Abstract
Amphidromous fish could be transported to other habitats during the marine pelagic larval phase by ocean currents, but extent and mechanism of the larval dispersal is poorly known. Since island stream habitats are generally small and fragile, it is very important to understand the connectivity between streams and between islands from the perspective of biodiversity conservation. In the present study, we investigated the genetic connectivity among populations of an amphidromous goby species, Luciogobius ryukyuensis, on four islands (Okinawa, Kume, Ishigaki, and Iriomote) of the Ryukyu Archipelago, Japan. The genome-wide SNPs analyses revealed that the populations of L. ryukyuensis from the four islands are genetically distinguishable. This suggested that larval transport of this species between these four islands is very limited. On the other hand, the mitochondrial genome analysis clearly divided the species into only two populations (Okinawa + Kume and Ishigaki + Iriomote). This may indicate that there was more recent genetic exchange between the two neighboring islands than between more distant islands. The populations on Kume, Ishigaki, and Iriomote islands are particularly small, with only two or three very small habitats on each island. Maintaining all existing habitats will be critical for conservation of the present diversity of the species.
Similar content being viewed by others
Introduction
The large number of small connected and remote islands in the Indo-Pacific explains the impressive biodiversity of the region. Most island ecosystems are limited in size and therefore freshwater-fish habitats are often very small. Such habitats are usually predominated by amphidromous fish, which migrate between freshwater and marine environments1,2,3. The pelagic larvae of amphidromous fish could be transported to other rivers or islands by ocean currents4,5. If the larvae are transported, then the entire connected population should be considered for conservation. On the other hand, even for amphidromous fishes, larval dispersal could be limited, and the species or population could be endemic to a small range6,7. If the population is very small and independent from others, conservation measures for that small population would be urgently needed from the perspective of biodiversity conservation, as it is important to keep the whole genetic diversity of existing species8. It is, therefore, very important to understand the connectivity between islands.
The Ryukyu Archipelago is an island chain in southern Japan, which is composed of several large and many small islands. The large islands usually have many freshwater streams, while the smaller islands have only a few small freshwater streams9. Fresh water on small islands is generally in high demand by local people and is used for drinking, domestic, and agricultural purposes. The freshwater environments of small islands are fragile due to their size and are often affected by dam construction, agricultural land development in the watershed, and urbanization. Many fish species are distributed in the inland water systems of the Ryukyu Archipelago, and most of these species spend their early life stages in the sea3,10,11, but little is still known about genetic connectivity between islands.
Luciogobius ryukyuensis is an amphidromous goby species endemic to the Ryukyu Archipelago (Okinawa and Kagoshima prefectures), southern Japan, which has an elongated body and small eyes. This species inhabits the gravel bottoms in the shallow rapids of the lower freshwater reaches of small streams and in brackish estuaries forming shallow rapids during low tide3,12,13. The eggs are deposited on the undersurface of stones, and the hatched larvae are transported to the sea immediately after hatching12,13. After about one month of pelagic larval life, the larvae migrate back to the stream and metamorphose into benthic juveniles12,13,14. Luciogobius ryukyuensis performs spawning migrations to congregate and reproduce in specific spawning grounds (the lowermost rapid in the brackish water area). This migration pattern differs slightly from that of typical amphidromous species, which only performs one downward and one upward migration in their lifetime. However, it can be considered amphidromous in terms of migration pattern between streams and the sea12,13. Opportunities for gene flow are restricted to colonisation during larval dispersal since the individuals do not return to the sea after the larvae migrate to the stream. However, details on this goby’s larval habitat and larval dispersal are unknown.
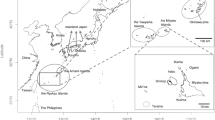
In Okinawa Prefecture, located in the southwestern Ryukyu Archipelago, L. ryukyuensis is distributed on four islands; Okinawa, Kume, Ishigaki, and Iriomote (Fig. 1). In the northern part of Okinawa Island it is found in many streams, however on the other three islands their habitat is restricted to two to three small streams, respectively. The species itself is evaluated as “Vulnerable” in the Red Lists of Okinawa Prefecture and Japanese governments, but the populations of the small islands should be considered critically endangered if they are isolated from others15,16. In the present study, genetic connectivity among populations of L. ryukyuensis on the four islands was determined using mitochondrial genome sequences and single nucleotide polymorphisms (SNPs) detected by a genome-wide screening using double digest restriction-site associated DNA sequence (ddRAD-seq; see details in Peterson et al.17).
