolate, white. Inflorescence terminal or axillary, sometimes internodal umbel, 8–10 flowered. Flowers ebracteate, ebracteolate, complete, bisexual, dichlamydeous, actinomorphic, hypogynous, yellowish green, 0.9–1.5 cm long; pedicellate, pedicel terete, sericeous, 4–6 mm long. Sepals united to form calyx tube, 9–10 × 1.4–2 mm, persistent, with 4 or 5 free lobes at apex, lobes lanceolate to oblong-lanceolate, obtuse, spreading, pubescent, valvate, yellowish green. Petaloid appendages 8–10, erose, inserted at the base of calyx lobes, 1–2 mm long, obtuse, valvate, pubescent, yellowish green. Stamens 8–10, free, epipetalous, slightly exerted, introrse; sessile; anther-lobes 2, dorsifixed, lobes c. 1 mm, dehiscence longitudinal, yellow. Carpel 1, ovoid, 1–2 mm long, densely pubescent; ovary ovoid, sessile, superior, 2-chambered, with single bitegmic ovule per locule; style cylindric, length c. 1 mm; stigma capitate, c. 1 mm thick. Fruits loculicidal capsules, obovate to oblanceolate, 3–4 × 1–1.5 cm, compressed, green, pubescent, bilocular, 1 seed per locule, base cuneate, apex obtuse to acute; seeds ovoid to ellipsoid, 1–1.2 × 4–6 mm, base attenuate, elongated into a slender appendage, appendage 0.8–1 cm long, dark brown, pubescent, apex acuminate.
Flowering: May–July, Fruiting: July–October.
Habitat
The species is found in the subtropical evergreen forest of Assam and Meghalaya with up to 1300 m elevation. The plant is found along with Acronychia pedunculata (L.) Miq., Castanopsis tribuloides (Sm.) DC., Calophyllum polyanthum Wall. ex Choisy, Camellia caudata Wall., Cinnamomum tamala (Buch. -Ham.) T. Nees & C.H. Eberm., Eurya acuminata DC., Garcinia anomala Planch. & Triana, Litsea elongata (Nees) Hook.f., Lithocarpus dealbatus (Hook.f. & Thomson ex Miq.) Rehder, Macaranga denticulata (Blume) Müll.Arg., Syzygium tetragonum (Wight) Kurz10 and also with Lagerstroemia speciosa (L.) Pers. and some species of ferns.
Distribution
INDIA [Meghalaya, Assam (reported here)]; endemic.
Etymology: The generic name originated from the Latin Aquila, ae “an eagle” or from the Akkadian eklu “dark: said of the day,” ekelu “to be dark: said of the sun, the day”16. The specific epithet is after its type locality, i.e., ‘Khasia’ (now in the Khasi Hills division of Meghalaya).
Additional specimens examined
INDIA. Meghalaya: Umteswar forest, 7.7.1935, S.R. Sarma 12036 (ASSAM); Umteswar, 28.8.1936, S.R. Sarma 13539 (ASSAM); Umteswar forest, 28.8.1936, S.R. Sarma 13540 (ASSAM); Umsaw, 16.7.1942, S.R. Sarma 21453 (ASSAM); Cherrapunjee, c. 1219 m, 18.7.1952, R.C. Thakur 6146 (MICH, barcode 1508045, L barcode 2472286, digital image); Cherrapunjee, c. 1219 m, 14.8.1952, W.N. Koelz 31115 (MICH, barcode 1508044, digital image, L barcode 2472285, digital image); Mawsynram, Mawkasain, 20.9.2016, A.H. Mir 90340 (ASSAM), Mawsynram, Mawkasain, 1142 m, 08.11.2023, B. Mallick, T. Shil & R. Shaw 93819 (CAL); Mawsynram, Mawkasain, 1142 m, 14.06.2024, R. Layola M.R. & B. Mallick 95387 (CAL). Assam: Jeypore Reserve Forest, 175 m, 31.10.2023, S. Sengupta 93764 (CAL); Jeypore Reserve Forest, 178 m, 25.5.2024, S. Sengupta 101066 (CAL). Assam/ Meghalaya: Griffith s.n. (K, barcode K000357720, digital image).
Note
Another herbarium sheet has been found at K with a single barcode (K000357720, image! ) which is a heterogeneous mixture of two species, viz. Aquilaria khasiana and A. malaccensis. Only the fruiting specimens mounted on the sheet are of A. khasiana, whereas the flowering specimens are of A. malaccensis. The label data (East Bengal, Khasia, Griffith 4381) associated with the K-sheet most probably corresponds to the specimen of A. malaccensis, not to A. khasiana.
According to the protologue, the number of calyx lobe and petaloid appendages are 4–5 and 8–10 respectively. However, 5 calyx lobes and 10 petaloid appendages were observed in all the flowers we dissected during our study. The photo plate provided by Mir et al.10 also portrays the presence of 5 calyx lobes.
Phylogenetic position of Aquilaria khasiana and confirmation of its extended distribution
The molecular analysis showed that the samples collected from the Mawkasain, East Khasi Hills district of Meghalaya and Dibrugarh district of Assam are identical to A. khasiana in all the markers except a single nucleotide difference in the 75th position of the matK region. All four accessions clustered in a single cluster confirming its conspecificity and extended distribution in the Dibrugarh district of Assam. However, the DNA sequences of the samples did not show affinity with any other species of Aquilaria, instead, it nested in an entirely different Clade with the only one Gyrinops sp. present in India and Sri Lanka i.e., G. walla Gaertn. (Fig. 3).
