Abstract
Staphylococcus epidermidis is an opportunistic commensal cutaneous biofilm-producing agent frequently causing musculoskeletal infections (MSI) with/without implants. This study was design to evaluate phenotypic and genomic relatedness between commensal skin and MSI S. epidermidis isolates and to assess patient-related and microbial markers in the outcome of patients after one-year follow-up. Demographics, clinical data and monomicrobial S. epidermidis isolates of MSI patients (n = 31) and healthy individuals (n = 15) were analyzed. Phenotypic profile was assessed by susceptibility tests by broth microdilution and biofilm formation. Phylogenetic relationships, resistome characterization, and virulome analysis were carried out by complete genome sequencing (n = 46). Overall, MSI-derived isolates were significantly more strong/moderate biofilm producers and depicted higher rates of resistance to methicillin (MRSE), ciprofloxacin, gentamicin and rifampicin. Demographics and clinical characteristics did not significantly affect MSI patients’ outcomes. In the whole-genomic sequencing (WGS) phylogeny, most MSI-derived isolates were grouped into the pathogenic-associated clonal complex CC2, and significantly higher prevalence of mecA gene and pathogenic marker IS256. Following multivariate analysis, MSI-derived isolates carrying transposable element IS256 are more likely to develop a persistent infection (odds ratio [OR], 8.00, [95% confidence interval (CI), 1.06 to 60.31], P = 0.044), while weaker biofilm producer were protectors (OR, 0.070, 95%CI, 0.005–0.979, P = 0.048), reducing the chance of recurrence by 93% (1–0.070). The logistic regression model’s performance was evaluated by Nagelkerke’s R, which resulted in 47.21%. In conclusion, S. epidermidis isolates producing MSI were phenotypically and genetically distinct from commensals, proven the association between independent genetic traits and patient’s outcome.
Similar content being viewed by others
Introduction
Musculoskeletal infections (MSIs) encompass numerous complex clinical conditions, such as haematogenous osteomyelitis, spondylodiscitis, infectious arthritis, fracture-related infections (FRIs), and periprosthetic joint infections (PJIs)1. These conditions share clinical aspects, including chronic pain, delayed healing, and reduced functionality of the affected limb. While the global burden of MSIs varies between developed and developing countries, in developed settings, the increasing number of elective joint replacements in the ageing and obese population contributes to the increasing incidence of MSIs1,2. In contrast, developing nations have a higher prevalence of MSIs associated with emergency orthopaedic surgeries for trauma stabilization because of the high burden of injuries and fractures due to interpersonal violence and road traffic accidents (approximately 178 million new fractures annually)1,2. These procedures often involve the insertion of orthopaedic implants, thereby increasing the risk of MSIs1,2,3.
Gram-positive cocci, especially staphylococci, are frequently implicated in MSIs, accounting for up to 75% of all MSIs, regardless of the infection acquisition phase4. This is related to microorganisms that inhabit the skin surface, including coagulase-negative staphylococci (CoNS). Despite being frequently considered “contaminants” owing to their low pathogenic potential, CoNS are commonly causative agents of MSIs after orthopaedic surgeries with implant placement3,4,5,6,7. Most MSIs are initiated mainly by the introduction of commensal skin microorganisms during trauma or surgery, leading to contamination of the surgical field and subsequent implant colonization8. Among CoNS species, Staphylococcus epidermidis (S. epidermidis) stands out as the predominant species in chronic infections, accounting for more than 50% of cases, which are likely associated with determinants of pathogenicity, particularly their ability to adhere to and capacity to form biofilms on the surface of orthopaedic implants8. The adhesion of Staphylococci to host tissue and abiotic surfaces (orthopaedic implants) is a critical step in establishing infection. It is mediated by a complex mechanism including unspecific forces involving surface charges, hydrophobicity, and a range of cell wall-anchored (CWA) surface proteins that bind to specific host tissues and molecules, such as fibrinogen, fibronectin, collagen, elastin, and vitronectin6,7,8,9.
Notably, biofilm-encapsulated microorganisms exhibit significantly greater antibiotic tolerance than their planktonic counterparts do, rendering commonly prescribed systemically administered antibiotics often ineffective at achieving therapeutic levels within the biofilm10,11,12. Consequently, successful management of MSIs necessitates a multimodal medical approach combining surgical debridement and targeted antimicrobial therapy towards biofilm-forming pathogens7.
In the last decade, significant progress has been made in terms of relevant insights into the molecular basis of the pathogenic potential of S. epidermidis through preclinical studies utilizing phenotypic and genotypic techniques, including next-generation sequencing (NGS)9. Nevertheless, knowledge gaps remain regarding the full spectrum and complexity of virulence and resistance factors in this opportunistic pathogen, especially their impact on clinical outcomes13. Indeed, the employment of NGS for S. epidermidis in a few studies has allowed for the molecular characterization of isolates by identifying genetic determinants of clinical importance that may even help predict the clinical response to treatment14,15,16. In this context, methicillin-resistant S. epidermidis (MRSE), a broad range of adhesion proteins, and biofilm formation ability are associated with pathogenicity15. Mobile genetic elements (MGEs), such as pathogenicity islands, plasmids, and bacteriophages, commonly carry these virulence determinants9,14,15,17. For example, previous studies have highlighted the association between the presence of the mecA gene (methicillin resistance) in the staphylococcal chromosomal cassette mec (SCCmec), the ica locus (associated with biofilm formation), and the insertion sequence IS256 (related to genomic plasticity and invasion) with pathogenic isolates of S. epidermidis causing MSI18. In addition, a new recently described microbial surface component recognizing adhesive matrix molecules (MSCRAMMs) subfamily SesJ is likely to play a role as a CWA protein in S. epidermidis virulence19. However, the discriminatory efficacy of these markers has been questioned11,20. Furthermore, MRSE exhibits a dynamic and continuous global dissemination pattern comprising sequence types (STs) 2, 5, and 23, often leading to clinically challenging infections20,21.
With the significant increase in the number of orthopaedic surgeries and medical device-related infections caused by the SoCN, there is a need to develop studies with advanced molecular basis techniques to fill in the gaps of knowledge regarding the genetic mechanisms through which S. epidermidis flexibly adapt and transit their behaviour to an invasive state depending upon several conditions, such as the microbial environment and/or type of implanted medical device22,23,24,25. Moreover, insight into genetic predictors of invasiveness and outcome is potentially valuable for clinical practice.
The present study aimed to evaluate the phenotypic and genomic characteristics of clinical isolates of S. epidermidis, correlating the presence of specific phenotypes and pathogenicity-related genes potentially associated with microbial invasiveness with the clinical outcomes of patients with MSI undergoing clinical treatment and orthopaedic surgeries. The study compared a collection of commensal control isolates from healthy individuals and patients presenting S. epidermidis MSI with and without implants that were prospectively followed for a minimum period of 12 months.
