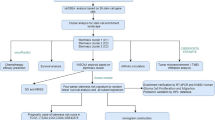
Abstract
Transmembrane (TMEM) proteins play a pivotal role in cancer progression, with TMEM150A specifically implicated in tumorigenesis. Despite its association with cancer, the precise role of TMEM150A in glioma remains underexplored. The expression of TMEM150A in glioma and its correlation with prognosis and the immune microenvironment were systematically analyzed. Prognostic models were developed using nomograms to establish the relationship between TMEM150A expression and patient survival. Bioinformatic analyses identified pathways and networks associated with TMEM150A co-expressed genes, while in vitro assays (CCK-8, migration, and invasion assays) examined the impact of TMEM150A inhibition on glioma cell proliferation and metastasis. TMEM150A was markedly overexpressed in glioma tissues and strongly associated with clinical features such as 1p/19q codeletion, age, IDH mutation status, histological subtype, WHO grade, and poor prognosis (P < 0.05). ROC analysis revealed high diagnostic accuracy, with area under the curve values of 0.962 and 0.896 for TCGA and XENA datasets, respectively, indicating the strong diagnostic potential of TMEM150A. Overexpression of TMEM150A was identified as a risk factor for poor prognosis. The constructed nomogram demonstrated that TMEM150A expression was predictive of survival, with time-dependent AUCs for 1, 3, and 5 years exceeding 0.75, confirming its prognostic relevance. TMEM150A co-expressed genes were linked to immune responses, necrotic cell death, antigen processing and presentation, cell differentiation, mast cell activation, endothelial cell migration, wound healing, and cell proliferation. Inhibition of TMEM150A expression suppressed U251 cell growth, migration, and invasion via epithelial-mesenchymal transition (EMT). Additionally, TMEM150A expression correlated with immune scores (r = 0.585), stromal scores (r = 0.498), ESTIMATE scores (r = 0.565), macrophage infiltration (r = 0.523), aDC (r = 0.473), and neutrophil presence (r = 0.453). Overexpression of TMEM150A serves as an independent prognostic marker in glioma and is intricately linked to the tumor’s immune microenvironment. Targeting TMEM150A could inhibit glioma progression through EMT modulation, presenting a potential novel therapeutic avenue.
Similar content being viewed by others
Introduction
Transmembrane (TMEM) proteins play pivotal roles in signal transduction, substance transport, and cell communication. These proteins recognize extracellular molecules and trigger various intracellular signaling pathways, making them valuable targets for drug screening. Transport-related TMEM proteins facilitate transmembrane substance movement and regulate organismal metabolism. Though their specific functions may vary, each TMEM protein is essential for maintaining cellular homeostasis. Numerous studies have highlighted the significant biological roles of TMEM proteins in cancer progression1,2,3,4. For instance, Li et al. identified TMEM158 as overexpressed in glioma, with its elevated expression correlating with poor overall survival (OS) in patients. Silencing TMEM158 inhibited glioma cell proliferation, migration, and invasion3. Similarly, Yang et al. observed significantly higher levels of TMEM60 in gliomas compared to normal brain tissues. Increased TMEM60 expression was associated with worsened prognosis in patients with glioma, and silencing TMEM60 reduced glioma cell viability, proliferation, and colony formation4.
In recent years, public databases have provided valuable insights and new cancer targets for researchers5,6,7,8. For example, Chen et al. reported that killer cell lectin-like receptor B1 (KLRB1) was downregulated in lung adenocarcinoma, with lower KLRB1 expression correlating with poor prognosis, T stage, N stage, and distant metastasis. Overexpression of KLRB1 inhibited cell proliferation and metastasis while promoting apoptosis, positioning KLRB1 as a potential therapeutic target for lung adenocarcinoma5. Shao et al. found that coactosin-like F-actin binding protein 1 (COTL1) was highly expressed in glioblastoma multiforme (GBM), with its increased expression linked to recurrence and poor prognosis. In vitro and in vivo studies confirmed that COTL1 promoted cell proliferation and GBM tumor growth in nude mice8. Among these proteins, TMEM150A has garnered attention due to its emerging roles in cancer9,10. For instance, inhibition of TMEM150A expression increased lipopolysaccharide-induced cytokine secretion and transcription. In lung epithelial cells, TMEM150A inhibition led to elevated cytokine levels under unstimulated conditions, suggesting its involvement in maintaining cellular homeostasis9. Furthermore, TMEM150A was found to be overexpressed in GBM, with its silencing inhibiting GBM growth and metastasis10. However, the precise role of TMEM150A in glioma remains incompletely understood. This study aims to explore the function of TMEM150A in glioma and its underlying mechanisms through bioinformatic analysis, offering potential new therapeutic strategies and candidate molecules for glioma treatment.