Results
Mitochondrial genome analyses
In the maximum likelihood phylogenetic tree using 16,303 bp of aligned mitochondrial genomes, all 10 species of Luciogobius, including L. ryukyueneis, formed each monophyletic clade supported by 100% bootstrap replicates (Fig. 2).
Maximum likelihood phylogenetic tree with 100 bootstraps using the aligned 16,303 bp of mitochondrial genomes in Luciogobius with additional taxa. Material sequenced in the present study are shown with the sample IDs (beginning with Lu) and sequences of Tridentiger, Eucyclogobius, Gillichthys, Gymnogobius, and Chaenogobius are shown with the accession numbers. The capital letters to the right of the symbols represent the sampling sites from which L. ryukyuensis specimens were collected and correspond to those in Fig. 1.
The L. ryukyuensis clade was subdivided into two subclades, supported by 100% bootstrap replicates. One subclade was composed of four specimens from Ishigaki Island and four specimens from Iriomote Island. Specimens from these two neighboring islands were intermixed within the subclade (Fig. 2).
Another subclade was composed of specimens from Okinawa and Kume islands. Three specimens from Kume Island were monophyletic with 100% bootstrap support, but 10 specimens from Okinawa Island were paraphyletic. Specimens from Okinawa Island were collected at three different sites (two on the west coast, and another on the east coast), but none of them formed a monophyletic geographic subclade (Fig. 2).
Specimens of Luciogobius spp. 7 and 8 were collected from Okinawa and Miyako islands. In both species, specimens from each island formed monophyletic subclade supported by 100% bootstrap replicate (Fig. 2).
Genome-wide SNPs analyses
Specimens of L. ryukyuensis from four islands were clearly separated from each other in the phylogenetic network (Fig. 3). Specimens of Okinawa Island were collected at four different sites and none of them formed geographic subclades, except for the Teima River (site B in Fig. 1) where only one specimen was analysed. In Iriomote Island, specimens were collected from two sites. They were intermixed within the clade.
Neighbor-net phylogenetic networks based on Nei’s genetic distances calculated from the 3929 SNPs. Scale bar indicates substitutions per site. The capital letters beside the symbols represent the sampling sites from which L. ryukyuensis specimens were collected and correspond to those in Fig. 1.
ADMIXTURE and PCA analyses further supported genetic divergence among the four islands. In the ADMIXTURE analysis, when K = 4, populations from the four islands formed distinct clusters, and no individual with shared ancestry was observed between the populations (Fig. 4). The lowest cross-validation (CV) error was observed at K = 3, where populations grouped into three geographic clusters: Kume Island, Okinawa Island, and the Yaeyama Islands (including Ishigaki and Iriomote) (Fig. 4). Increasing the number of clusters did not result in population structuring by river within each island. The PCA analysis, PC1–PC2 plot also indicated genetic differentiation among the four islands (Fig. 5). Two neighboring islands, Ishigaki and Iriomote showed relatively high similarity but exhibited slight separation along PC1.
ADMIXTURE results showing K = 2–6 genetic clusters based on the 3929 SNPs among four islands populations. The cross-validation errors are shown in parentheses below the K values. The alphabets below the bars represent the sampling sites from which L. ryukyuensis specimens were collected and correspond to those in Fig. 1.
Discussion
Population structure and larval dispersal
The genome-wide SNPs analyses revealed that the populations of L. ryukyuensis from the four islands are genetically distinguishable (Fig. 3). The ADMIXTURE analysis did not detect any samples with shared ancestry between populations (Fig. 4), indicating that none of the examined individuals have recently interbred with other populations. The results suggested that their larvae are currently not transported between these four islands, and consequently there is no gene flow between these populations. On the other hand, the mitochondrial genome analysis clearly divided the species into only two populations (Okinawa-Kume and Ishigaki-Iriomote). Okinawa and Kume belong to the Okinawa Islands, and Ishigaki and Iriomote belong to the Yaeyama Islands. Each of the two islands is closer together than the other two, and historically they have been connected by land until more recently9,18 (see below). It is highly probable that there was genetic exchange within each of the Okinawa and Yaeyama islands until more recently, which may have prevented the complete sorting of the mitochondrial lineages.