Results
Study population, clinical characteristics, and clinical outcomes of patients with musculoskeletal infections (MSIs)
This study included 31 patients with monomicrobial S. epidermidis-associated MSIs and 15 S. epidermidis-colonized healthy individuals. Overall, 60.9% of the patients were male (28/46), with a mean age of 43.39 years (SD ± 17.76). Among the 31 patients with MSI, 52,2% were male, with a mean age of 48.1 years (SD ± 18.21). In terms of preoperative conditions, FRI was observed in 16 (51.6%) patients, PJI in 12 (38.7%), bacterial septic arthritis in 2 (6.4%), and chronic osteomyelitis in one patient (3.2%). Lower limb infections were predominant, accounting for 93.5% of the cases. Implanted-related MSIs (FRIs and PJIs) are late infections. Following a minimum follow-up duration of one year, a successful treatment outcome was documented in 16 (51.6%) patients, whereas 15 (48.4%) experienced recurrence. The complete demographic and clinical details of the study population are summarized in Table 1.
Clinical aspects, origin, phenotypic characterization, and antibiotic sensitivity of S. epidermidis isolates, with correlations to clinical outcomes
The study included 31 (67.4%) S. epidermidis isolates obtained from symptomatic MSI patients through the sonication of surgically excised implants (n = 18) and cultures of peri-implant and soft tissue (n = 13). For comparative purposes, 15 S. epidermidis isolates were obtained from swab samples of colonized, healthy individuals (32.6%). Overall, MRSE was observed in 65.2% (30/46) of the isolates and was significantly more common among isolates from MSI patients (52.1%, 27/46) than among colonization isolates (13%, 6/46) (P = 0.012). Erythromycin resistance was identified in 60.9% (28/46) of the isolates, with a greater incidence among the MSI-derived isolates than among the colonization isolates (39.1% vs. 21.7%, P = 0.575). Ciprofloxacin resistance was present in 50% (23/46) of the isolates, with a significant difference observed between the MSI and colonization isolates (45.5% vs. 6.5%, P = 0.004). Resistance to gentamicin was also prominent and was detected in 41.3% (19/46) of the isolates, predominantly among those derived from MSI cases (34.8% vs. 6.5%, P = 0.041). Worryingly, while colonization isolates were universally sensitive to rifampicin, a resistance rate of 26% (12/46) was noted among MSI isolates (P = 0.004), underscoring potential implications for the clinical management of MSI. No resistance to vancomycin and linezolid was detected.
Patient- and microbiological-related factors that were investigated for possible associations between infected (n = 31) and colonized patients (n = 15) with the S. epidermidis via univariate analysis are shown in Table 1. Notably, the MSI-high patients were male (52.2% vs. 8.7%, P < 0.001) and older (median age of 48.1 years vs. 33.7 years, P = 0.015). The groups were similar regarding comorbidities, except for higher rates of immunosuppression in the MSI patients (P = 0.040). In patients with MSI progressing to recurrence (n = 15) or cure (n = 16), no statistically significant differences were observed in terms of demographics or clinical characteristics (obesity, smoking habits, diabetes, and immunosuppression), whereas FRI patients were significantly more likely to progress to cure than those with other diagnoses were (P = 0.009). S. epidermidis isolates resistant to methicillin and at least two other classes of antibiotics, were more common in patients with infection recurrence than in those who were cured (80% vs. 62.5%, P = 0.433). On the other hand, MRSE strains were significantly more common in recurrent MSI patients (14/31) than in cured patients (10/31) (45.1% vs. 32.3%, P = 0.040). However, no significant difference was observed between cured and recurrent MSI patients treated with erythromycin, ciprofloxacin, or gentamicin (Table 2). The sensitivity profile using MICs of the 46 S. epidermidis isolates is described in Table 2.
Ability to form a biofilm
All 46 S. epidermidis isolates could form biofilms, although strong/moderate and weak biofilm producers accounted for 80.6% and 19.4%, respectively. In total, strong/moderate biofilm-producing isolates were identified in 73.9% (34/46) of patients with MSI and 26.1% (12/46) of colonized individuals (P = 0.040). However, not statistically significant, strong/moderate biofilm producers were more frequently detected in patients who progressed to infection recurrence (14/31) than in those considered cured (11/31) after one year of follow-up (P = 0.083).
Clonality profile and genomic characteristics associated with resistance and virulence in S. epidermidis isolates via next-generation sequencing (NGS)
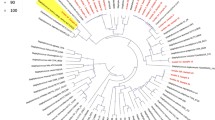
All 46 S. epidermidis isolates were subjected to NGS. The classification of S. epidermidis isolates for Multi Locus Sequence Typing (MLST) is based on the nucleotide sequences of seven loci (arcC, aroE, gtr, mutS, GlpF, pyrR, tpiA, and yqiL). In this study, all the STs of all the isolates were characterized, including the identification of two new STs (ST1200 and ST1201). Overall, 25 different STs were characterized. All isolates of ST2, ST6, ST23, ST885, and ST1199 were associated with MSI. There was significant variability in ST2 between isolates from MSIs and colonized healthy individuals (32.2% vs. 0%, P = 0.019) (Fig. 1 and Supplementary Table S2). Importantly, most of the isolates (37%, n = 17/46) were grouped into clonal complex 2 (CC2) (ST2, ST23, ST5, ST73, and ST89) by PubMLST. Resistance genes were identified via next-generation sequencing (NGS). The blaZ and mecA genes, which confer resistance to β-lactam antibiotics, were present in 84.8% (39/46) and 56.5% (26/46) of the isolates, respectively (Supplementary Table S3). Indeed, SCCmec type IV (2B) harbouring mecA gene (MRSE isolates) predominated in the MSI isolates (54.8%) (Supplementary Table S4). A mutation in the parE gene (D432V) was found in 13% of the isolates (all in patients with MSI). The identification of aminoglycoside resistance genes varied from 6.5% (ant(9)-Ia) to 37% (AAC(6’)-Ie-APH(2’’)-Ia), being almost exclusively carried by MSI-causing isolates (P < 0.001) (Supplementary Table S3). Importantly, rifampicin resistance develops through a single-step mutation in the rpoB gene. Specific H481N and I527M mutations were detected; the latter, previously mentioned as the most common cause of rifampicin resistance in hospital-origin S. epidermidis within the ST2/CC2 and ST23/CC2 lineages, was detected only in MSI-derived isolates21. (Fig. 1 and Supplementary Table S2).
Phylogenomic analysis and resistance and virulence genes of S. epidermidis strains using WGS associated with clinical outcome. In the MSI isolates, ST2, ST6, ST23, ST885, and ST1199 predominated; while all ST2 originated only from pathogenic isolates (32.2% vs. 0%, P = 0.019). Clonal complex 2 (CC2) (ST2, ST23, ST5, ST73, and ST89) were 37%. MecA gene predominated in pathogenic isolates (50% vs. 6.5%, P < 0.001), without significant impact in the outcome. The complete icaADBC operon was carried mainly in pathogenic isolates (39.1% vs. 19.6%, P = 0.90), without significant impact in the outcome. Genetic element IS256, was carried by 41.3% of pathogenic isolates, and a significant predictor of recurrence (OR, 8.00, 95%CI, 10.6–60.3, P = 0.044).
The assessment of the presence of accessory virulence genes revealed that virtually all 46 isolates carried genes associated with biofilm formation (atl), adhesion (ebh, sdrG, sdrH), cytotoxic enzymes (hlb, hld, lip, geh), and neutrophil evasion (nuc). Additionally, the icaADBC operon associated with biofilm formation was identified in 58.7% (27/46) of the isolates, being more common in the MSI isolates (39.1%) than in the colonized isolates (19.6%) (P = 0.90). Notably, the genetic element IS256, previously described in studies as a molecular marker that can differentiate commensal from pathogenic isolates11,37, was carried by 41.3% (19/46) of the isolates, exclusively those originating from MSI cases (Fig. 1).