The maximum likelihood analysis resulted in a paraphyletic tree. Even though some species of Gyrinops are placed in Aquilaria and the other way round, each resulting clades were comprised of species that were of the same geographic distribution. Species in Clade I (A. malaccensis¸ A. microcarpa, A. beccariana, A. cumingiana, and G. versteegii) and Clade IV (A. rostrata and G. caudata) included species belonging to the Malayan peninsular region. The Clade II [A. agallochum (= A. malaccensis, collection from India), A. malaccensis (present collections from India), A. rugosa and A. yunnanensis], Clade III (A. sinensis, A. crassna, and A. subintegra), and Clade V (A. khasiana and G. walla) comprised of species occurring in the upper Southeast Asia, China, India and Sri Lanka. The phylogenetic tree was well supported with high bootstrap value in most of the major branches. Conspicuously, A. agallochum (MH134137) (synonym of A. malaccensis, Indian element) along with our present collections of A. malaccensis (vouchers 93711, 93712, 93825, and 93828) and A. malaccensis (Malayan element) were placed in separate clades (Clade II and Clade I respectively) with other species of respective origin.
The phylogenetic tree resulted from the maximum likelihood (ML) and Bayesian inference (BI) analysis using the chloroplast genome of Aquilaria and Gyrinops spp. as ingroup. Phaleria macrocarpa, Wikstroemia ridleyi, and Gonystylus bancanus of the same family are used as outgroups. The molecular placement of Aquilaria khasiana is shown, which is nested along with Gyrinops walla in clade V. Bootstrap support values (BS ≥ 50) from the ML analysis and posterior probability values (PP ≥ 0.5) from the BI analysis are provided in the respective branches of the tree. The GenBank accession/ voucher number of the ITS marker is prefixed with the species’ name for reference.
Chloroplast genome characteristics of Aquilaria khasiana
The DNA integrity in the sample assessed on Genomic DNA screen tape showed a DNA Integrity Number (DIN) value of 7. The length of the raw Nanopore data generated for the sample was ~ 4.68 GB. The adapter-free Nanopore long reads were mapped against the reference chloroplast genome of Aquilaria sinensis. A total of 47,933 (5.02%) uniquely mapped reads were obtained and used to assemble the chloroplast genome of Aquilaria khasiana. The assembly resulted in a single contig with 178,834 bp genome length. The GC content of the genome was estimated as 37%. The NCBI nucleotide blast result reported maximum homology (99% coverage and identity) of the assembled chloroplast sequence against Gyrinops and Aquilaria species chloroplast genomes.
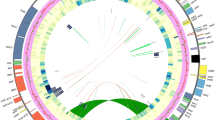
The annotation of assembled chloroplast sequence predicted protein-coding and rRNA genes based on the identification and mapping of the most similar, full-length protein, cDNA and rRNA sequences by integrating results from Blastx, Blastn, protein2genome and est2genome programs. It also identified tRNA genes and inverted repeats (IR) using tRNA scan, ARAGORN and Vmatch 2.3.1 respectively. A total of 90 protein-coding genes, 38 tRNAs and 8 rRNAs were predicted for the chloroplast genome of A. khasiana (Table 3). The circular genome map showed all the predicted features of the chloroplast genome (Fig. 4).
Chloroplast genome map of Aquilaria khasiana showing the Large Single Copy (LSC) regions, Small Single Copy (SSC) regions, and Inverted Repeats (IR) of the genome with relative positions of predicted genes. Genes marked outside the external circle are transcribed anticlockwise direction, while genes marked inside the external circle are transcribed clockwise direction. Colour represents the functional group of the genes.
Comparison of the chloroplast genome structure of Aquilaria khasiana and related species
The comparative chloroplast genome structure analysis of A. khasiana with fifteen related species showed minor differences (Fig. 5 ). The rpl2 gene is found completely in the IRA and IRB region of all the species, whereas in A. khasiana, the rpl2 was extended in IRA and IRB by 45 and 46 bases respectively. Similarly, the rps19 gene is restricted to LSC in A. khasiana, while it is extended towards the IRB region in all other species. All other junction sites (IRB/SSC and SSC/IRA) were similar in all species with few base pair differences.
Chloroplast sequence-based phylogeny analysis of Aquilaria khasiana and related species
Phylogenetic analysis using the complete chloroplast genome of 15 Aquilaria spp., 8 Gyrinops spp., and 4 Outgroup species resulted in a tree with well branch support. Four distinct clades were formed in the ingroup taxa, of which most of the Aquilaria spp. were nested within the clades I and II. Whereas most of the Gyrinops spp. were nested within the clades III and IV. However, here also, A. khasiana was placed consistently within the Gyrinops clade (clade IV) showing close affinity with G. walla. This clade represented the basal clade in the phylogenetic tree. Similarly, G. caudata was nested within the Aquilaria cluster. Nevertheless, the four clades were in accordance with the geographic origin of the species where the members of the clades I and IV are of upper southeast Asian, Chinese, and Indian/Sri Lankan origin. Clades II and III represented the species with Malayan peninsular origin (Fig. 6).
The phylogenetic tree resulted from the maximum likelihood (ML) and Bayesian inference (BI) analysis using the chloroplast genome of Aquilaria and Gyrinops spp. as ingroup. Daphne kiusiana, Stellera chamae, Phaleria macrocarpa, Wikstroemia ridleyi, and Gonystylus bancanus of the same family are used as outgroups. The molecular placement of Aquilaria khasiana is shown, which is nested along with Gyrinops walla in clade V. Bootstrap support values (BS ≥ 50) from the ML analysis and posterior probability values (PP ≥ 0.5) from the BI analysis are provided in the respective branches of the tree. The GenBank accession/ voucher number of the ITS marker is prefixed with the species’ name for reference.