Materials and methods
Data from patients with glioma
Transcriptome data from 706 cancer tissues and 5 normal tissues, along with clinical data from patients with cancers, were retrieved from the TCGA official website. Additionally, transcriptome data from 1152 normal tissues from the GTEx database, and data from 689 cancer tissues and 5 normal tissues from the TCGA database, were obtained from the XENA database. These datasets, provided in TPM format, were publicly accessible in the Xiantao Academic Website and did not require Ethics Committee approval for use.
Expression of TMEM150A and its relationship with clinical features in glioma
Differential expression analysis was performed to assess the expression levels of TMEM150A in normal and cancer tissues using TCGA and XENA databases. Clinical data, including WHO grade, IDH status, age, histological subtype, and 1p/19q codeletion from the TCGA database, were cleaned and processed. Statistical significance between groups was determined using one-way ANOVA or the Wilcoxon rank-sum test.
Diagnostic and prognostic values of TMEM150A determined using ROC analysis
Receiver operating characteristic (ROC) analysis was conducted to evaluate the diagnostic value of TMEM150A by analyzing the area under the ROC curve (AUC) for normal and cancerous tissues in both TCGA and XENA datasets. To further assess its prognostic significance, time-dependent ROC analysis was performed to examine the 1-year, 3-year, and 5-year survival AUCs for patients with glioma. An AUC value closer to 1 indicated greater diagnostic value.
Relationship between TMEM150A overexpression and prognosis of glioma
Following the cleaning and processing of patient survival data, disease-specific survival, and disease progression time from the TCGA database, survival analysis was conducted to investigate the relationship between TMEM150A expression levels and prognosis, as well as cancer progression in patients with glioma. Subgroup survival analysis was performed to explore the prognostic implications of TMEM150A overexpression or underexpression in patients with glioma, stratified by WHO classification, IDH status, age, histological subtype, and 1p/19q codeletion. The cohort was divided into TMEM150A overexpression and under-expression groups based on the median expression level of TMEM150A.
COX regression analysis and nomogram visualization
Univariate Cox regression analysis was performed to investigate the association between various clinical factors, including WHO grade, IDH status, age, histological subtype, 1p/19q codeletion, and TMEM150A expression with prognostic indicators in patients with glioma. Based on the median expression of TMEM150A, patients were divided into TMEM150A overexpression and TMEM150A under-expression groups. P-values derived from these analyses were used to inform the inclusion of variables in the construction of a prognostic nomogram. The final model incorporated WHO classification, IDH status, 1p/19q codeletion, age, and TMEM150A overexpression as key predictors of patient prognosis.
Function and protein network involved in TMEM150A co-expressed genes
The co-expressed genes of TMEM150A were identified through Spearman correlation analysis, with correlations having an absolute coefficient greater than 0.6 considered significant. Bioinformatic analysis was performed to explore the functions and protein interactions of TMEM150A co-expressed genes using the STRING database, and a protein–protein interaction (PPI) network was constructed with a binding confidence score greater than 0.7. The network was then visualized and refined using Cytoscape software for clearer presentation.