Additionally, the pangenome analysis comparing isolates from carriers and musculoskeletal infections was performed to search for potential new gene markers associated with infection. Roary’s recovery analysis showed greater conservation of gene content among colonization isolates. In contrast, the infection-associated isolates exhibited greater variability in gene content, both in the presence of accessory genes and in the total number of genes, suggesting greater genomic plasticity associated with the infectious environment. Hypothetical gene clusters (group_2760, group_4529, group_3460, group_3389) were identified in at least one colonized isolate, which may be potential markers of colonization, but not with invasion. (Supplementary Figure S1 and S1).
In summary, the genomic traits of MSI isolates showed frequently associated with the CC2 (mainly ST2 and ST23) with no clear significant association with clinical outcomes, while also carried mecA and single-point mutation rpoB conferring methicillin- and rifampicin-resistance, but also a broader range of genes mediating resistance to β-lactams, quinolones, aminoglycosides, MLSb (Macrolides, Lincosamides and Group B Streptogramins), fusidic acid and trimethoprim.
Associations between virulence and resistance genes of S. epidermidis isolates and clinical outcomes in patients with MSI
The 46 genomes of S. epidermidis were analysed for a specific group of genes previously associated with virulence factors and other genetic components that could be characterized as preponderant in their pathogenicity10,38,39. With respect to bacterial resistance genes, isolates carrying the mecA gene were found in patients with MSI and colonized 50% (23/46) and 6.5% (3/46), respectively (P < 0.001), but no significant differences were found between isolates identified in the MSI patients cured (10/31) and those with recurrence (13/31) (P = 0.220). No statistically significant differences were found for the following resistance genes: blaZ, norA, gyrA, gyrB, ant(9)-Ia, AAC(6’)-Ie-APH(2’’)-Ia, aadD, ermA, ermC, msr(A), mph(C), and the following mutations: parE_D432V, rpoB_I527M and rpoB_H481N (Supplementary Table S3).
In contrast, the presence of virulence genes varied between MSI- and colonization-isolates. For example, insertion sequence 256 (IS256), which was exclusively found in MSI isolates, was significantly more prevalent in patients who experienced infection recurrence (38.7% vs. 22.6%, P = 0.038) (Table 1). Additionally, the complete ica operon (composed of icaA, icaD, icaB, and icaC) was more common in MSI isolates than in colonized isolates (39.1% vs. 19.6%, P = 0.900) and in patients with recurrent MSI (32.2% vs. 25.8%, P = 0.347).
When performing logistic regression model with the predictor’s age, biofilm, and IS256 to predict recurrence, biofilm and IS256 were statistically significant. Following this result, the stepwise method was executed. Except for age, all the differences were statistically significant (Supplementary Table S5). The IS256 gene was identified as a significant predictor, with an odds ratio (OR) of 8.002, a 95% confidence interval (CI) between 1.062 and 60.315, and a P value of 0.044 (Table 1). Weak biofilms were protective, reducing the chance of recurrence by 93% (1–0.070). These results suggest that the presence of the IS256 gene and biofilm are significant predictors of the outcome of recurrence. The model demonstrated substantial predictive capability with a Nagelkerke’s R² value of 0.472, indicating that it explains approximately 47.2% of the variability of the outcome analysed, which is a good ability to discriminate between patients with higher or lower probabilities of recurrence. Nagelkerke’s R² values close to 1 suggest a highly explanatory model, especially in logistic models, where values between 0.2 and 0.4 are often considered acceptable for a good fit40. For the predicted probability cut-off point of 0.5, the model showed a sensitivity of 73% and a specificity of 81%, demonstrating a high capacity to correctly identify both positive and negative cases, minimising classification errors. This accuracy is reinforced by the positive (79%) and negative (76%) predictive values, suggesting a high probability of the test providing true results for both positive and negative diagnoses. Additionally, the internal calibration curve (Supplementary Figure S3), along with the GiViTI test (p = 0.4343), supported the internal adequacy of the model, suggesting that its predictions are well-aligned with observed rates (Supplementary Figure S4). The area under the ROC curve (AUC) was 0.842 (95% CI: 0.698 to 0.985), suggesting that the model is effective in correctly distinguishing individuals with and without recurrence (Supplementary Figure S5). These results reinforce that, even in an exploratory context, the model presents consistent and reliable performance. Based on the predicted probabilities, it is possible to identify patients with a higher likelihood of treatment failure.
Discussion
Although S. epidermidis is associated primarily with its commensal nature and is predominantly present on the skin and mucous membranes41, there has been growing scientific interest in investigating its pathogenic characteristics owing to its typical epidemiological association with implant-related MSIs11,29,37. Indeed, in cases of FRI and delayed- and late-onset PJI (3–12 months and > 12 months, respectively), CoNS, particularly S. epidermidis, is the most commonly identified pathogen23,37. In a recently published French cohort of patients with bone and joint infections assessing nearly 12,000 cases of PJI, CoNS represented 25% of the cases11. Moreover, there is limited knowledge concerning the impact of S. epidermidis, previously known as a critical phenotypic and genomic determinant of invasiveness, on the clinical outcome of patients treated with MSIs. By comparing phenotypic and genomic data from a collection of commensal and MSI-associated S. epidermidis isolates, we assessed the importance of adaptive processes likely involved in the transition from a commensal nature to a state of infection. We attempted to provide insights into the molecular basis of MSI caused solely by invasive S. epidermidis, thus excluding MSIs caused by associations with pathogens. Polymicrobial peri-implant tissue, synovial and sonication fluid cultures yielding other skin and mucous-colonizing pathogens, such as S. aureus and gram-negative bacilli, have been described in up to 30% of MSIs2,3,4,5,6,7, which hampers the inclusion of a greater number of patients. Despite the limited number of MSI patients prospectively followed, biofilm formation and methicillin resistance were the key phenotypes, whereas the presence of IS256 was a genomic independent marker of poor outcomes.
In the present study, S. epidermidis isolates were obtained from 31 patients with different MSIs, including patients with FRI, PJI, bacterial septic arthritis, and chronic osteomyelitis. Importantly, all patients were prospectively followed for one year to evaluate their clinical outcomes accurately. FRI cases were the most commonly evaluated (n = 16) due to the high rates of trauma observed in Brazil. In a recent prospective cohort study conducted by our group, the overall FRI rate was 15.9%42. Among surgically removed orthopaedic implants, knee arthroplasty is the most common intervention. According to the findings of Lourtet-Hascoët et al.43, implant-associated complications occurred mainly in the knees (54.2%), followed by the hips (39.1%) and other areas (6.7%). We did not identify any other associations between MSI type, infection location, and clinical outcome in patients with MSI caused by S. epidermidis.