Assessment of conservation status
Aquilaria khasiana was known from some historical collections by Hooker & Thomson (in between 1848 and 1851), and Griffith (most probably during 1837–1838) from ‘Khasia’ (Meghalaya)/ erstwhile ‘Assam’ (includes the present Khasi Hills division, Meghalaya). Later, the species was further collected from Umteswar forest, Khasi Hills division, Meghalaya in 1935 and 1936, Umsaw forest, Khasi Hills division, Meghalaya in 1942, Cherrapunjee, Khasi Hills division, Meghalaya in 1952. However, there is no record of the collection of this species after 1952 until Mir et al.10 rediscovered it in 2016 from Mawkasain, Meghalaya. Mir et al.10 reported nine individuals (including one mature individual) of A. khasiana at Mawkasain, Meghalaya. During the first field survey in Mawkasain, Meghalaya in November 2023, seven individuals (including one mature individual) were observed. The number got reduced to five (with the same mature individual) when revisited after six months, i.e. in June 2024. The existing single mature individual was reported to be cut down in 20168, however, the plant is still surviving by its coppices. The new population located in Jeypore Reserve Forest, Assam was represented by around 800 individuals (including 210 mature individuals) as observed in October 2023 and May 2024. The quality and extent of habitat in Mawkasain, Meghalaya is on continuous decline due to forest fires, and anthropogenic activities like timber extraction, gathering of non-timber forest products, extraction of plants as an alternative source of agarwood, and collection by villagers for firewood. The single mature individual was found affected by the larvae of Heortia vitessoides Moore (Fig. 1c, d). However, the habitat quality in Jeypore Reserve Forest, Assam is not much affected, though extraction of some plants in past has been reported by the local villagers. The overall observed, inferred and suspected population reduction was more than 80% in the past where the causes of reduction have not ceased and are not reversible. Therefore, the species meets the threshold for the Critically Endangered category under criterion A (A2cd)26,27. For criteria B, only two locations (Mawkasain and Jeypore; Fig. 7) where the species is presently found are considered, because the species could not be located in previously known locations (Umteswar, Umsaw, Cherrapunjee) despite several surveys. The species is now present in two distant locations. As there are less than 3 locations, a minimum convex polygon cannot be created covering both locations for calculating the Extent of Occurrence (EOO). The EOO estimation by creating a ‘buffer’ will not be accurate, and therefore, the EOO is not considered here for assessing the conservation status. The Area of Occupancy (AOO) is estimated at 8 km2 by considering a 2 × 2 km grid size, i.e. 4 km2 for each location. However, as the Assam subpopulation (i.e. majority of the overall population) is within a reserve forest and less affected by threats, the requirements for the subcriteria under criteria B2 are not met at present (Kathryn Fowler, pers. com. in October 2024). Since the population is small with very few mature individuals (< 250) and continually declining in terms of the percentage of mature individuals in one subpopulation, it meets the criterion C2a(ii) of IUCN26,27 for CR. Hence, A. khasiana is assessed as Critically Endangered [CR A2cd; C2a(ii)] in the present study.
Distribution map of A. khasiana generated using ArcGIS version 10.7.1 (https://www.esri.com/en-us/arcgis/products/arcgis-desktop/overview).
Discussion
Extended distribution of Aquilaria khasiana
Aquilaria khasiana was first described by Hallier11 based on Hooker and Thomson’s collection (as ‘Gyrinops?’) from ‘Khasia’, Meghalaya, India between 1848 and 1851. But it has been observed that its first collection was made by Griffith as ‘Aquilaria agallocha’, most probably during 1837–1838, though the precise location in the erstwhile ‘Assam’ which includes the present-day Meghalaya, and date/ year of collection were not mentioned in label-data of the collected specimen (Griffith s.n., K000357720 image! ). Aquilaria khasiana was later collected from Umteswar forest in 1935 (S.R. Sarma 12036, ASSAM! ) and 1936 (S.R. Sarma 13539, 13540, both at ASSAM! ), Umsaw forest (S.R. Sarma 21453, ASSAM! ) in 1942, Cherrapunjee (W.N. Koelz 31115, L image!, MICH image!; R.C. Thakur 6146, L image!, MICH image! ) in 1952. Though Mir et al.10 reported its rediscovery in 2016 from Mawkasain after a lapse of almost ‘74 years’ by considering its last collection in 1942, they overlooked the specimens at L and MICH which were collected in 1952 from Cherrapunjee. All these reported locations of A. khasiana presently come under the Khasi Hills division of Meghalaya. During the present study, the species is found only in two locations, i.e. in Mawkasain, East Khasi Hills district, Meghalaya (the last known location) and Jeypore Reserve Forest, Dibrugarh district, Assam (new location). The identity of the samples collected from the new location was confirmed by morphological and molecular methods. Hence the present study reports a new distribution record for A. khasiana in Assam, India.
Characterization of the chloroplast genome
The sequencing and annotation of the A. khasiana chloroplast genome resulted in the assembly of a single contig with 178,834 bp length. The assembly length was almost equal to that of A. malaccensis (174,832), A. agallochum (174,866), A. sinensis (174,907) and G. walla (175,130)28. However, GC content was 37 while all other species had 36.733,49,50,51. The genome contained 90 protein-coding genes, 38 tRNAs and 8 rRNAs. The number of tRNAs and rRNAs in Aquilaria and Gyrinops were reported to be 38 and 8 respectively. However, the number of coding regions varied from species to species (89–96)33,49. The comparative analysis of the junction sites of IRs, SSC, and LSC showed minor differences in the position of rpl2 and rps19 genes as compared to other related species. The change in the chloroplast genome structure mainly takes place by the loss, shrinkage or expansion of the IR regions19. Such changes might have taken place in the genome of A. khasiana that may imparted the structural variations. The IR regions in land plants generally range from 15 to 30 kb and typically contain rRNA genes, tRNA genes, and many other genes that result from lineage-specific contractions and expansions. Shifts in the IR boundaries are found very minimal in closely related species. However, large-scale expansions in the IR regions have led to the transfer of genes from SSC to IRs in angiosperms18,30,31.