Since no results were obtained indicating genetic isolation between sites within each island, it is thought that there is genetic exchange between habitats within the islands due to larval dispersal. However, because samples from the northeastern site of Ishigaki Island (Ôura River) could not be used for Genome-wide SNPs analyses the relationship between this site and other sites in the Ishigaki-Iriomote group is unknown.
Miyako Island is located between Okinawa-Kume and Ishigaki-Iriomote (Fig. 1). However, Miyako is a relatively flat island composed of limestone, with underdeveloped rivers on the surface, and L. ryukyuensis does not inhabit this island. Therefore, there is currently no habitat for this species between Okinawa-Kume and Ishigaki-Iriomote, a distance of 320 km from the Urachi River on Kume Island to the Ôura River on Ishigaki Island.
The habitat of L. ryukyuensis on Okinawa Island is limited to the northern part of the island, with its southern limit on the west coast at Nago City and on the east coast at Kin Town16,19. The L. ryukyuensis habitat on Kume Island is 90 km from the nearest point on Okinawa Island (Naha Airport in the southern part), and 120 km from the nearest occupied habitat in the northern part of Okinawa Island (streams in Motobu Town). This goby is likely unable to migrate beyond this distance.
The shortest distance between Ishigaki Island and Iriomote Island is only 15 km, and the two islands share a lagoon (Sekisei Lagoon) between them. The distance between habitats of L. ryukyuensis is about 23 km (between Nagura Bay on Ishigaki and Honera River on Iriomote; G and H in Fig. 1). We had assumed that there was genetic exchange between the populations of these two islands, but the results of the genome-wide SNPs analyses refuted this. The results suggest that the larvae are not transported between the two islands. The dispersal distance for this species is likely smaller than expected and this should be kept in mind when considering conservation.
Based on the mitochondrial genome sequences, it was estimated that the clades of the Okinawa Islands clade (Okinawa and Kume) and Yaeyama Islands clade (Ishigaki and Iriomote) of L. ryukyuensis diverged approximately 0.91 million years ago (mya) (Supplementary Information 2). It is considered that many islands in the Ryukyu Archipelago had been connected during the Calabrian (1.80–0.77 mya) to form a land bridge. After that, during the Chibanian (0.77–0.13 mya), the land bridge was divided by the sea and separated into small islands9,18,20. The divergence time of the two clades roughly coincides with the beginning of the Chibanian, suggesting that the two lineages may have been separated by a marine transgression at that time. During the late Pleistocene (0.13–0.01 mya), the landmasses expanded again, but the Kerama gap, a deep structural rift between the Okinawa Islands and Miyako Islands, did not become land, and a complete land bridge was never formed21. The two lineages did not migrate between the Okinawa and Yaeyama regions during this period. On the other hand, each of Okinawa and Kume in Okinawa Islands and Ishigaki and Iriomote in Yaeyama Islands were connected during the Last Glacial Maximum (0.021 mya) of the late Pleistocene (when sea levels dropped by about 80–140 m)18. Although it is difficult to estimate the divergence time of both the Okinawa and Kume populations, and Ishigaki and Iriomote populations of this species based on the data from this study, it is inferred from the geological history that they diverged after the Last Glacial Maximum.
Mitochondrial genome analysis indicated monophyly of each of the 10 species of Luciogobius found in Okinawa Prefecture (one species, Luciogobius sp. 10, could not be included in the analysis). In the two species (Luciogobius sp. 7 and sp. 8) for which specimens from Miyako Island were analysed in addition to Okinawa Island, the two island populations were divided into separate subclades (Fig. 2). This may suggest that other species also lack larval dispersal between islands, as seen in L. ryukyuensis, but only one specimen of Luciogobius sp. 7 from Miyako Island was analysed. It will be necessary to analyse more samples in the future.