The mechanisms involved in the evolution from the commensal to the pathogenic (invasive) form of S. epidermidis include horizontal gene transfer (HGT), which is associated with pathogenic characteristics such as adhesion to tissues and host implants, biofilm formation, cellular toxicity, and antimicrobial resistance18,44,45,46. However, few studies have focused on the correlation between clinical outcomes after treatment and the molecular epidemiology (clonality) of S. epidermidis isolates causing MSI. In this study, the molecular epidemiology analysis of the 46 S. epidermidis isolates subjected to NGS revealed an expected predominance of the ST2 clade (21.7%) exclusively associated with MSI20. These ST2 isolates, belonging to CC2, which is the most widespread CC worldwide, appear to be the most pathogenic. In the study by Méric and colleagues, 415 S. epidermidis isolates from colonized and MSI samples were evaluated, where the increasing presence of accessory genes in clinical samples suggested the dissemination of genes by HGT between colonizing and invasive isolates47. As expected, there was significant variability in the frequency of sequence types (STs). However, most were grouped in the clonal complex (CC2). The model published in these studies effectively demonstrated how a commensal isolate of S. epidermidis can become invasive. Another molecular epidemiology study of S. epidermidis in PJIs conducted in Sweden revealed that nearly 300 S. epidermidis isolates from PJI and nasal commensals presented distinct genomic characteristics. PJI isolates were exclusively identified as MRSE ST2 strains (43%), ST215 strains (25%), and ST5 strains (9%)20. Additionally, Trobos et al. reported that in 50 S. epidermidis isolates from PJI, genotypic and phenotypic characteristics were positively associated with treatment failure, especially in ST2 isolates (49%)48. In the study by Sánchez et al., S. epidermidis ST2 isolates were predominant in 44% of PJIs, followed by ST640 and ST523. Despite the variability of STs in our study (25 different varieties), ST2, ST23, ST6, ST885, and ST1199 were identified exclusively in MSI, and 37% of the isolates were grouped in CC2 (ST2, ST23, ST5, ST73, and ST89), which is associated with bacterial resistance and strong biofilm production. Sanchez et al. reported that the combination of pathogenic factors may contribute to the adaptation and survival of invasive clones, such as ST2, to specific physiological conditions during infection, thus overcoming the host’s natural and antibiotic-induced immunological protection23.
Recent studies have demonstrated that invasive S. epidermidis isolates expressing antimicrobial-resistant phenotypes to β-lactams or multidrug-resistant and strong/moderate biofilm formers harbour the mecA gene, mobile genetic elements (IS256, SCCmec) and other genes (ica locus) associated with biofilm formation20,23,38,39,47,48,49,50. Frequently, these pathogenic isolates belong to the ST2 or ST23 lineage of global and hospital spread and multidrug resistance, containing mutations in the rpoB gene that confer resistance to rifampicin and glycopeptides such as teicoplanin21. In the present study, isolates resistant to methicillin were significantly associated with MSI and, interestingly, with infection recurrence (univariate analysis). Indeed, the blaZ and mecA genes were carried by 84.8% and 56.5% of the isolates, respectively. Isolates resistant to quinolones and aminoglycosides were also significantly more common in MSIs. Quinolone resistance was associated with a mutation in the parE gene (D432V) found in 13% of the MSI isolates. Similarly, aminoglycoside resistance genes varied from 6.5% (ant(9)-Ia) to 37% (AAC(6’)-Ie-APH(2’’)-Ia), being found almost exclusively in MSI isolates. Additionally, the rpoB gene, which confers resistance to rifampicin, was present in 21.7% of the isolates and was found in only patients with MSI. Rifampicin resistance develops through a single-step mutation in the rpoB gene encoding the β subunit of bacterial DNA-dependent RNA polymerase. We identified specific mutations D471E and I527M in the rpoB gene, the latter associated with the multidrug-resistant global dissemination clone of hospital-origin S. epidermidis (MRSE), previously referred to as the most common cause of rifampicin resistance, which comprises the ST2 and ST23 lineages21. Indeed, five of the six S. epidermidis isolates presenting the I527M mutation in the rpoB gene presented MICs > 128 µg/mL for rifampicin. Rifampicin resistance is concerned owing to the clinical impact of this drug on MSI treatment. Recently, Lazarines et al.49 demonstrated a significant increase in the risk of treatment failure in PJIs caused by rifampicin-resistant S. epidermidis. The findings of this study are regionally essential and unprecedented in Brazil, as they demonstrate that this MRSE lineage is adapted to the local hospital environment and is often associated with a worse clinical prognosis. According to Lee and colleagues, rifampicin resistance in MSI is currently needed, as these two concurrent mutations in rpoB (D471E and I527M) can lead to concomitant heteroresistance to vancomycin and teicoplanin21,49.
S. epidermidis expresses genes that produce numerous surface proteins capable of adhering to multiple substrates and forming biofilms11,12,37,51. These characteristics have been attributed to the primary virulence mechanisms of this species52. Indeed, the well-adapted mechanisms of adhesion and biofilm formation of S. epidermidis contribute to the high frequency of chronic infections associated with implants.
In our results, the presence of IS256 (regulating biofilm formation and genes encoding resistance to aminoglycosides) in the S. epidermidis isolates was a strong independent predictor of MSI with poor outcomes, with an odds ratio of 8.002. Moreover, weak biofilm production in the S. epidermidis isolates was protective, reducing the chance of MSI recurrence by 93% (1–0.070). These results suggest that the IS256 mobile genetic element and weak biofilm formation are significant genomic markers predictors of MSI recurrence caused by S. epidermidis, which may have future implications to the clinical setting. In practical terms, the application of this model can be envisaged in the stratification of patients into risk groups. Based on these predicted probabilities (IS256), it is possible to identify patients with a higher likelihood of treatment failure. These individuals could be candidates for more aggressive therapeutic interventions or more frequent clinical follow-ups, while low-risk patients would be managed less intensively. This personalised approach, when refined by future studies, can optimise resources and improve clinical outcomes. Considering what is currently known, S. epidermidis expresses a striking genetic flexibility by easily repurposing itself depending upon local challenging environmental conditions through continuous generation of novel phenotypic and genotypic variants to switch from a colonizing to an invasive state9,39,50,51. Nosocomial S. epidermidis strains typically harbour multiple copies of the highly active IS256 in their genomes affecting the expression of virulence and drug resistance genes and represents an obvious driving force for the flexibility of the S. epidermidis genome18. It has been speculated by previous studies and confirmed by our results, that the acquisition of readily IS256 mobile genetic element is correlated with increased biofilm formation, while it has been regarded as a positive molecular marker of invasive hospital-origin isolates and associated with unfavourable prognoses after MSI treatment11,12,20,23. In fact, our results provide evidence that IS256 is not only a genetic marker of invasive isolates but also strongly associated with the clinical outcome of infection recurrence, as we identified that this mobile genetic element was carried by 41.3% of the isolates, exclusively those derived from MSI. Meanwhile, several other genetic traits which have not been tested in the present study is likely important to the multifactorial processes of infection adaptation and would also play a role in the outcome of MSI such as enhanced growth in iron-free and nutrient-poor media, reduced production of hemolysins and mutation in the acetate kinase (ackA) and the β-subunit of the RNA polymerase (rpoB) gene50.
The presence of the complete ica operon (icaA, icaB, icaD, icaC) was more common in MSI isolates (39.1%) and in patients with recurrent MSI (32.2% vs. 25.8%); however, the difference was not statistically significant between these groups, which may be associated with the small number of isolates evaluated in the present study. While previous studies have successfully associated the complete ica operon (associated with biofilm formation) harbouring the insertion sequence IS256, with invasive S. epidermidis isolates53 our findings are in agreement with others study’s conclusions, from which ica operon is not a useful diagnostic marker of the invasive capacity of S. epidermidis in MSI10,24,54,55.