Function and protein network involved in TMEM150A co-expressed genes
Additionally, immune fractions and levels of various immune cell types—such as aDC, B cells, CD8 T cells, cytotoxic cells, DC, eosinophils, iDC, macrophages, mast cells, neutrophils, NK cells, T cells, and others—were calculated using Estimate and ssGSEA analyses. Spearman correlation was applied to evaluate the relationship between TMEM150A expression, glioma immune scores, and immune cell abundance.
Culture of U251 cells and construction of down-regulated cell model of TMEM150A
Glioma U251 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (from the Typical Culture Preservation Committee of the Chinese Academy of Sciences cell bank). Once cells showed optimal growth, siRNA and transfection reagents were added, and a fresh medium was replaced after 24 h of incubation. Total RNA and protein from both control and transfected groups were then collected to assess TMEM150A mRNA and protein expression using RT-PCR and Western blotting.
RT-PCR and Western blotting
The RT-PCR and Western blotting procedures followed methods described in our previous study10. The TMEM150A primers used were: 5’-GCCTGTCAGCGTTCTCCATCAC-3’ (forward) and 5’-AGGACTCGTTGTAGGACCAGTTCTC-3’ (reverse), sourced from Shanghai Biological Engineering Co., Ltd. The siRNA interference sequence employed was GUGGAGAACUGGUCCUACATT. Western blotting was performed according to standard procedures. Before transferring the membrane, we have cut the membrane to the corresponding proportion, and then incubate the primary and secondary antibodies. The antibody dilutions were as follows: TMEM150A (1:1000), GAPDH (1:3000), E-cadherin (1:1500), and Snail (1:2000). All antibodies were purchased from Proteintech, Wuhan.
Cell proliferation experiment
To inhibit TMEM150A expression, cells were cultured under optimal conditions, and cell counts were performed. CCK-8 solution (10 µL) was added to the adherent cells, and absorbance was measured at 0 h, 24 h, 48 h, and 72 h post-adherence. Three replicate wells were used for each group.
Cell migration and invasion experiments
The expression of TMEM150A was inhibited while maintaining optimal growth conditions for glioma cells. Cell counting was then performed to prepare a cell suspension. A 200 µL serum-free cell suspension was added to the upper chamber of a transwell, while an 800 µL serum-containing medium was added to the lower chamber. The chamber was incubated for 24 h, and an invasion assay was conducted with Matrigel coating. After fixation and washing, cell images were captured.
Statistical analysis
Statistical significance between groups was assessed using the t-test and Wilcoxon rank-sum test. The t-test was used to compare means between two groups, while the Wilcoxon rank-sum test was applied when necessary. ROC analysis was performed to evaluate the diagnostic value, with an AUC closer to 1 indicating higher diagnostic value. A P-value of < 0.05 was considered statistically significant in this study.
Results
The overexpression of TMEM150A was significantly correlated with WHO grade, IDH status, age, histological subtype, and 1p/19q codeletion of patients with glioma
Based on the analysis of TCGA and XENA databases, TMEM150A was significantly overexpressed in glioma tissues (Fig. 1A,B). Specifically, TMEM150A expression was higher in G3 and G4 glioma tissues compared to G2 glioma, showing a gradual increase in expression with tumor grade (Fig. 1C). Moreover, TMEM150A was more highly expressed in IDH wild-type glioma tissues compared to IDH mutant glioma tissues (Fig. 1D). A similar trend was observed when comparing 1p/19q codeletion gliomas, where TMEM150A was overexpressed in tissues of patients with non-codeletion compared to those with codeletion (Fig. 1E). Additionally, TMEM150A expression was higher in glioma tissues from patients aged > 60 years compared to those ≤ 60 years (Fig. 1F). Statistically significant differences were also observed in TMEM150A expression across various glioma histological subtypes (Fig. 1G).
Expression of TMEM150A in glioma tissues based on data analysis. (A, B) Comparison of normal tissues vs. glioma tissues from TCGA and XENA databases; (C) Expression across WHO grades; (D) Expression in relation to IDH status; (E) Expression in relation to 1p/19q codeletion status; (F) Expression in relation to age; (G) Expression across histological subtypes.