Phylogenetic analysis
Generation of genomics data and super barcode-based phylogenetic studies are very useful in the forensic studies of CITES-listed plants32. The present phylogenetic studies using three barcode markers and chloroplast genome showed that both Aquilaria and Gyrinops are paraphyletic in origin, where some of the Aquilaria spp. are found nested within the Gyrinops clade and vice versa. The results are incongruent with the previous phylogenetic analysis conducted by Eurlings and Gravendeel13Farah et al.14Feng et al.33 and Lee et al.23.
In both phylogenetic analyses, A. khasiana was placed within the Gyrinops Clade and showed a close affinity with G. walla, the only Gyrinops sp. found in the Western Ghats and Sri Lanka biodiversity hotspot. So far, A. khasiana was represented in only one phylogenetic analysis using trnL-trnF sequence data which also placed A. khasiana with G. walla. Both these taxa formed the basal clade of the phylogenetic tree13. Even though A. khasiana bears Aquilaria-specific morphological characters, in all the phylogenetic analyses, it was clustered with the Gyrinops Clade. A similar condition was also observed in the phylogenetic study of Indonesian Agarwood producing taxa where A. cumingiana was nested within the Gyrinops clade34. In their study, they separated Aquilaria and Gyrinops based on the colour of the mature fruit. The fruit colour of Aquilaria is green, while Gyrinops is orange/ yellow. However, A. cumingiana is characterized by orange to brownish mature fruits instead of green fruit. This led to the proposal of retaining A. cumingiana in the Gyrinops genus even though it has 10 stamens (characteristic of Aquilaria) suggesting that this single characteristic (10 stamens) difference in Aquilaria and Gyrinops can also be disputed13. Similarly, we observed deviation in the number of stamens i.e., 8 to 10 in A. khasiana and 10 to 12 in A. malaccensis. Hence we suggest that the number of stamens should not be considered as the only character for this generic delimitation.
Lee et al.23based on plastome-based phylogeny, proposed two suggestions viz. both Aquilaria and Gyrinops should be treated as two natural groups or both Aquilaria and Gyrinops should be treated as separate genera. In the former suggestion, Gyrinops should be merged into Aquilaria (considering its nomenclature priority) while in the latter suggestion, G. caudata (which was nested within Aquilaria-specific clade) should be treated as A. caudata to resolve the molecular classification. However, we agree with the second suggestion of Lee et al.23 in which both Aquilaria and Gyrinops should be treated as separate genera. Therefore, the name A. khasiana can be retained even though it is nested within Gyrinops clade as Gyrinops is paraphyletic to Aquilaria. However, using an integrative approach (combining morphological, molecular, and biogeographic) using more samples from a wide distributional range is required in Aquilaria and Gyrinops to resolve their phylogenetic relationships and refine the taxonomy of these closely related genera.
Conservation status assessment
Harvey-Brown et al.8 assessed A. khasiana as Critically Endangered [CR A2c; B1ab(i, ii) + 2ab(i, ii); D]. In the present study also, this species is assessed as Critically Endangered [CR A2cd; C2a(ii)], but with updated criteria, even after its new distributional record in Assam. The criteria B1, B2 and D considered by Harvey-Brown et al.8 are not applicable due to the existence of the species in only two locations and the increase in total number of mature individuals due to its extended distribution in Assam. As the species is endemic, no down-listing or up-listing of the IUCN category26 has been done with the final assessment. More intense survey and habitat management is therefore recommended for its future conservation.
Conclusions
Molecular and morphological analyses show that Aquilaria and Gyrinops are closely related, with Aquilaria khasiana closely linked to Gyrinops walla (formerly Aquilaria walla Hallier. f.). Given their paraphyletic origin, both genera should be treated as separate rather than natural groups. Broader sampling and integrative analyses are needed to further clarify the taxonomy of Aquilaria and Gyrinops. The report of new distributional records and the updated assessment of conservation status will be highly useful for the conservation management of this endemic and Critically Endangered [CR A2cd; C2a(ii)] species. The subpopulation in Mawkasain is under high threat since the location is not under any designated protected area. Further, only one mature plant exists in Mawkasain and the number of remaining young plants is also declining due to anthropogenic activities and infection by the larvae of Heortia vitessoides. The state forest department is suggested to protect the remaining plants by imposing strict legislation and monitoring, preferably by implementing an effective species recovery programme. Though the newly located subpopulation of A. khasiana in Jeypore is better protected due to its occurrence within a reserve forest, still there are some informal reports of the collection of this species as an alternate source of agarwood. There is a high need to conserve the species both by ex-situ and in-situ methods.
Materials and methods
Taxon sampling
A total of four samples each of A. khasiana (wild) and A. malaccensis (wild and cultivated) were collected during our recent field surveys with permissions from the forest departments of Assam and Meghalaya states, India. In addition, two samples of A. sinensis (cultivated) were also collected from Karnataka state. The collected materials included fresh young leaf material for genomic DNA isolation for the DNA barcode-based phylogenetic study and corresponding voucher specimens for herbarium preparation and morphological study. The identity of the samples was confirmed by A. Bhattacharjee and R. Layola M.R.