The most significant limitation of the present study is the small number of patients evaluated, and isolates tested, especially those isolated from only 31 patients with musculoskeletal infections. However, the study design favoured the inclusion of patients with MSIs caused solely by S. epidermidis isolates, which could demonstrate the isolation and real pathogenic role of this agent. MSIs identified in S. epidermidis are often accompanied by other gram-positive and gram-negative bacteria. Additionally, the prospective observation of patients with MSI for at least one year to reduce the bias of underdiagnosis of recurrence in late-diagnosed infections was also a concern of the authors. On the other hand, this study advanced the search for independent associations of the phenotypic and genotypic characteristics of infected S. epidermidis isolates with those of colonized isolates and evaluated the clinical outcomes of infection recurrence and cure after one year of follow-up. Logistic regression models optimized with the stepwise method were used to test the accuracy of the independent forces of the genomic variables (virulence and resistance genes and IS256) in predicting the clinical outcome. Additionally, the model’s performance was evaluated via Nagelkerke’s R, which resulted in 47.21% confidence in the model. The objective of the predictive analysis of recurrence in this study was exploratory and investigative, acknowledging the limitations imposed by the current sample size. Although our findings are promising, we emphasise that they should be tested in new populations before a safe clinical application can be developed and implemented. Nevertheless, the robust metrics obtained reinforce the potential of the model as a clinical decision support tool, particularly in the management of patients with treatment for S. epidermidis-MSI. In the final model, the variables IS256 and biofilm production were significant independent predictors of the recurrence outcome. Another limitation of the study is the performance of nasal swabs to identify control isolates in healthy individuals, which is not related to patients who evolve to infection. In contrast the ideal approach would be to analyses isolates from the skin/mucosa of patients undergoing orthopaedic surgeries, such as arthroplasty or fracture correction, that do not result in infection. However, to include only isolates from patients who underwent orthopaedic surgery that evolved to infection, collecting samples from thousands of patients before hospitalization for more than ten years would be necessary. A relevant limitation is the decision to evaluate only a single colony of S. epidermidis from each patient and healthy individual, as the infection could be caused by several clones (polyclonal), while expressing significant spatiotemporal diversity and genomic flexibility dependent on environmental selective pressures within the same individual55. Another technical limitation concerns the clonal relatedness analysis of commensal and invasive isolates by applying MLST, which is based on sequence comparisons of seven core genome loci, allowing classifications into global lineages rather than the gold standard to determine clonal relatedness in staphylococci via pulsed-field gel electrophoresis (PFGE), which is more discriminatory than MLST54. Finally, we acknowledge that certain clinical variables, such as the type of antibiotic therapy administered to MSI patients, were not included in the analytical model evaluating independent factors for treatment failure in MSI patients with S. epidermidis. This omission may have influenced the outcomes of our multivariate analysis. Nonetheless, it is worth mentioning that all patients received antibiotic treatments guided by the standard bacterial sensitivity profiles (EUCAST/ BRCAST) and were administered for the appropriate durations.
In conclusion, genomic analysis revealed evidence of the presence of specific phenotypes and genomic traits associated with the pathogenicity and risk of treatment failure in MRSE isolates causing MSI. Commensal and invasive S. epidermidis isolates are not the same. Differences between invasive isolates and those derived from colonization are associated with pathogenic characteristics such as strong/moderate biofilm formation and bacterial resistance to multiple antibiotics, such as β-lactams, aminoglycosides, and quinolones. These traits should be considered important risk factors to guide the microbiological diagnosis and clinical treatment of MSIs. The detection of a pathogenic MRSE lineage causing MSI reinforces recommendations for the implementation of rigorous measures to control the spread of MDR strains in the hospital environment. Future studies should include a greater number of patients with MSI in a prospective design to reinforce the clinical relevance and impact of findings related to genetic variants and virulence factors associated with treatment failure.
Methods
Study population and design
This study is an experimental and observational cohort investigation of patients diagnosed with MSI with implants (FRI or PJI) or without implants (osteomyelitis and septic arthritis) caused by S. epidermidis, which were compared with strains from the skin swabs of healthy individuals. Polymicrobial infections, in which S. epidermidis was diagnosed in conjunction with other pathogens, were excluded. Patients who underwent clinical and surgical treatment between January 2019 and December 2021 were included. Patients with hip and knee PJI underwent two-stage revision surgery, with the first stage consisting of surgical removal of all infected tissues and prosthesis, followed by the insertion of a vancomycin-impregnated spacer for a period varying between 6 and 10 weeks, subsequent reimplantation using a new sterile component, and systemic antibiotic therapy for up to 12 weeks. Patients with FRI underwent a standardized surgical strategy consisting of two stages: thorough debridement, saline irrigation, soft tissue coverage, and implant exchange. After local tissue sampling, systemic antibiotic therapy lasted up to 12 weeks. Bacterial septic arthritis was managed with arthroscopic debridement and irrigation, followed by a six-week course of antibiotics, and chronic osteomyelitis was treated with local debridement and systemic antibiotic therapy for six weeks. The participants were followed for at least one year to assess infection recurrence, which was carried out at a specialized Brazilian orthopaedic trauma centre, where similar surgical and clinical approaches (antibiotic therapy) were employed. All patient outcomes were obtained from electronic medical records collected during hospitalization and outpatient follow-up, ensuring patient anonymity. Informed consent was obtained from all patients. All experiments were performed in accordance with relevant guidelines and regulations. This study was performed in accordance with the Declaration of Helsinki. The project received ethical approval from the Institutional Review Board of Universidade Federal de São Paulo (CEUA/UNIFESP) ethics committee under number 0595/2019.
Diagnostic criteria for osteoarticular infections and MRSE
In accordance with previously established standards, patients fulfilled the criteria for the diagnosis of FRI according to the definition recently published by an international expert group26. For PJI diagnosis, the requirements of the EBJIS (European Bone and Joint Infection Society) were applied27. Confirmatory criteria for S. epidermidis infections require the identification of at least two tissue culture samples and/or synovial fluid containing the same phenotypic profile or one tissue culture combined with the same phenotypic profile in sonication fluid. Aseptic implant loosening is defined as prosthetic failure without these criteria28. Multidrug-resistant Staphylococcus epidermidis is a strain resistance to methicillin and more antimicrobial drugs across three or more antimicrobial categories16,28.
Criteria for treatment failure or recurrence
Patients were considered infection-free if they showed no clinical, laboratory, or radiological signs of bone infection at the last medical visit (1-year follow-up), with no need for reoperation or new cycles of antibiotic therapy for bone infection at the same site, followed by treatment discontinuation. Treatment failure or recurrence was defined as infection at the same surgical site, previously controlled, requiring surgery and/or a second cycle of intravenous antibiotic therapy.
Tissue samples
Bacterial isolates from patient tissue cultures were isolated at the Microbiology Laboratories of the participating hospitals (Hospital São Paulo and Santa Casa de São Paulo) following recommended protocols and sent to the Special Laboratory of Clinical Microbiology (LEMC) at EPM/UNIFESP. During orthopaedic surgeries, four to six bone and peri-implant tissue samples were collected in sterile thioglycolate medium, sealed, labelled, and transported to the microbiological laboratory within 2 h for culturing. To ensure bacterial colony viability, the isolates were recovered from blood agar plates, identified, and stored at -20 °C in cryotubes containing tryptic soy broth (TSB-Oxoid, Basingstoke, England) with 20% glycerol (v/v).