Amphidromous fish can be transported to other habitats during their pelagic larval phase, but the extent of their dispersal varies among taxa. For example, in many sicydiine goby species22,23 and Eleotris24,25, there are no genetic structure over a wide area within the distribution range, suggesting widespread dispersal. On the other hand, species of the genus Rhinogobius show genetic differences over relatively small ranges7,26. Maeda et al.27 and Okinawa Prefectural Government16 speculated that larvae of the genus Luciogobius stay near the river mouth and usually do not disperse far. The results of the present study clearly supported this view. The pelagic larval duration of Luciogobius is approximately one month13,14, which is considerably shorter than those of the Sicydiine goby and Eleotris (two months or longer4,28,29). However, considering that the strong Kuroshio Current flows along the Ryukyu Archipelago and can rapidly transport goby larvae5, a one-month pelagic phase may not be short enough to explain the level of isolation of L. ryukyuensis observed in this study. The newly hatched Luciogobius larvae are more developed than those of the Sicydiine goby and Eleotris and can swim against the flow13,27, so the larvae probably remain near the river mouth. This may instead have a stronger influence on the lack of dispersal of Luciogobius. At present the details of their larval habitat are unknown, and it is not yet certain whether the larvae really remain near the river mouth and therefore are not absorbed by open ocean currents such as the Kuroshio Current. To elucidate this retention mechanism, it is necessary to clarify the habitat and behaviour of the larvae.
Conservation
This study has important implications for conservation, as our results revealed that the genetic diversity of L. ryukyuensis is more vulnerable than previously thought. This species is also distributed in the Amami Islands (Amami-oshima, Kakeroma-jima, and Tokunoshima) of Kagoshima Prefecture, located northeast of Okinawa Island16,30. Although the Amami Islands population was not analysed in this study, the suggested lack of gene flow between the four islands of Okinawa Prefecture makes it highly unlikely that the Amami Islands population is connected to the Kume, Ishigaki, and Iriomote populations, bypassing the neighboring Okinawa Island population.
Kume, Ishigaki, and Iriomote islands have only a few very small habitats for L. ryukyuensis. The Urachi River is the only habitat on Kume Island reported so far31,32, and samples collected there were analysed in this study (site E in Fig. 1). The adult habitat of L. ryukyuensis (the area where we were able to find adults in 2008 and 2018) is less than 10 m square (Fig. 6). The extent of the larval habitat is unknown, but it is assumed to be part of the harbor area around the river mouth. The habitat (for adults, larvae, and migration routes between them) is surrounded by urban structures, and the upper watershed is developed as agricultural land. Therefore, it could easily be affected by human life, agriculture, and fisheries. On Kume Island, one individual of L. ryukyuensis has been found in the Shirase River adjacent to the Urachi River, which flows into the same harbour area (600 m east of the Urachi River mouth; Toshifumi Saeki, personal communication). Although the details of this habitat have not yet been investigated, we believe that the habitat of Shirase River is also very small. If there is a serious impact on these habitats and the population becomes extinct, it is almost impossible to expect a larval supply from other islands. In addition, the Kume population is genetically differentiated and is a valuable component of the genetic diversity of this species. Maintaining these two habitats is, therefore, critical for the conservation of the whole genetic diversity of this species.
On Ishigaki Island, this species has been found in three places; creeks on Nagura Bay (Hayashi & Ito33 and this study; site G in Fig. 1), Shitafuki River on Kabira Bay33, and Ôura River on Ibaruma Bay (this study; site F in Fig. 1). On Nagura Bay, we found L. ryukyuensis at the mouths of two creeks 500 m apart in 2010. Shiiu River, a habitat reported in Hayashi & Itoh33, is probably one of them (or another creek between these two). These are very small creeks flowing through agricultural land, and we only found juveniles along the creeks on the tidal flats seaward of the seaside road. There may be other habitats in this area, but even if so, they would be few and small. The Ôura River is a small stream in the northeastern part of the island. We obtained only one specimen there, and the details of the habitat are unknown. Shitafuki River is listed in Hayashi & Itoh33 as a collection site, but the details are unknown, too. Because a long coastline and many headlands separate Nagura Bay, Kabira Bay, and Ibaruma Bay, it seems difficult for larvae to migrate between these three places. There is an urgent need to understand the overall habitat of Ishigaki Island and the genetic relationships among the habitats in order to assess the status of this species in Ishigaki.
On Iriomote Island, L. ryukyuensis is known from three streams in the eastern part of the island; Aira River (site I in Fig. 1), Maira River, and Honera River (site H in Fig. 1)34 (Honera River was referred to as “Nishifonera-gawa” in Chen et al.34). The Aira and Maira rivers are nearby streams that flow into the same bay. Although specimens from the Maira River were not examined in this study, the finding that the populations of the Aira River and the more distant Honera River were not genetically distinct suggests that there is no genetic structure among the gobies from these three streams. All populations on Iriomote Island are small and vulnerable to habitat alteration. There are many streams and rivers on this island, but this species has never been found in streams other than these three, and other major habitats are unlikely to exist.