Fresh young leaves were kept in a layer of tissue paper inside a ziplock cover and self-indicating silica gel (mesh size – 6–20). The silica gel removes the water content present in the leaf tissue and protects it from decaying and fungal infection resulting in the preservation of DNA during extended field study. Simultaneously, the leaf tissue of one accession of A. khasiana collected from Meghalaya was washed and immediately transferred to dry ice and sent for plastid genome sequencing. Hence a total of 10 samples belonging to 3 species were newly used for the present study.
The herbarium voucher specimens were prepared and deposited at CAL. The details of plant collections such as specific location, date of collection, collector, elevation etc. were tabulated (Table 1). To construct a comprehensive DNA barcode-based phylogenetic tree, sequences of agarwood-producing species of Aquilaria and Gyrinops available in the NCBI databases were retrieved (Table 2). The plastid genomes of Aquilaria and Gyrinops spp. used for the chloroplast genome-based phylogenetic study is also tabulated (Table 4).
Taxonomic study
A thorough literature survey of A. khasiana was done including consultation of the protologue, revisionary and floristic studies, checklists, regional accounts and other authentic literature. The accepted name and nomenclature were determined in reference to the ICN35and by consulting international databases like International Plant Names Index36, Plants of the World Online9, TROPICOS37, World Flora Online38 etc. Protologues and relevant literature have been searched from BHL and other offline and online libraries from India and abroad. Herbarium specimens of A. khasiana available at ASSAM, BM, GH, K, L, and MICH, were studied to get the distributional record, phenology in detail and the range of variation in morphological characters. Detailed descriptions were prepared by studying the morphology of live specimens. A digital illustration was prepared by photographing the habit and capturing macro-microscopic images of the dissected floral parts.
DNA barcode-based phylogenetic analysis
The genomic DNA was extracted from the silica-dried young leaves using DNeasy Plant Pro Kit (QIAGEN). Extracted DNA was quantified using a NanoDrop Lite Spectrophotometer (Thermo Scientific). The PCR amplification was carried out using nuclear (ITS2) and chloroplast (rbcl and matK) regions. The PCR amplification was carried out as per Lee et al.29. The DNA extraction, PCR amplification and bioinformatic study were conducted at the Molecular Biology Laboratory, Central National Herbarium, Howrah. The purification and sequencing of the amplified products were outsourced at GeneSpec Pvt. Ltd., Kerala, India.
All the sequences were aligned and contigs were generated using BioEdit v.7.239. All the newly generated sequences along with the sequence set from the NCBI GenBank were aligned using ClustalW40 multiple alignment programme in BioEdit v.7.239. From the ITS sequences retrieved from the NCBI (MH134137 – MH134154), only the ITS2 region was extracted after alignment. The sequences of all three markers were then concatenated, and a combined sequence set was made for the phylogenetic analysis (ITS2, rbcl, and matK). Maximum likelihood (ML) analysis was carried out using IQ-TREE v.1.6.1241. The best-fit substitution model (TVM + F + G4) for the ML analysis was selected using Model Finder42 based on BIC (Bayesian Information Criterion). The ML analysis was run with selected substitution models and 1,000 ultrafast bootstrapping method43. All other parameters were set to default.
The Bayesian Inference (BI) analysis was carried out using the MrBayes v.3.2.744. A mixed type (4 by 4) nucleotide substitution model was selected for the run with 20 million Markov chain Monte Carlo (MCMC) simulations, 1000 sample frequency, and 1000 print frequency. The data were sampled in every 1000 generations and the first 25 trees were omitted as burn-in. The resulting phylogenetic tree of ML and BI analysis was visualized in FigTree v.1.4.345. The tree topology and branch support values of both trees were checked individually and a final consensus tree showing ML bootstrap (BS) support and BI posterior probability (PP) values were manually annotated on an ML majority consensus tree.
Chloroplast sequencing, assembly, and annotation
For chloroplast genome sequencing, A. khasiana collected from Meghalaya was used. DNA was isolated from the fresh leaf using a Blood and cell culture DNA mini kit (Qiagen, Cat no. 13323). The concentration and purity of genomic DNA were quantified using a Nanodrop Spectrophotometer. The integrity of DNA in the samples was assessed on Genomic DNA screen tape (Cat no. 5067–5365). DNA concentration was quantified using a Qubit dsDNA HS assay kit (Cat no. Q32854).
For library preparation, the genomic DNA was end-polished and A-tailed (NEBNext Ultra II end repair kit, New England Biolabs, MA, USA). The end-prepared samples were barcoded using Blunt TA ligase master mix (M0367L). The equimolar concentration of the barcoded sample was pooled and sequencing adapters (SQK-NBD114.96) were ligated onto double-stranded DNA fragments using NEB Quick T4 DNA Ligase (New England Biolabs, MA, USA). After purification with AMPure XP beads, the prepared library was checked with Qubit for quantification. The libraries were pooled and sequenced on the Nanopore PromethION system, (PromethION P24 and Data Acquisition Unit, ONT, Oxford, UK) using a PromethION flow cell (FLO-PRO114M).
A total of ~ 4.68 GB of nanopore long reads data was generated for the plant sample. The average quality score distribution and total yield-related statistics were generated using the NanoPlot v1.41.61 tool. The nanopore long reads were trimmed for adapter removal using Porechop v0.2.42. The adapter-free processed reads were mapped against the reference genome of Aquilaria sinensis (GenBank ID: NC029243) using Minimap2 v2.26-r11753 and pre-processing of alignment data was done using Samtools v1.184 to filter out chloroplast genome-specific reads. The mapped reads were used for de novo assembly of the sample through RA assembler v0.2.15. The annotation of the assembled chloroplast genome was carried out using the CPGAVAS2 v0.036 tool46. The circular genome map was generated using the OGDRAW747.