Overexpression of TMEM150A was negatively correlated with diagnosis and poor prognosis in patients with glioma
Diagnostic analysis using TCGA and XENA databases revealed that the area under the ROC curve for TMEM150A expression in glioma tissues was 0.962 and 0.896, respectively (Fig. 2A,B), suggesting a strong diagnostic potential for TMEM150A as a biomarker for glioma. Survival analysis further confirmed that TMEM150A overexpression was associated with poor prognosis in patients with glioma (Fig. 2C–E). Specifically, TMEM150A overexpression correlated with shorter overall survival (HR = 3.38, 95% CI = 2.61–4.38, P < 0.001), disease-specific survival (HR = 3.48, 95% CI = 2.65–4.58, P < 0.001), and disease progression (HR = 2.51, 95% CI = 2.01–3.12, P < 0.001).
Overexpression of TMEM150A was negatively correlated with poor prognosis in patient subgroups
Further subgroup survival analysis, stratified by clinical characteristics, showed that TMEM150A overexpression was negatively correlated with survival outcomes in various groups, including G2-3, G3, G3-4, G4, IDH mutation, non-1p/19q codeletion, male, female, and age ≤ 60 and age > 60 patients with glioma. In all subgroups, the HR values remained significantly greater than 1 (Figs. 3, 4, 5).
Overexpression of TMEM150A was a risk factor for poor prognosis in patients with glioma
The time-dependent ROC analysis for TMEM150A showed the following AUC values for predicting survival outcomes at 1-year, 3-year, and 5-year intervals: 0.796, 0.813, and 0.776 for overall survival; 0.792, 0.813, and 0.797 for disease-specific survival; and 0.728, 0.767, and 0.764 for disease progression (Fig. 6A–C). Cox regression analysis further indicated that WHO classification, IDH status, 1p/19q codeletion, age, and TMEM150A overexpression were all identified as risk factors for shorter overall survival, disease progression, and disease-specific survival in patients with glioma (Tables 1, 2, 3). Based on the results of the Cox regression, a nomogram was constructed to predict the prognosis of patients with glioma, incorporating WHO classification, IDH status, 1p/19q codeletion, age, and TMEM150A overexpression as key prognostic indicators (Fig. 7).
Function and protein network of TMEM150A co-expressed genes
Based on a phase relationship coefficient exceeding 0.6, 502 genes were identified as co-expressed with TMEM150A. Nine genes—GUSB, DTX2, TCIRG1, SRA1, CTSA, TP53I13, CASP6, CNPY4, TPRA1, and EDEM2—demonstrated the strongest correlation coefficients, as shown in Fig. 8. These TMEM150A-associated genes were implicated in various biological processes, including immune response, necrotic cell death, myeloid leukocyte activation, antigen processing, leukocyte differentiation, mast cell activation, cell adhesion, protein processing, endothelial migration, wound healing, and cell proliferation (Fig. 9A,B). Figure 10 illustrates the protein network interactions among these genes.
Inhibition of TMEM150A expression could inhibit the growth and metastasis of glioma cells
Inhibition of TMEM150A expression in glioma U251 cells was achieved through siRNA technology, as demonstrated by the reduced mRNA and protein levels (Fig. 11A,B), with statistically significant differences observed. CCK-8 assays confirmed that TMEM150A knockdown significantly suppressed U251 cell proliferation (Fig. 11C). Transwell assays further revealed that TMEM150A silencing impeded cell migration and invasion (Figs. 11D, 12A,B). Additionally, reduced TMEM150A expression led to a downregulation of Snail protein and an upregulation of E-cadherin during the epithelial-mesenchymal transition (EMT) pathway (Fig. 12C), suggesting that TMEM150A influences glioma progression via EMT.