Thus, the habitats on Kume, Ishigaki, and Iriomote islands are all very small, few in number, and fragile. Given the suggested lack of larval dispersal among the islands, it is crucial to conserve each habitat.
Okinawa Island has more habitats and larger populations than the three other islands. The distance between a habitat on the west coast of Nago City and a habitat on the east coast of Kin Town is about 150 km via the coastline around the northern tip of the island, which is farther than the distance between the habitats on Okinawa Island and Kume Island (120 km). However, there are many habitats between these localities, which are thought to link these habitats. On Okinawa Island, the habitats of this species are often destroyed by dredging of the stream bed to improve drainage16,35. The population may recover through larval recruitment from neighboring streams after the habitat is restored (usually it takes several years). Still, it should be noted that recovery may be difficult if the habitats are destroyed simultaneously in neighboring streams because their larvae are not likely to disperse far.
Because amphidromous fish are generally considered to be dispersive, the importance of each population tends to be underestimated. In the case of a species with low dispersal like L. ryukyuensis, the conservation of the habitats of the isolated small island’s population is extremely important. Even for relatively large populations such as the Okinawa Island population, attention should be paid to the fragmentation of populations due to habitat loss, which may lead to population instability.
Methods
Sampling and DNA extraction
Specimens of L. ryukyuensis were collected using hand nets on Okinawa, Kume, Ishigaki, and Iriomote islands in Okinawa Prefecture, southwestern part of the Ryukyu Archipelago. Specimens of other species of Luciogobius collected on Okinawa and Miyako islands were also used for mitochondrial genome analysis. After being euthanised with 2-phenoxyethanol solution, the specimens were fixed in 99.5% ethanol or 10% formalin. In the latter case, the right pectoral fin was cut off before the body was put into formalin and that pectoral fin was preserved in 99.5% ethanol for genetic analyses. The specimens were identified as L. ryukyuensis because their morphology matched that of L. ryukyuensis described by Chen et al.34.
In addition to L. ryukyuensis, 10 species of Luciogobius are distributed in Okinawa Prefecture16. The taxonomic status of the nine of these species, except for L. griseus, remains unclear. They may be undescribed species, but the taxonomic issue is not the subject of this study and will not be pursued. We follow Maeda et al.14 for the tentative codes (Luciogobius spp. 3–6) of four of the nine species, since they described morphologies of these species. In the present study, the remaining five species are designated Luciogobius spp. 7–11, and their morphologies are described in Supplementary Information 1. In this study, nine of the 10 species were included in the mitochondrial genome analysis, excluding Luciogobius sp. 10 for which DNA samples were not available.
The total genomic DNA of 60 specimens of L. ryukyuensis and 33 specimens of other species of the genus Luciogobius was extracted from right pectoral and/or caudal fins (for two specimens, piece of right opercle was used in addition to the pectoral and caudal fins), using Maxwell RSC Blood DNA Kit (Promega, Fitchburg, Wisconsin, USA) following manufacturer’s instructions. Sample IDs and data for each specimen are given in Supplementary Table S1 online.
Mitochondrial genome analyses
Whole-genome shotgun sequencing libraries were prepared using a KAPA HyperPlus Kit (KAPA Biosystems, Wilmington, Massachusetts, USA). PCR amplification of sequencing libraries (15 cycles) was performed for sample ID Lu1 and Lu42-49. The shotgun libraries were then sequenced on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, California, USA) in Rapid Run mode version 1 using a TruSeq Rapid PE Cluster Kit and a TruSeq Rapid SBS Kit (v1) (Illumina) for Lu1 and Lu42-49 (137 bp paired-end) or in high output mode using HiSeq PE Cluster Kit v4 cBot and HiSeq SBS Kit v4 (Illumina) for other libraries (163 bp paired-end) following manufacturer’s instructions.