Comparative genome structure analysis of A. khasiana and related species
The GC content of the chloroplast genome was estimated using Emboss (https://www.bioinformatics.nl/cgi-bin/emboss/geecee) and compared with that of the related species. The structural organization of the inverted repeats (IRA and IRB), short single copy (SSC) and long single copy (LSC) junction sites were visualised and compared with the other Aquilaria and Gyrinops species (except A. crassna, A. microcarpa, A. subintegra, and A. yunnanesis which were unable to visualize) used in the study using IRscope (https://irscope.shinyapps.io/irapp/). The genome plots of 16 species including A. khasiana were manually combined and represented.
Chloroplast genome-based phylogenetic analysis
Chloroplast-based phylogenetic analysis was carried out using the assembled sequence of A. khasiana and the sequences of 23 taxa (19 ingroup and 4 outgroup taxa) obtained from the NCBI (Table 4). All the sequences were aligned using MAFFT v.748. The phylogenetic analysis was done using both ML and BI methods separately. ML analysis was carried out using IQ-TREE v.1.6.1241 with 1,000 ultrafast bootstraps. All other parameters were kept as default. The best-fit model for the ML analysis was TVM + F + R2.
The Bayesian Inference (BI) analysis was carried out using the MrBayes v.3.2.744. A mixed type (4 by 4) nucleotide substitution model was selected for the run with 10 million Markov chain Monte Carlo (MCMC) simulations, 1000 sample frequency, and 1000 print frequency. The data were sampled in every 1000 generations and the first 25 trees were omitted as burn-in. The resulting phylogenetic tree of ML analysis was visualized in FigTree v.1.4.345 and the final tree was generated the same as in the case of barcode-based phylogenetic analysis.
Assessment of conservation status
Distribution data recorded from literature, herbarium records and field surveys were tabulated in Excel. Geo-coordinates were assigned for each location from GPS and Google Earth. Mapping and calculation of Area of Occupancy (AOO) and Extent of Occurrence (EOO) was done using ArcGIS version 10.7.1 (https://www.esri.com/en-us/arcgis/products/arcgis-desktop/overview), and GeoCAT49. To assess the conservation status, guidelines for using the IUCN Red List Categories and Criteria26 were followed.
Data availability
The complete chloroplast genome and annotations are available at the NCBI database (Aquilaria khasiana: PQ576418).
References
Chakrabarty, K., Kumar, A. & Menon, V. Trade in Agarwood (WWF-India/ TRAFFIC-India, 1994).
Yaacob, S. Agarwood: Trade and CITES Implementation in Malaysia (Unpublished report prepared for TRAFFIC Southeast Asia, 1999).
TRAFFIC. The trade and use of agarwood in Taiwan, Province of China. East Asia-Taipei: TRAFFIC & Southeast Asia: TRAFFIC (PC15 Inf. 7). (2005).
Bhattacharjee, A. et al. Non-detriment Findings (NDFs) of Aquilaria malaccensis Lam. (Agarwood) in India (Botanical Survey of India, 2024).
Gratzfeld, J. & Tan, B. Agarwood-saving a precious and threatened resource. BG J. 5, 27–29 (2008). https://www.jstor.org/stable/24810587
United Nations Office on Drugs and Crime (UNODC). Trafficking in protected species Report. (2016). https://www.unodc.org/
Lee, S. Y. & Mohamed, R. The origin and domestication of Aquilaria, an important Agarwood producing genus. Pp. 1–20 : (ed Mohamed, R.) Agarwood: Science behind the Fragrance. Singapore: Springer. https://doi.org/10.1007/978-981-10-0833-7_1. (2016).
Harvey-Brown, Y., Mir, A. H. & Upadhaya, K. Aquilaria khasiana. IUCN Red List. Threatened Species. 2018, eT88305771A88305775. Accessed on 14 August 2024. https://doi.org/10.2305/IUCN.UK.2018-1.RLTS.T88305771A88305775.en
POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. (2024). http://www.plantsoftheworldonline.org/ (accessed 1 Sep 2024).
Mir, A. H., Roy, D. K. & Upadhaya, K. Taxonomy, recollection and conservation implications of Aquilaria khasiana (Thymelaeaceae): an endemic and threatened species of India. Rheedea 27 (2), 85–89. https://doi.org/10.22244/rheedea.2017.27.2.14 (2017).
von Hallier, H. Beiträge Zur Kenntnis der thymelaeacean under ihrer natürlichen umgrenzung. Meded Rijks-Herb. 44, 1–31 (1922).
Hou, D. Thymelaeaceae. Pp. 1–48 in: Steenis, C. G. G. J. van & Steenis-Kruseman, M. J. van (eds.), Flora Malesiana – Series I, Spermatophyta, Volume 6. Djakarta: Noordhoff-Kolff. (1960).
Eurlings, M. C. M. & Gravendeel, B. TrnL-trnF sequence data imply paraphyly of Aquilaria and Gyrinops (Thymelaeaceae) and provide new perspectives for Agarwood identification. Pl Syst. Evol. 254, 1–12. https://doi.org/10.1007/s00606-005-0312-x (2005).