Overexpression of TMEM150A was significantly correlated with the microenvironment of glioma
Correlation analyses revealed a significant positive association between TMEM150A overexpression and the glioma estimate score (R = 0.565), immune score (R = 0.585), and stromal score (R = 0.498) (Fig. 13A–C). A positive correlation was also observed between TMEM150A expression and various immune cell types, including Macrophages (R = 0.523), aDC (R = 0.475), Neutrophils (R = 0.453), Eosinophils (R = 0.438), iDC (R = 0.319), Cytotoxic cells (R = 0.301), NK CD56dim cells (R = 0.293), T cells (R = 0.264), Th17 cells (R = 0.239), Th2 cells (R = 0.205), NK cells (R = 0.146), and T helper cells (R = 0.139). Conversely, a significant negative correlation was observed with TFH (R = − 0.323), Tcm (R = − 0.312), Mast cells (R = − 0.264), NK CD56bright cells (R = -0.23), Tgd (R = − 0.182), Tem (R = − 0.141), B cells (R = − 0.096), Th1 cells (R = − 0.087), and pDC (R = -0.084), as shown in Fig. 14.
Discussion
During glioma progression, numerous abnormal molecular expressions occur, which are closely associated with prognosis and disease progression in patients with cancers11,12,13,14,15. For instance, SRPK1 is overexpressed in glioma, and its elevated levels correlate with poor prognosis and higher glioma grade. SRPK1 overexpression inhibits cell proliferation, migration, and invasion, induces G2/M phase arrest, triggers mitotic mutations, and promotes apoptosis, potentially through the inactivation of the Wnt/β-catenin and JAK-2/STAT-3 signaling pathways14. Furthermore, m6A modifications are strongly linked to GBM progression. IGF2BP3 is also overexpressed in gliomas, with high IGF2BP3 expression correlating with significantly reduced survival. Inhibition of IGF2BP3 expression suppresses glioma cell proliferation, migration, and invasion, and impedes xenograft tumor growth in mice, possibly through the downregulation of CDK115. Our study further identified TMEM150A as a key factor with substantial clinical relevance in glioma. TMEM150A is overexpressed in glioma tissues, and its expression strongly correlates with various clinicopathological features, including WHO grade, IDH status, age, histological subtype, and 1p/19q codeletion. ROC analysis showed that TMEM150A has significant diagnostic potential, with an area under the curve greater than 0.8. Overexpression of TMEM150A is linked to shorter survival, poorer disease-specific survival, and accelerated disease progression in patients with glioma, and is inversely associated with prognosis in subgroups of patients with glioma. These findings suggest that TMEM150A overexpression is a risk factor for poor survival and disease progression in glioma, underscoring its important prognostic value.
TMEM150A co-expressed genes are implicated in several key biological processes, including necrotic cell death, cell–cell adhesion, migration, wound healing, and proliferation. Inhibiting TMEM150A expression in glioma U251 cells significantly reduces cell proliferation, invasion, and migration. Similar results were observed in glioma U118 cells, where TMEM150A inhibition suppressed growth and metastasis, further confirming its role as an oncogene10. Literature reviews highlighted the involvement of EMT in cancer metastasis16,17,18. For example, PSMA2 expression is significantly elevated in gliomas, with higher levels correlating with advanced glioma stages and poorer survival. Inhibition of PSMA2 expression attenuates glioma cell proliferation and migration, while PSMA2 promotes EMT protein expression in glioma cells17. Building on this, the relationship between TMEM150A and EMT was examined. Inhibiting TMEM150A expression in glioma cells significantly reduced vimentin and Snail protein levels during EMT. This suggests that TMEM150A may promote glioma progression by facilitating the EMT process.
Over the past decade, immunotherapy has emerged as a prominent research focus, offering hope for improving the long-term prognosis of patients with cancers19,20,21,22. For example, in oral squamous cell carcinoma (OSCC), a high proportion of cDC1 cells was found in metastatic lymph nodes. Elevated PD-L1 expression was associated with an increased risk of metastatic or recurrent OSCC. Furthermore, metastatic lymph nodes showed higher levels of memory CD8( +) T cells and terminally exhausted PD-1( +) CD8( +) T cells compared to healthy lymph nodes. These findings suggest that metastatic lymph nodes harbor cells capable of triggering an anti-tumor adaptive response, and PD-L1 expression on dendritic cells in these lymph nodes could serve as a biomarker for poor prognosis in patients with OSCC22. Our study revealed a significant positive correlation between TMEM150A overexpression and various prognostic factors, including the estimate score, immune score, and stromal score. Additionally, a positive correlation with immune cells such as Macrophages, aDC, Neutrophils, Eosinophils, iDC, Cytotoxic cells, NK CD56dim cells, T cells, Th17 cells, Th2 cells, NK cells, and T helper cells was observed, and alongside a negative correlation with TFH, Tcm, Mast cells, NK CD56bright cells, Tgd, Tem, B cells, Th1 cells, and pDC. These findings further support the notion that TMEM150A may influence glioma progression through immune escape mechanisms.