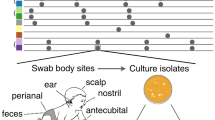
Swab samples from colonized volunteers
Following informed consent, nasal and axillary swabs were collected from healthcare workers’ volunteers, who were nurses and physicians working at the same institution not infected at the time of sampling to recover commensal S. epidermidis isolates. Swabs were cultured on blood agar plates to recover these commensal microorganisms. Species with morphologies similar to those of Staphylococcus were sent for species identification via MALDI-TOF MS on a Microflex LT spectrometer (Bruker Daltonics, MA, USA).
Surgical removal of implant samples
Orthopaedic implants (osteosynthesis or joint prosthesis) surgically removed under suspicion of infection were placed in sterile containers with up to 250 mL of Ringer’s lactate or saline, sealed (autoclaved at 121 °C for 15 min), and stored for up to 6 h in the refrigerator for sonication. The sample container underwent vigorous vortexing for 30 s to homogenize the sample, followed by ultrasonic bath sonication using a BactoSonicTM ® device (Bandelin electronic, Berlin, Germany) at 40 Hz and a power of 0.22 W/cm2 for 5 min, followed by vortexing again for 30 s. The sonicated fluid was directly inoculated into thioglycolate broth and trypticase soy broth (TSB) and streaked (10 µL) via a calibrated loop technique (allowing colony counting) on two blood agar plates. TSB and one blood agar plate were incubated at 35 ± 2 °C under aerobic conditions, whereas the thioglycolate broth and the other blood agar plate were incubated in an anaerobic jar with 5% CO2. If turbidity was observed, the samples were restaked on blood agar. The remaining sonication mixture (15 mL) was stored and frozen at -80 °C. The plates were inspected daily for microbial growth. In accordance with the characteristic morphology of the species, all the plates whose growth was greater than 50 CFU were considered positive29.
Bacterial species identification by MALDI-TOF MS
Following the manufacturer’s recommendations, S. epidermidis was identified via MALDI-TOF MS via a Microflex LT spectrometer (Bruker Daltonics, Massachusetts, USA). Total protein extraction from each isolate was performed according to the manufacturer’s standards (Bruker Daltonics, Billerica, MA, USA). Spectra were obtained in triplicate for each pathogen, and bacterial identification was based on comparing the spectra with those in the MALDI Biotyper 3.3 software database (Bruker Daltonics). According to this software, a score ≥ 2.3 indicates reliable genus and species identification; a score between 2.0 and 2.29 indicates reliable genus identification and probable species identification; and a score between 1.7 and 1.9 indicates probable genus identification. Scores below 1.69 indicate unreliable identification and should be repeated.
Biofilm mass quantification test
Biofilm mass quantification assays were conducted in a polystyrene microplate, as described by Stepanovic et al.30. The mixture was cultivated in TSB and then subcultured in fresh TSB containing 1% glucose to approximately 1 × 106 CFU/ml. A total of 20 µL of the bacterial suspension was incubated in 96-well flat-bottom polystyrene microtiter plates with 180 µL of TBS containing 1% glucose for 24 h at 37 °C. The plates were washed three times with 200 µL of 0.9% saline solution. Then, 200 µL of absolute methanol was added to each well to fix and incubated for 15 min before the next step, which was to aspirate the methanol and stain it with 200 µL of Hucker’s crystal violet solution (2%) for 5 min. After this step, the plates were washed with running water and dried at room temperature. Then, 160 µL of 33% acetic acid was added, and the absorbance was read on an ELISA reader. The optical density (OD) was measured as the absorbance at 550 nm. All the isolates were tested in triplicate in three independent experiments. Each microtiter plate contained negative controls (wells without bacterial inoculation). The average OD value was calculated for each isolate and the negative control. The results were evaluated via the scale described by Stepanovic et al.29, whereby isolates can be categorized into four categories: no biofilm producers, weak biofilm producers, intermediate biofilm producers, and strong biofilm producers. Based on the OD a and the cut-off value (OD c), which is defined as three standard deviations (SDs) above the average OD of the negative control, OD c = average OD of the negative control + (3 × SD of the negative control). The strength of biofilm production for each isolate was calculated as follows: OD a ≤ OD c = no biofilm producer, OD c < OD a ≤ 2 × OD c = weak biofilm producer, 2 × OD c < OD a ≤ 4 × OD c = moderate biofilm producer, and 4 × OD c < OD a = strong biofilm producer. The reference strain S. epidermidis RP12 (ATCC 35983) was used as a control for biofilm production.
Minimum inhibitory concentration (MIC) sensitivity testing
The minimum inhibitory concentrations (MICs) of all 46 selected isolates were determined via broth microdilution based on the criteria and recommendations of the Brazilian Committee on Antibiotic Susceptibility Testing-BrCAST and the European Committee on Antibiotic Susceptibility Testing-EUCAST (BrCAST) (http://brcast.org.br/Accessed in August 2022)/EUCAST (https://www.eucast.org/Accessed in August 2022). The following antimicrobial agents were tested: oxacillin, erythromycin, gentamicin, ciprofloxacin, rifampicin, linezolid and vancomycin. Antimicrobial salts were acquired from Sigma‒Aldrich (Sigma‒Aldrich, St. Louis, USA). Quality control was performed via the standard strains S. aureus ATCC 29,213, Enterococcus faecalis ATCC 29,212, and Escherichia coli 25,922.
Next-generation sequencing (NGS)
S. epidermidis isolates were cultured on TSA plates at 37 °C for 24 h. Individual colony cultures were resuspended in 3 mL of TSB medium and incubated at 37 °C with shaking overnight. The genomes of all the isolates were sequenced. Total DNA from the isolates was extracted via a total DNA kit (Zymo®) and quantified via a Qubit 3.0 fluorometer (Thermo Fisher Scientific®, Waltham, MA, USA). Species determination and phylogenetic relationships (PubMLST), as well as resistome characterization (ResFinder), characterization of mobile genetic elements (MobileElementFinder) (https://www.genomicepidemiology.org/), and virulome (VFDB) (http://www.mgc.ac.cn/VFs/main.htm) were performed using sequences obtained through whole-genome sequencing (Ion Torrent Thermo Fisher®). Genome processing was performed on the BV-BRC platform (https://www.bvbrc.org/) with Ridom SeqSphere + v9.0 software (Münster, Germany). The reads were submitted to the “Comprehensive Genome Analysis” pipeline, which performs assembly, annotation, and phylogenetic analysis via Unicycler v0.4.9, RASTk, and RAxML software, respectively. A pangenome analysis comparing isolates from carriers and musculoskeletal infections was performed using Roary v3.13.0, and statistical association with clinical outcomes was evaluated using Scoary v1.6.16. The whole-genome sequences were deposited in the GenBank database under the accession numbers shown in supplementary Table S1.