Farah, A. H. et al. Genome size, molecular phylogeny, and evolutionary history of the tribe Aquilarieae (Thymelaeaceae), the natural source of Agarwood. Front. Pl Sci. 9, 712. https://doi.org/10.3389/fpls.2018.00712 (2018).
Neuhaus, H. E. & Emes, M. J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant. Biol. 51 (1), 111–140. https://doi.org/10.1146/annurev.arplant.51.1.111 (2000).
Jansen, R. K. & Ruhlman, T. A. Plastid genomes of seed plants, in Genomics of Chloroplasts and Mitochondria, vol. 35. Eds (eds Bock, R. & Respiration, V.). and Dordrecht, Netherlands, Springer, 103–126. (2012).
Sugiura, M. The Chloroplast genome. Plant. Mol. Biol. 19, 149–168. https://doi.org/10.1007/BF00015612 (1992).
Sun, Y. X. et al. Complete plastid genome sequencing of Trochodendraceae reveals a significant expansion of the inverted repeat and suggests a Paleogene divergence between the two extant species. PLoS One. 8(4), e60429. https://doi.org/10.1371/journal.pone.0060429 (2013).
Bock, R. & Knoop, V. (eds). Genomics of Chloroplasts and Mitochondria , Vol. 35 (Springer, 2012).
Lei, W. et al. Intraspecific and heteroplasmic variations, gene losses and inversions in the Chloroplast genome of Astragalus Membranaceus. Sci. Rep. 6, 1–13. https://doi.org/10.1038/srep21669 (2016).
Zhou, S. M. et al. Phylogenomics and plastome evolution of Indigofera (Fabaceae). Front. Plant. Sci. 14, 1186598. https://doi.org/10.3389/fpls.2023.1186598 (2023).
Hishamuddin, M. S. et al. Comparison of eight complete Chloroplast genomes of the endangered Aquilaria tree species (Thymelaeaceae) and their phylogenetic relationships. Sci. Rep. 10, 13034. https://doi.org/10.1038/s41598-020-70030-0 (2020).
Lee, S. Y. et al. Phylogenetic relationships of Aquilaria and Gyrinops (Thymelaeaceae) revisited: evidence from complete plastid genomes. Bot. J. Linn. Soc. 200 (3), 344–359 (2022b).
Rieseberg, L. H. & Soltis, D. E. Phylogenetic consequences of cytoplasmic gene flow in plants. Evo Trend Plant. 5, 65–84 (1991).
Dunning, L. T. et al. Hybridisation and Chloroplast capture between distinct Themeda triandra lineages in Australia. Mol. Ecol. 31 (22), 5846–5860. https://doi.org/10.1111/mec.16691 (2022).
IUCN Standards and Petitions Committee. Guidelines for Using the IUCN Red List Categories and Criteria, Version 16. Prepared by the Standards and Petitions Committee. Downloadable from (2024). https://www.iucnredlist.org/documents/RedListGuidelines.pdf
IUCN. IUCN Red List Categories and Criteria, Version 3.1 (Second Edition) (IUCN: Gland and Cambridge, 2012).
Chen, J. et al. The complete Chloroplast genome of Walla patta, Gyrinops walla (Thymelaeaceae), an agarwood-producing tree species from Sri Lanka. Mitochondrial DNA Part. B. 6 (6), 1699–1701. https://doi.org/10.1080/23802359.2021.1926362 (2021).
Lee, S. L. et al. DNA databases of a CITES listed species Aquilaria malaccensis (Thymelaeaceae) as the tracking tools for forensic identification and chain of custody certification. Foren Sci. Int. Gen. 57 https://doi.org/10.1016/j.fsigen.2021.102658 (2022).
Downie, S. R. & Jansen, R. K. A comparative analysis of whole plastid genomes from the apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40 (1), 336–351. https://doi.org/10.1600/036364415X686620 (2015).
Zhu, A., Guo, W., Gupta, S., Fan, W. & Mower, J. P. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New. Phytol. 209 (4), 1747–1756. https://doi.org/10.1111/nph.13743 (2016).
Dasgupta, P., Senthilkumar, S., Barthwal, S., Sasidharan, K. R. & Ghosh Dasgupta, M. Characterization of the chloroplast genome of Dalbergia latifolia (Fabaceae) and its phylogenetic relationship with other Dalbergia species. Nordic J. Bot. 2023(7): p.e03811. (2023). https://doi.org/10.1111/njb.03811
Feng, T. et al. Phylogenetic analysis of Aquilaria Lam. (Thymelaeaceae) based on DNA barcoding. Holzforschung 73 (6), 517–523 (2019). https://doi.org/10.1515/hf-2018-0127
Lee, S.Y., Turjaman, M. and Mohamed, R., 2018. Phylogenetic relatedness of several agarwood-producing taxa (Thymelaeaceae) from Indonesia. Trop. Life Sci. Res. 29 (2), 13–28. https://doi.org/10.21315/tlsr2018.29.2.2
Turland, N. J. et al. (eds) International Code of Nomenclature for Algae, fungi and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017 (Koeltz Botanical Books, 2018). Regnum Vegetabile 159.
IPNI. International Plant Names Index. The Royal Botanic Gardens, Kew, Harvard University Herbaria & Libraries and Australian National Herbarium. (2024). Available from: http://www.ipni.org (accessed 7 Aug 2024).
TROPICOS. Tropicos v3.4.2. Published online by Missouri Botanical Garden. (2024). http://www.tropicos.org/ (accessed 1 Sep 2024).
WFO. World Flora Online. Published on the Internet (2024). http://www.worldfloraonline.org. (accessed 1 Sep 2024).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98 (1999).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acid Res. 22, 4673–4680. https://doi.org/10.1093/nar/22.22.4673 (1994).