Although TMEM150A plays a critical biological role in glioma, several limitations in our study warrant further investigation. First, additional studies are needed to verify the broader mechanisms of TMEM150A, including exploring its role in tumor formation in nude mice. Furthermore, while bioinformatics analysis suggests a link between TMEM150A and the immune microenvironment, this phenomenon requires further validation. In conclusion, our findings demonstrate that TMEM150A is significantly overexpressed in glioma and correlates with key clinical features such as 1p/19q codeletion, age, IDH mutation status, histological subtype, WHO grade, and poor prognosis, indicating its potential as a diagnostic biomarker. TMEM150A overexpression also serves as a prognostic risk factor for glioma and is associated with the immune microenvironment. Notably, inhibition of TMEM150A expression could suppress glioma progression via EMT, positioning it as a promising therapeutic target and offering a new theoretical foundation for the treatment of patients with glioma.
Data availability
The study data are available upon request from the corresponding authors.
References
Zhang, L., Wu, F. & Zhao, J. Transmembrane protein 45A regulates the proliferation, migration, and invasion of glioma cells through nuclear factor kappa-B. Anticancer Drugs. 31(9), 900–907. https://doi.org/10.1097/CAD.0000000000000890 (2020).
Liu, J., Liu, Y., Ren, Y., Kang, L. & Zhang, L. Transmembrane protein with unknown function 16A overexpression promotes glioma formation through the nuclear factor-κB signaling pathway. Mol. Med. Rep. 9(3), 1068–1074. https://doi.org/10.3892/mmr.2014.1888 (2014).
Li, J. et al. TMEM158 promotes the proliferation and migration of glioma cells via STAT3 signaling in glioblastomas. Cancer Gene Ther. 29(8–9), 1117–1129. https://doi.org/10.1038/s41417-021-00414-5 (2022).
Yang, F., Zhang, X., Wang, X., Xue, Y. & Liu, X. The new oncogene transmembrane protein 60 is a potential therapeutic target in glioma. Front. Genet. 20(13), 1029270. https://doi.org/10.3389/fgene.2022.1029270 (2023).
Chen, J. L. et al. Down-regulation of KLRB1 is associated with increased cell growth, metastasis, poor prognosis, as well as a dysfunctional immune microenvironment in LUAD. Sci. Rep. 14(1), 11782. https://doi.org/10.1038/s41598-024-60414-x (2024).
Khan, A. B. et al. CXCR4 expression is associated with proneural-to-mesenchymal transition in glioblastoma. Int. J. Cancer 152(4), 713–724. https://doi.org/10.1002/ijc.34329 (2023).
Guo, Q. et al. Evaluation of the prognostic value of STEAP1 in lung adenocarcinoma and insights into its potential molecular pathways via bioinformatic analysis. Front. Genet. 20(11), 242. https://doi.org/10.3389/fgene.2020.00242 (2020).
Shao, S., Fan, Y., Zhong, C., Zhu, X. & Zhu, J. Coactosin-Like protein (COTL1) promotes glioblastoma (GBM) growth in vitro and in vivo. Cancer Manag. Res. 30(12), 10909–10917. https://doi.org/10.2147/CMAR.S246030 (2020).
Romanet, J. L., Cupo, K. L. & Yoder, J. A. Knockdown of transmembrane protein 150A (TMEM150A) results in increased production of multiple cytokines. J. Interferon. Cytokine Res. 42(7), 336–342. https://doi.org/10.1089/jir.2022.0063 (2022).