Bioinformatics analyses were conducted using the Bactopia pipeline (v3.2.0) followed by TORMES pipeline (v1.3.0), specifically designed for bacterial genome analysis, to perform quality filtering, genome assembly, and annotation (using SPAdes and Prokka). Species determination and phylogenetic relationships were verified via PubMLST, while antimicrobial resistance genes, virulence factors, and mobile genetic elements (MGEs) were identified using ResFinder, VFDB, and MobileElementFinder, respectively. Additional comprehensive genome analysis was performed on the BV-BRC platform, incorporating assembly validation (Unicycler v0.4.9), annotation (RASTk), and phylogenetic reconstruction (RAxML).
The phylogenetic tree was generated using the Bacterial Genome Tree service of the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) [https://www.bv-brc.org/], employing the Codon Tree method with default parameters. This method selects single-copy PGFams shared among the input genomes, aligns their proteins with MAFFT, and infers the phylogeny using RAxML. A total of 51 Staphylococcus epidermidis genomes were analyzed alongside two references: S. epidermidis ATCC 12,228 and S. aureus RN4220 (used as an outgroup). From 1000 target genes, 931 single-copy genes were recovered and included in the final tree, which was exported in SVG and Newick formats.
Statistical analysis
The characteristics of 31 subjects with MSI and 15 healthy individuals are summarized as frequencies and percentages or means (ranges) and standard deviations (SDs). The associations between continuous and categorical variables (bacterial phenotypes, clades, and the presence or absence of genes) in healthy colonized individuals and MSI patients were statistically analysed via Pearson’s chi-square tests or Fisher’s exact tests and Mann‒Whitney tests when appropriate. The chi-square test was used to test the null hypothesis (strains are homologous in clinical outcome or resistance phenotype). For statistical analysis, follow-up was defined as the interval between the date of the first medical consultation and the date of remission or treatment failure of MSI, considering at least one year of follow-up. A difference was considered statistically significant for P values less than 0.05. Logistic regression models were used to test the accuracy of the independent forces of the genomic variables (virulence and resistance genes and IS256) in predicting the clinical outcome. The dependent variable is the binary outcome (0: cure; 1: recurrence). This model was optimized via the stepwise method for logistic regression. To select the predictor variables for the logistic regression model, only variables with P < 0.100 in the bivariate tests were selected. All analyses were conducted via version 4.4.1 of the R software (R Core Team, 2024). The figure of the models was created with the ggplot2 package31. Logistic regression was performed with the glm() function from the Stats package32, with the assistance of the car33, Easystats34, Jtools35, and MASS36 packages.
Data availability
The reads used for the assembly of the 49 S. epidermidis genomes that support the findings of this study have been deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) under the primary accession code PRJNA938510, and the whole-genome sequences were deposited in the GenBank database under the accession numbers shown in supplementary Table 1. The versions described in this paper are the first versions.
References
Moriarty, T. F. et al. Fracture-related infection. Nat. Rev. Dis. Primers. 8, 67. https://doi.org/10.1038/s41572-022-00396-0 (2022).
Depypere, M. et al. Pathogenesis and management of fracture-related infection. Clin. Microbiol. Infect. 26, 572–578. https://doi.org/10.1016/J.CMI.2019.08.006 (2020).
Izakovicova, P., Borens, O. & Trampuz, A. Periprosthetic joint infection: current concepts and outlook. EFORT Open. Rev. 4, 482–494. https://doi.org/10.1302/2058-5241.4.180092 (2019).
Triffault-Fillit, C. et al. Microbiologic epidemiology depending on time to occurrence of prosthetic joint infection: a prospective cohort study. Clin. Microbiol. Infect. 25, 353. https://doi.org/10.1016/j.cmi.2018.04.035 (2019).
Morgenstern, M. et al. Staphylococcal orthopaedic device-related infections in older patients. Injury 47, 1427–1434. https://doi.org/10.1016/j.injury.2016.04.027 (2016).
Benito, N. et al. The different microbial etiology of prosthetic joint infections according to route of acquisition and time after prosthesis implantation, including the role of multidrug-resistant organisms. J. Clin. Med. 8, 573. https://doi.org/10.3390/jcm8050673 (2019).
Li, C., Renz, N. & Trampuz, A. Management of periprosthetic joint infection. Hip Pelvis. 30, 138–146. https://doi.org/10.5371/hp.2018.30.3.138 (2018).
Brescó, M. S. et al. Pathogenic mechanisms and host interactions in Staphylococcus epidermidis device-related infection. Front. Microbiol. 8, 1401. https://doi.org/10.3389/fmicb.2017.01401 (2017).
Becker, K., Both, A., Weißelberg, S., Heilmann, C. & Rohde, H. Emergence of coagulase-negative Staphylococci. Expert Rev. Anti-Infect Ther. 18, 349–366. https://doi.org/10.1080/14787210.2020.1730813 (2020).
Arciola, C. R. et al. Antibiotic resistance in exopolysaccharide-forming Staphylococcus epidermidis clinical isolates from orthopaedic implant infections. Biomaterials 26, 6530–6535. https://doi.org/10.1016/j.biomaterials.2005.04.031 (2005).
Post, V. et al. Comparative genomics study of Staphylococcus epidermidis isolates from orthopedic-device-related infections correlated with patient outcome. J. Clin. Microbiol. 55, 3089–3103. https://doi.org/10.1128/JCM.00881-17 (2017).
Montanaro, L. et al. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 6, 1329–1349. https://doi.org/10.2217/fmb.11.117 (2011).
Wildeman, P. et al. Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci. Rep. 10, 62751. https://doi.org/10.1038/S41598-020-62751-Z (2020).
Wolska-Gębarzewska, M., Międzobrodzki, J. & Kosecka-Strojek, M. Current types of Staphylococcal cassette chromosome mec (SCCmec) in clinically relevant coagulase-negative Staphylococcal (CoNS) species. Crit. Rev. Microbiol. https://doi.org/10.1080/1040841X.2023.2274841 (2023).
Widerström, M., Wiström, J., Sjöstedt, A. & Monsen, T. Coagulase-negative staphylococci: update on the molecular epidemiology and clinical presentation, with a focus on Staphylococcus epidermidis and Staphylococcus saprophyticus. Eur. J. Clin. Microbiol. Infect. Dis. 31, 7–20. https://doi.org/10.1007/s10096-011-1270-6 (2012).
Siciliano, V., Passerotto, R. A., Chiuchiarelli, M., Leanza, G. M. & Ojetti, V. Difficult-to-treat pathogens: A review on the management of multidrug-resistant Staphylococcus epidermidis. Life 13, 1126. https://doi.org/10.3390/LIFE13051126 (2023).
Beck, C., Krusche, J., Elsherbini, A. M. A., Du, X. & Peschel, A. Phage susceptibility determinants of the opportunistic pathogen Staphylococcus epidermidis. Curr. Opin. Microbiol. 78, 102434. https://doi.org/10.1016/J.MIB.2024.102434 (2024).
Schoenfelder, S. M. K. et al. Success through diversity – how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 300, 380–386. https://doi.org/10.1016/j.ijmm.2010.04.011 (2010).
Arora, S., Uhlemann, A. C., Lowy, F. D. & Hook, M. A novel MSCRAMM subfamily in coagulase-negative Staphylococcal species. Front. Microbiol. 7, 540. https://doi.org/10.3389/fmicb.2016.00540 (2016).