Sequencing data from each library was assembled using IDBA_UD assembler version 1.1.136 with different kmer lengths (60, 80, 100). Identification of complete mitochondrial genomes from assembled contigs was performed by: (1) comparing them with the complete mitochondrial genome of Stiphodon alcedo (accession: AB613000.1) (BLASTN e-value B 1e-100); and (2) confirming that 100 bp of both head and tail DNA sequences of a contig were identical, indicating that the sequence was circular. Complete mitochondrial genomes were aligned using MAFFT v7.24437 and all positions with gaps were removed using trimAl38. We performed molecular phylogenetic analyses of the aligned mitochondrial genomes of Luciogobius species along with mitochondrial genome sequences of Tridentiger kuroiwae (LC653489 and LC653490) sequenced in Maeda et al.39, Eucyclogobius newberryi (NC_028288.1), Gillichthys seta (NC_012908.1), Gillichthys mirabilis (NC_012906.1), Gymnogobius urotaenia (NC_028432.1), Gymnogobius petschiliensis (NC_008743.1), Chaenogobius annularis (NC_063745.1), and Chaenogobius gulosus (NC_027193.1) retrieved from the NCBI RefSeq database as outgroup taxa, using the GTR + I + Gamma model and performed a maximum likelihood (ML) analysis using RAxML-NG with 100 bootstrap replicates40.
Genome-wide SNPs analyses
The library for ddRAD-Seq17 was created using the method described in Sakaguchi et al.41 with slight modifications, in which BglII and EcoRI was used as restriction enzymes. The library was sequenced with 150-bp paired-end reads in an Illumina HiSeq X. The reads were adapter-trimmed, and low-quality (mean quality < 5) and short (< 50 bp) reads were filtered out using fastp version 0.21.042.
For 51 individuals of the L. ryukyuensis, genotyping was performed using the Stacks version 2.41 denovo_map.pl pipeline program43. Denovo-loci were built using default settings except for the minimum of 10 reads to create a stack. Loci were filtered to only include those found in all populations (four islands) and in all individuals within each population. Genotype outputs were created in VCF format, including only the first SNP per locus, resulting in a total of 3929 SNP sites.
This VCF file was converted to a PED file using PLINK version 1.944. ADMIXTURE version 1.3.045 was then run for 2–9 clusters (K). Statistical support for the different numbers of clusters was evaluated using the cross-validation techniques implemented in ADMIXTURE. Principal component (PC) analyses were also conducted for PC2 to PC9 with the R package SNPRelate version 1.16.046, using the VCF file of the 3929 SNPs. Additionally, an individual-based phylogenetic network was constructed using SplitsTree v.4.14.847. The network was built using the Neighbour-Net method based on p-distances between individuals, which were calculated from the VCF with VCF2Dis version 1.46 (https://github.com/BGI-shenzhen/VCF2Dis).
Ethics declarations
All methods were carried out in accordance with relevant guidelines and regulations and reported in accordance with the ARRIVE guidelines 2.0. The procedures used to handle live fish specimens in this study were approved by the Animal Care and Use Committee of the Okinawa Institute of Science and Technology Graduate University, Japan. No permission was required from the national or local government to collect fish for this study. Although Kumejima Town Ordinance prohibits unauthorized collection of wildlife on Kume Island from 2022, the specimens from Kume Island used in this study were collected in 2008 and 2018.
Data availability
The assembled mitochondrial genome sequences with gene annotations are available in the DNA Data Bank of Japan (DDBJ) under accession numbers: LC859275–LC859328. The ddRAD-Seq reads were stored in DDBJ Sequence Read Archive under the accession numbers DRR641339–DRR641389. Accession numbers for each specimen are given in Supplementary Table S1 online.
References
Keith, P. Biology and ecology of amphidromous Gobiidae of the Indo-Pacific and the Caribbean regions. J. Fish Biol. 63, 831–847 (2003).
McDowall, R. M. Ancestry and amphidromy in island freshwater fish faunas. Fish Fish. 5, 75–85 (2004).
Maeda, K. & Tachihara, K. Fish Fauna in the Teima Stream, Okinawa Island. Biol. Mag. Okinawa 44, 7–25 (2006) (in Japanese with English abstract).
Maeda, K., Yamasaki, N. & Tachihara, K. Size and age at recruitment and spawning season of sleeper, genus Eleotris (Teleostei: Eleotridae) on Okinawa Island, southern Japan. Raffles Bull. Zool. Suppl. 14, 199–207 (2007).
Iida, M., Zenimoto, K., Watanabe, S., Kimura, S. & Tsukamoto, K. Larval transport of the amphidromous goby Sicyopterus japonicus by the Kuroshio Current. Coast. Mar. Sci. 34, 42–46 (2010).
