Abstract
Nepal’s unique geography and diverse biodiversity present a significant opportunity to discover new actinomycetes species that may lead to developing novel compounds with pharmaceutical applications. Fifty-nine actinomycetes strains were isolated from fifty-six soil samples collected from different altitudes and locations in Nepal. These actinomycetes strains were classified through cultural, morphological, partial 16 S rRNA gene sequence analysis, and phylogenetic analysis and were identified as belonging to the genera Streptomyces, Kitasatospora, Nocardia, Nocardiopsis, Nocardioides, Arthrobacter, Amycolatopsis, Promicromonospora, and Rhodococcus. The genus Streptomyces appeared to be the most abundant, comprising 44 isolates. Promicromonospora was isolated for the first time from high-altitude soils. Of the 59 isolated actinomycetes, 29 strains exhibited antimicrobial activity against the tested microorganisms. The highest activity was observed against fungi and Gram-negative bacteria, followed by Gram-positive bacteria. The crude extracts of Streptomyces olivaceus (13081) and Nocardia thailandica (13105) showed broad-spectrum antimicrobial activity against the tested bacteria and fungi. Ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) analysis of these two extracts revealed several antimicrobial compounds such as Neoaspergillic acid, Okaramine D, Antimycin A3, Tricycloalternarene A, Nocardimicin B, 4-O-methylmelleolide, Flocculosin, and Microcolin A. The study revealed the culture-based diversity of actinomycetes in Nepal and provided insights into their potential to produce bioactive compounds.
Similar content being viewed by others
Introduction
Actinomycetes are Gram-positive, aerobic, and filamentous bacteria with high G + C content1,2,3. Although they are typically found in soil and aquatic environments, they could also be found in other substrates, including plants and animals4. This group of bacteria is recognized as an established source of many available drugs, including antibiotics and antitumor compounds5,6.
Actinomycetes account for more than 45.0% of bioactive compounds obtained from microbes7. Streptomyces, the largest genus within actinomycetes, produces the majority of secondary metabolites8 accounting for about 75.0% of the currently available compounds9. In addition to Streptomyces, other actinomycetes such as Micromonospora, Kitasatospora, Nocardia, Nocardiopsis, and others also contribute to the production of secondary metabolites10. Some of the antibacterial and antifungal antibiotics Nocardia produces are Nocardicin A and B, Brasilinolide A and B, Erythromycin E, Tubelactomicin A, and Nargenicin11. Similarly, Streptomyces are also well known for producing antimicrobials like Chloramphenicol, Erythromycin, Streptomycin, Neomycin, Nystatin, and Tetracycline12.
Currently, the world is facing an increased prevalence of antibiotic resistance, which has become a serious problem for both humans and animals13. The major causes of antibiotic resistance include overuse of antibiotics, incorrect antibiotic prescriptions, extensive antibiotic use in agriculture and veterinary sectors, and limited discovery and availability of new antibiotics14. The unavailability of new classes of antibiotics that are effective against multidrug-resistant bacteria15 has limited the scope of antibiotic therapy against infections16. To address this problem, there is an immense need for the discovery and production of new and efficient antimicrobials with novel mechanisms of action17.
Nepal has a unique geographical niche8 and climatic diversity18 experiencing altitude variations from 70 to 8848 m.a.s.l. However, limited data on actinomycetes’ diversity and antimicrobial potential are available. In this study, we isolated and identified actinomycetes from soil samples from high altitudes in Nepal and investigated their antibacterial and antifungal activities. Our research provides a comprehensive culture-based diversity of actinomycetes with future opportunities to discover new antimicrobials to combat antibiotic resistance in favor of public health.
Materials and methods
Soil sample collection
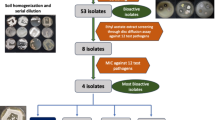
Fifty-six soil samples collected in 2018 and 2019 from various locations in Nepal, ranging in altitudes from 957 to 5550 m above sea level (m.a.s.l), were processed for use in this study (Fig. 1). GPS data, collection dates, sampling locations, and altitudes for all 56 soil samples were recorded during field collection (Table S1). The GPS data were generated using the Quantum Geographic Information System (QGIS), and the GIS map used to pinpoint the sampling locations was obtained from the Survey Department, Government of Nepal.
The soil samples collected 5–10 cm below the ground surface were placed in sterile plastic bags and containers using clean spatulas. Subsequently, they were barcoded in the laboratory, stored at room temperature, and air-dried before isolating the actinomycetes.
Isolation and morphological characterization of actinomycetes
To isolate actinomycetes, 1 gm of soil was suspended in 10 mL sterile Milli-Q water in duplicates. Two separate 10− 1 dilutions were prepared and plated for each soil sample on two Actinomycetes Isolation Agar (AIA) plates. Aliquots of 0.1 mL from the 10− 1 of soil were spread-plated on actinomycete isolation agar (AIA, g/L: L-asparagine 0.1, dipotassium phosphate 0.5, ferrous sulfate 0.001, magnesium sulfate 0.1, sodium caseinate 2, sodium propionate 4, Bacto Agar 15)19. The agar was supplemented with 50 µg/mL each of cycloheximide and nalidixic acid. The inoculated AIA plates were air-dried for 5 min to evaporate excess fluid before incubating at 30 °C for 7–10 days. After incubation, colonies exhibiting similar morphological characteristics across both plates were considered duplicates, and unique colonies were selected for further isolation and characterization. Colonies exhibiting a dry, chalky, and powdery appearance on AIA were picked using a sterile needle. The strains were purified through standard microbiological agar streaking to obtain single colonies. The axenic cultures were morphologically characterized based on their growth on the AIA plate, including recording the color of the colony and its diffusible pigments20.