Månsson, E., Johannesen, B., Nilsdotter-Augustinsson, T., Söderquist, Å., Stegger, M. & B. & Comparative genomics of Staphylococcus epidermidis from prosthetic-joint infections and Nares highlights genetic traits associated with antimicrobial resistance, not virulence. Microb. Genom. 7, 000504. https://doi.org/10.1099/mgen.0.000504 (2021).
Lee, J. Y. H. et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 3, 1175–1185. https://doi.org/10.1038/s41564-018-0230-7 (2018).
Otto, M. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol. 34, 201–214. https://doi.org/10.1007/S00281-011-0296-2 (2012).
Sánchez, A. et al. Pathogenesis of Staphylococcus epidermidis in prosthetic joint infections: can identification of virulence genes differentiate between infecting and commensal strains? J. Hosp. Infect. 105, 561–568. https://doi.org/10.1016/j.jhin.2020.04.026 (2020).
Arciola, C. R., Campoccia, D. & Montanaro, L. Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409. https://doi.org/10.1038/s41579-018-0019-y (2018).
Centers for Disease Control and Prevention. National Center for Health Statistics, United States Census Bureau (USCB). United States National Hospital Discharge Survey. (2009).
Metsemakers, W. J. et al. Fracture-related infection: A consensus on definition from an international expert group. Injury 49, 505–510 (2018).
Mcnally, M. et al. Infographic: The EBJIS definition of periprosthetic joint infection. Bone Joint J. 103-B(1), 16–17. https://doi.org/10.1302/0301-620X.103B1.BJJ-2020-2417 (2021).
Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x (2012).
Yano, M. H. et al. Improved diagnosis of infection associated with osteosynthesis by use of sonication of fracture fixation implants. J. Clin. Microbiol. 52, 4176–4182 (2014).
Stepanovic, S., Vukovic, D., Dakic, I. & Savic, B. Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of Staphylococcal biofilm formation. J. Microbiol. Methods. 40, 175–179 (2000).
Wickham, H. ggplot2 (Springer, 2016).
R Core Team. A Language and environment for statistical computing. Computing 1, (2006).
Fox, J. & Weisberg, S. An R Companion To Applied Regression (SAGE, 2011).
Lüdecke, D., Ben-Shachar, M. S., Patil, I. & Wiernik, B. M. & Makowski, D. easystats: framework for easy statistical modeling, visualization, and reporting. CRAN Reposit. (2022).
Long, J. A. Jtools: analysis and presentation of social scientific data. R Package Version. 2, 2 (2022).
Ripley, B. et al. Package ‘MASS’. CL R. 538, 113–120 (2013).
Tande, A. J. & Patel, R. Prosthetic joint infection. Clin. Microbiol. Rev. 27, 302–345 (2014).
Salgueiro, V. C., Iorio, N. L. P., Ferreira, M. C., Chamon, R. C. & Santos, K. R. N. D. Methicillin resistance and virulence genes in invasive and nasal Staphylococcus epidermidis isolates from neonates. BMC Microbiol. 17, 1 (2017).
Cherifi, S. et al. Genetic characteristics and antimicrobial resistance of Staphylococcus epidermidis isolates from patients with catheter-related bloodstream infections and from colonized healthcare workers in a Belgian hospital. Ann. Clin. Microbiol. Antimicrob. 13, 20 (2014).
Nagelkerke, N. J. D. A note on a general definition of the coefficient of determination. Biometrika 78, 691–692 (1991).
Becker, K., Heilmann, C. & Peters, G. Coagulase-negative Staphylococci. Clin. Microbiol. Rev. 27, 870–926 (2014).
Prebianchi, S. et al. Type of antibiotic but not the duration of prophylaxis correlates with rates of fracture-related infection. Eur. J. Orthop. Surg. Traumatol. 33, 987–992 (2023).
Lourtet-Hascoët, J. et al. Species and antimicrobial susceptibility testing of coagulase-negative Staphylococci in periprosthetic joint infections. Epidemiol. Infect. 146, 1771–1776 (2018).
Rohde, H. et al. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 42, 5614–5619 (2004).
Rolo, J., de Lencastre, H. & Miragaia, M. Strategies of adaptation of Staphylococcus epidermidis to hospital and community: amplification and diversification of SCCmec. J. Antimicrob. Chemother. 67, 1333–1341 (2012).
Rohde, H. et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720 (2007).
Méric, G. et al. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat. Commun. 9, 5034 (2018).
Trobos, M. et al. Genomics of Staphylococcus aureus and Staphylococcus epidermidis from periprosthetic joint infections and correlation to clinical outcome. Microbiol. Spectr. 10, e02181–e02121 (2022).
Lazarinis, S., Hailer, N. P., Järhult, J. D. & Brüggemann, A. Incidence of rifampicin resistance in periprosthetic joint infection: a single-centre cohort study on 238 patients. Antibiotics 12, 1499 (2023).
Both, A. et al. Distinct clonal lineages and within-host diversification shape invasive Staphylococcus epidermidis populations. PLoS Pathog. 17, e1009304 (2021).
Otto, M. Staphylococcus epidermidis - the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567 (2009).
Heilmann, C. et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091 (1996).
Kozitskaya, S. et al. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43, 4751–4757 (2005).
Koskela, A., Nilsdotter-Augustinsson, A., Persson, L. & Söderquist, B. Prevalence of the Ica Operon and insertion sequence IS256 among Staphylococcus epidermidis prosthetic joint infection isolates. Eur. J. Clin. Microbiol. Infect. Dis. 28, 655–660 (2009).
Zhou, W. et al. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell 180, 454–470e18 (2020).
Acknowledgements
We thank the students of the LEMC and ALERTA laboratories and the ORTOINFECTO group (UNIFESP) for their support in conducting this work. We thank the funding agency FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) for the research funding (Grant FAPESP 2019/20959-7). We are grateful to the Coordination for the Improvement of Higher Education Personnel (CAPES) for the doctoral scholarship awarded to INMS (grant number: 88887.817948/2023-00).
Funding
This study received funding from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) under the number 2019/20959-7. In addition, the Brazilian agency Coordination for the Improvement of Higher Education Personnel (CAPES) provided doctoral scholarship to INMS (grant number: 88887.817948/2023-00). These Governmental Agencies were not involved in the conceptualization, design, data collection, analysis, decision to publish, or preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
I.N.M.S. and M.J.S. conceived the experiment(s); I.N.M.S., F.A.-L., M.F.C.B., and M.N.L.K. conducted the experiment(s); I.N.M.S., F.A.-L., and M.J.S. analyzed the results; C.C.C., L.S.S., and T.S.D., F.B.d.R., A.K.A.E., G.S.C., J.d.A., E.L.D., performed the whole-genome sequencing; I.N.M.S., F.A.-L., G.B.K., and M.J.S. contributed to the interpretation of data and provided critical feedback. All authors reviewed the manuscript. I.N.M.S. and F.A.-L. contributed equally to this work as first co-authors. The order of their names was agreed upon between both authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Santos, I.N.M., Alberto-Lei, F., Santos, F.F. et al. Comparative genomic analysis of resistance and virulence genes in staphylococcus epidermidis and their impact on clinical outcome of musculoskeletal infections. Sci Rep 15, 23506 (2025). https://doi.org/10.1038/s41598-025-09061-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09061-4