Liao, T.-Y. et al. Amphidromous but endemic: Population connectivity of Rhinogobius gigas (Teleostei: Gobioidei). PLoS ONE 16(2), e0246406 (2021).
Takagi, M. et al. Genetic structure of freshwater goby Rhinogobius sp. DL widely distributed in the Ryukyu arc, Japan. Bull. Biogeogr. Soc. Jpn. 70, 123–130 (2015) (in Japanese with English abstract).
Garcia-Dorado, A. & Caballero, A. Neutral genetic diversity as a useful tool for conservation biology. Conserv. Genet. 22, 541–545 (2021).
Nishida, M., Shikatani, N. & Shokita, S. The Flora and Fauna of Inland Waters in the Ryukyu Islands (Tokai University Press, 2003) (in Japanese).
Yoshigou, H. Annotated checklist and bibliographic records of inland water fishes of the Ryukyu Archipelago, Japan. Fauna Ryukyuana 9, 1–153 (2014) (in Japanese with English abstract).
Maeda, K. & Tachihara, K. Larval fish fauna of a sandy beach and an estuary on Okinawa Island, focusing on larval habitat utilization by the suborder Gobioidei. Fish. Sci. 80, 1215–1229 (2014).
Maeda, K. What is amphidromy? A discussion of various migration patterns. Aquabiology 38, 350–355 (2016) (in Japanese with English abstract).
Kondo, M., Maeda, K., Yamasaki, N. & Tachihara, K. Spawning habitat and early development of Luciogobius ryukyuensis (Gobiidae). Environ. Biol. Fish. 95, 291–300 (2012).
Maeda, K., Yamasaki, N., Kondo, M. & Tachihara, K. Occurrence and morphology of larvae and juveniles of six Luciogobius species from Aritsu Beach, Okinawa Island. Ichthyol. Res. 55, 162–174 (2008).
Ministry of the Environment. Red Data Book 2014; Threatened Wildlife of Japan, Volume 4, Pisces (Brackish and Freshwater Fishes) (GYOSEI Corporation, 2015) (in Japanese).
Okinawa Prefectural Government. Red Data Okinawa; Threatened Wildlife in Okinawa, 3rd Edition (Animals) (Okinawa Prefectural Government, 2017) (in Japanese).
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7, e37125 (2012).
Furukawa, M. & Fijitani, T. Comparative study of Pleistocene paleogeographic maps of Ryukyu Arc. Bull. Fac. Sci. Univ. Ryukyus 98, 1–8 (2014) (in Japanese with English abstract).
Inui, R. & Takemura, S. Hand Book for Nature of the Okukubi River, Gobies in the Estuary and Mangrove (Kin-cho Board of Education, 2016) (in Japanese).
Kimura, M. Quaternary paleogeography of the Ryukyu Arc. J. Geogr. 105, 259–285 (1996) (in Japanese with English abstract).
Watanabe, et al. Geological history of the land area between Okinawa Jima and Miyako Jima of the Ryukyu Islands, Japan, and its phylogeographical significance for the terrestrial organisms of these and adjacent islands. Prog. Earth Planet. Sci. 10, 40 (2023).
Lord, C. et al. From endemism to widespread distribution: Phylogeography of three amphidromous Sicyopterus species (Teleostei: Gobioidei: Sicydiinae). Mar. Ecol. Prog. Ser. 455, 269–285 (2012).
Lord, C., Maeda, K., Keith, P. & Watanabe, S. Population structure of the Asian amphidromous Sicydiinae goby, Stiphodon percnopterygionus, inferred from mitochondrial COI sequences, with comments on larval dispersal in the northwest Pacific Ocean. Life Environ. 65, 63–71 (2015).
Maeda, K., Mukai, T. & Tachihara, K. Newly collected specimens of the sleeper Eleotris acanthopoma (Teleostei: Eleotridae) from French Polynesia indicate a wide and panmictic distribution in the West and South Pacific. Pac. Sci. 65, 257–264 (2011).
Mennesson, M. I., Bonillo, C., Feunteun, E. & Keith, P. Phylogeography of Eleotris fusca (Teleostei: Gobioidei: Eleotridae) in the Indo-Pacific area reveals a cryptic species in the Indian Ocean. Conserv. Genet. 19, 1025–1038 (2018).