Nguyen, L. T., Schmidt, H. A., Haeseler, A., Von & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. https://doi.org/10.1093/molbev/msu300 (2015).
Kalyaanamoorthy, S. et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Meth. 14, 587–589. https://doi.org/10.1038/nmeth.4285 (2017).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. https://doi.org/10.1093/molbev/mst024 (2013).
Ronquist, F. et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Boil. 61 (3), 539–542. https://doi.org/10.1093/sysbio/sys029 (2012).
Rambaut, A. FigTree v1.3.1. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh. (2010). http://tree.bio.ed.ac.uk/software/figtree/
Liu, C. et al. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced Chloroplast genome sequences. BMC Gen. 13, 1–7. https://doi.org/10.1186/1471-2164-13-715 (2012).
Greiner, S., Lehwark, P. & Bock, R. Organellar genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucl. Acids Res. 47, 59–64. https://doi.org/10.1093/nar/gkz238 (2019).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. https://doi.org/10.1093/bib/bbk014 (2019).
Bachman, S., Moat, J., Hill, A. W., De La Torre, J. & Scott, B. Supporting Red List threat assessments with GeoCAT: geospatial conservation assessment tool. ZooKeys 150, 117–126. (2011). https://doi.org/10.3897/zookeys.150.2109
Zhang, Y. H., Huang, Y., Li, Z. M. & Zhang, S. D. Characterization of the complete Chloroplast genome of the vulnerable Agarwood tree, Aquilaria yunnanensis (Thymelaeaceae). Conserv. Genet. Resour. 11, 161–164. https://doi.org/10.1007/s12686-018-0989-0 (2019).
Lin, C. P. et al. The complete Chloroplast genome of Agarwood producing species, Aquilaria sinensis (Lour.) gilg: a species on IUCN red list. Mitochon DNA Part. B. 4 (2), 2992–2993. https://doi.org/10.1080/23802359.2019.1664954 (2019).
Deng, X. et al. Characterization of the complete Chloroplast genome of Aquilaria sinensis, an endangered agarwood-producing tree. Mitochon DNA Part. B. 5 (1), 422–423. https://doi.org/10.1080/23802359.2019.1703593 (2020).
Cho, W. B., Han, E. K., Choi, G. & Lee, J. H. The complete Chloroplast genome of Daphne kiusiana, an evergreen broad-leaved shrub on Jeju Island. Cons Gen. Res. 10, 103–106. https://doi.org/10.1007/s12686-017-0774-5 (2018).
Qian, S. J., Zhang, Y. H. & Li, G. D. The complete Chloroplast genome of a medicinal plant, Wikstroemia chamaedaphne (Thymelaeaceae). Mitochon DNA Part. B. 5 (1), 648–649. https://doi.org/10.1080/23802359.2019.1711228 (2020).
Acknowledgements
The present communication is part of the outcomes of two projects, i.e. ‘Non-detriment Findings (NDFs) of Aquilaria malaccensis Lam. (Agarwood) in India’, funded by the Ministry of Environment, Forest and Climate Change, Wildlife Division (MoEF&CC–WL), Government of India, New Delhi, and an in-house Action Plan project of Botanical Survey of India (BSI) entitled, ‘Morpho-molecular and phytochemical identification of 30 CITES listed plants in high international trade’, funded by MoEF&CC. Dr R.K. Gupta, the former Scientist ‘F’ and Head of Office (HoO), Central National Herbarium (CNH), BSI, and Dr Kanad Das, Scientist 'F' and HoO, CNH, BSI are thankfully acknowledged for providing research facilities. We acknowledge Genotypic Technology Private Limited, Bangalore for NGS sequencing and data annotation. We are grateful to the Principal Chief Conservator of Forests and Head of Forest Forces of Assam and Meghalaya for the forest entry permit (Assam: Permit No. 39/23/Agarwood/2004/Pt-II dated10.11.2023; Meghalaya: Permit No. FWC/Research/145/665-671 dated 23.05.2024) and the officials of the forest departments for their help during the surveys. We are thankful to Ms Emily Beech, Head of Conservation Prioritisation Botanic Gardens Conservation International (BGCI) and Head of BGCI’s IUCN Red List activities for the Global Tree Assessment (GTA), Richmond, Ms Kathryn Fowler, Conservation Officer (GTA), BGCI, Richmond and Mr Neil Cox, Manager of the Biodiversity Assessment Unit of IUCN, Cambridge, for their guidance in finalising the conservation status of A. khasiana. We are thankful to the reviewers and the editors for their valuable comments which improved the quality of the manuscript.
Author information
Authors and Affiliations
Contributions
A.B. and R.L.M.R. designed the research and confirmed the identity of the specimens. A.B., R.L.M.R., S.S., B.M., T.S., R.S., S.C., Sudipta S., O.C., Sayak C., and S.B., conducted a field survey. A.B., S.S., B.M., S.C., Sudipta S., O.C., and Sayak C. carried out a taxonomic study. A.B., S.S., O.C., and Sayak C. carried out the IUCN assessment. R.L.M.R did the computational analysis. R.L.M.R. and F.B. deposited sequences. A.B. and R.L.M.R. wrote the manuscript; A.A.M. provided inputs and edited the manuscript. All authors have read, reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Layola M.R., R., Sengupta, S., Mallick, B. et al. Phylogenetic analysis of the Critically Endangered Aquilaria khasiana (Thymelaeaceae) using barcode markers and chloroplast genome, with updated conservation status. Sci Rep 15, 21511 (2025). https://doi.org/10.1038/s41598-025-08606-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08606-x