Fan, S. T. et al. Overexpression of TMEM150A in glioblastoma multiforme patients correlated with dismal prognoses and compromised immune statuses. PLoS ONE 18(12), e0294144. https://doi.org/10.1371/journal.pone.0294144 (2023).
Cai, L. et al. TRIM37 interacts with EZH2 to epigenetically suppress PTCH1 and regulate stemness in glioma stem cells through sonic hedgehog pathway. J. Neurooncol. https://doi.org/10.1007/s11060-024-04726-y (2024).
Zhang, H. et al. Unveiling the therapeutic potential of IHMT-337 in glioma treatment: Targeting the EZH2-SLC12A5 axis. Mol. Med. 30(1), 91. https://doi.org/10.1186/s10020-024-00857-0 (2024).
Yan, T. et al. ZNF384 transcriptionally activated MGST1 to confer TMZ resistance of glioma cells by negatively regulating ferroptosis. Cancer Chemother. Pharmacol. https://doi.org/10.1007/s00280-024-04681-5 (2024).
Shi, M., Sun, D., Deng, L., Liu, J. & Zhang, M. J. SRPK1 promotes glioma proliferation, migration, and invasion through activation of wnt/β-catenin and JAK-2/STAT-3 signaling pathways. Biomedicines 12(2), 378. https://doi.org/10.3390/biomedicines12020378 (2024).
Zheng, X. et al. N6-methyladenosine reader IGF2BP3 as a prognostic Biomarker contribute to malignant progression of glioma. Transl. Cancer Res. 12(4), 992–1005. https://doi.org/10.21037/tcr-23-449 (2023).
Zhang, J. et al. LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients. J. Exp. Clin. Cancer Res. 37(1), 225. https://doi.org/10.1186/s13046-018-0864-6 (2018).
Zhang, Y. et al. PSMA2 promotes glioma proliferation and migration via EMT. Pathol. Res. Pract. 256, 155278. https://doi.org/10.1016/j.prp.2024.155278 (2024).
Zhang, J., Li, R., Zhang, H., Wang, S. & Zhao, Y. ITGA2 as a prognostic factor of glioma promotes GSCs invasion and EMT by activating STAT3 phosphorylation. Carcinogenesis 45(4), 235–246. https://doi.org/10.1093/carcin/bgad096 (2024).
Reck, M., Remon, J. & Hellmann, M. D. First-line immunotherapy for non-small-cell lung cancer. J. Clin. Oncol. 40(6), 586–597. https://doi.org/10.1200/JCO.21.01497 (2022).
Routy, B. et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: A phase I trial. Nat. Med. 29(8), 2121–2132. https://doi.org/10.1038/s41591-023-02453-x (2023).
Al Zein, M. et al. Immunotherapy and immunoevasion of colorectal cancer. Drug Discov Today. 28(9), 103669. https://doi.org/10.1016/j.drudis.2023.103669 (2023).
Eric, H. et al. High expression of PD-L1 on conventional dendritic cells in tumour-draining lymph nodes is associated with poor prognosis in oral cancer. Cancer Immunol. Immunother. 73(9), 165. https://doi.org/10.1007/s00262-024-03754-x (2024).
Author information
Authors and Affiliations
Contributions
Hao-Qiang Xu: Writing-original draft, Data curation, Visualization, and Investigation. Dan Zhao: Writing-original draft, Visualization, Investigation, and Data curation. Shi-Hao Xu: Investigation and Visualization. Rui Mao: Methodology and Data curation. Wen-Qiong Yang and Qi Han: Validation, Methodology, Formal analysis, Data curation, Writing-review & editing, and Conceptualization.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, HQ., Zhao, D., Xu, SH. et al. TMEM150A overexpression was associated with poor prognosis and cancer progression in glioma verified by comprehensive analysis and cell experiments. Sci Rep 15, 25017 (2025). https://doi.org/10.1038/s41598-025-08752-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08752-2