Yamasaki, Y. Y., Nishida, M., Suzuki, T., Mukai, T. & Watanabe, K. Phylogeny, hybridization, and life history evolution of Rhinogobius gobies in Japan, inferred from multiple nuclear gene sequences. Mol. Phylogenet. Evol. 90, 20–33 (2015).
Maeda, K., Iida, M. & Kondo, M. Diversity of larval dispersal strategies in freshwater and estuarine gobies in Early Life History of Fishes (ed. Mochioka, N., Kinoshita, I. & Minami, T.) 89–101 (Kouseisha-kouseikaku, 2015) (in Japanese).
Radtke, R. L., Kinzie, R. A. III. & Shafer, D. J. Temporal and spatial variation in length of larval life and size at settlement of the Hawaiian amphidromous goby Lentipes concolor. J. Fish Biol. 59, 928–938 (2001).
Teichert, N. et al. Pelagic larval traits of the amphidromous goby Sicyopterus lagocephalus display seasonal variations related to temperature in La Réunion Island. Ecol. Freshw. Fish 25, 234–247 (2016).
Wada, H. et al. First records of 122 fish species from Tokunoshima island, the Amami Islands, Kagoshima, Japan. Ichthy, Nat. Hist. Fish. Jpn. 7, 35–52 (2021).
Satou, F. Fishes and crustaceans around Kumejima Firefly Museum (lower reaches of Urachi-gawa) and freshwater fish on Kume Island. Kumejima Shizen Bunka Center Kiyou 5, 67–77 (2005) (in Japanese).
Yoshigou, H. Inland water fishes of the Kume Island, Ryukyu Islands, Japan. Misc. Rep. Hiwa Mus. Nat. Hist. 48, 25–51 (2007) (in Japanese with English abstract).
Hayashi, M. & Itoh, T. Gobioid fishes of Ryukyu Islands, southern Japan (I). Sci. Rep. Yokosuka City Mus. 24, 59–82 (1978) (in Japanese with English abstract).
Chen, I-S., Suzuki, T. & Senou, H. A new species of gobiid fish, Luciogobius from Ryukyus, Japan (Teleostei: Gobiidae). J. Mar. Sci. Technol. 16, 248–252 (2008).
Ebner, B. C. et al. Pebbled places preferred by people and pipefish in a World Heritage protected area. Cybium 47, 401–416 (2023).
Peng, Y., Leung, H. C., Yiu, S. M. & Chin, F. Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Maeda, K. et al. Two new species of Rhinogobius (Gobiiformes: Oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa 5068, 81–98 (2021).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Sakaguchi, S. et al. High-throughput linkage mapping of Australian white cypress pine (Callitris glaucophylla) and map transferability to related species. Tree Genet. Genomes 11, 121 (2015).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Rochette, N., Rivera-Colón, A. & Catchen, J. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 28, 4737–4754 (2019).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Huson, D. H. & Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2005).
Acknowledgements
We appreciate Masashi Kondo, Nozomi Hanahara, Katsunori Tachihara, Toshifumi Saeki, Masatomi Hosokawa, and Toshiyuki Suzuki for providing valuable samples and information, and/or their support to the fieldwork, Reo Koreeda and Hiroyuki Motomura (Kagoshima University Museum) for providing information about taxonomy of the genus and registration of the specimens, Kanako Hisata and Saori Miura (OIST) for assistance with sample preparation for sequencing and data handling, the OIST Sequencing Section staffs for preparing the libraries and sequencing for mitochondrial genome analysis, and Manon Mercader and Danielle Miller (OIST) for helpful suggestions to improve the manuscript.
Funding
This study was partially supported by a research grant of the Fujiwara Natural History Foundation, JSPS KAKENHI Grant Number 16K07492, and by the Okinawa Institute of Science and Technology Graduate University.
Author information
Authors and Affiliations
Contributions
KM conceived and designed the research, collected samples, analysed the data, and wrote and edited the manuscript. HK, TU, CS, RK, and AN performed DNA sequencing, analysed the data, and edited the manuscript. NS and VL supervised the research and edited the manuscript. All authors contributed to manuscript revisions and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Maeda, K., Kobayashi, H., Uchida, T. et al. Genetic isolation of an amphidromous goby species, Luciogobius ryukyuensis, highlights limited gene flow in small island populations. Sci Rep 15, 24193 (2025). https://doi.org/10.1038/s41598-025-09050-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09050-7