Phylogenetic analysis
The genomic DNA was isolated using a QIAGEN Gentra Puregene Yeast-Bacterial kit, following the Gram-positive protocol. The DNA concentration was measured with NanoDrop (Thermo Scientific) and adjusted to 10 ng/µL using sterile Milli-Q water. The amplification of the 16 S rRNA gene was performed using the universal primers 27 F and 1492R21. Following the manufacturer’s protocol, the PCR product was purified using a NucleoSpin PCR Clean-up kit (Macherey-Nagel). The DNA concentration was adjusted to 40 ng/µL and then sequenced using the same set of primers. The obtained 16 S rRNA gene sequences were de novo assembled before a nucleotide BLAST search to determine the closest related type species in NCBI.
The 16 S rRNA gene sequences of 44 Streptomyces and 15 non-Streptomyces isolates and those of their closest type species were used in the phylogenetic analysis. In MEGA22 software version 12, the sequences were aligned using the Clustal W algorithm23 and the phylogenetic trees were generated using the Maximum Likelihood method and Tamura-Nei (1993) model24 of nucleotide substitutions. The evolutionary history was also inferred using the Neighbor-Joining method25 and the Minimum Evolution method26 to provide independent distance-based assessments of topology, cross-validate clade stability, and mitigate algorithm-specific biases such as model mis-specification and long-branch attraction. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches27. The sequences of Cellulomonas massiliensis JC225T and Myxococcus xanthus ATCC 25232T were used as outgroups to root the tree in the Streptomyces and non-Streptomyces phylogenetic tree, respectively.
Secondary metabolite screening and antimicrobial testing
The screening for secondary metabolites was performed by cultivating the actinomycetes isolates in 200 mL ISP-2 broth supplemented with 2.0% (v/v) XAD-16 (Sigma) and inoculated with 2.0% (v/v) seed ISP-2 culture. Fermentation was carried out at 180 r.p.m. using rotary shaking at 30 °C for 10 days under dark conditions28. The cultures were harvested by centrifugation at 4000 r.p.m. for 10 to 15 min. The cell pellet and the resins were extracted with 80 mL acetone and concentrated in a vacuum using a rotary evaporator. The dried extract was dissolved and suspended in 2.2 mL methanol and stored in a -80 °C freezer until further use29.
The crude extracts were tested in sterile 96-well plates (Sarstedt) against the Gram-negative bacteria Escherichia coli BW25113 and E. coli JW0451-2, the Gram-positive bacterium Bacillus subtilis DSM 10T, and the fungi Wickerhamomyces anomalus DSM 6766T and Mucor hiemalis DSM 2656T. The bacterial strains were grown overnight in Mueller Hinton (Sigma), while test fungal strains were cultivated in MYC broth (w/v: 1.0% Difco Phytone peptone, 1.0% glucose, 50 mM HEPES, pH 7.0) for 24 h. Test bacterial and fungal cultures were to a concentration of OD600 0.01 and 0.05, respectively. The 96-well plates were filled with 150 µL of the respective medium and 150 µL of the test strain. The 20 µL test crude extract was added, establishing this as the 100% concentration (Well A), along with the other test extracts in different wells and the negative control (methanol). A 150 µL volume was serially diluted to the subsequent well (Well B) by mixing using a multi-channel pipette, making it a 50% (v/v) concentration. This process was repeated sequentially across the plate, halving the concentration at each step, until the eighth well (Well H), representing an 8-fold dilution (0.78%, v/v). The last 150 µL in the last row was discarded, and the plate was sealed with parafilm before being incubated overnight at 30 °C in a shaker incubator16.
Antimicrobial compounds from actinomycetes
The crude extract of Streptomyces olivaceus (13081) and Nocardia thailandica (13105), which showed broad-spectrum antimicrobial activity, was diluted 1:10 with methanol and centrifuged at 21,500x g for 5 min at 4 °C to eliminate any remaining cell debris and other solid fragments. The samples were analyzed using Ultra-performance liquid chromatography-Tandem mass spectrometry (UPLC-MS/MS). Measurements were performed using a Dionex Ultimate 3000 RSLC (Rapid Resolution Liquid Chromatography system) system (Thermo) with a BEH C18, 100 × 2.1 mm, 1.7 μm column (Waters GmbH, Germany) following the procedure described previously30. All Liquid Chromatography-Mass Spectrometry (LC-MS) data from samples were processed using the T-ReX in MetaboScape (Bruker Daltonics GmbH & Co. KG) with a minimum intensity threshold of 5000 counts and a minimum peak length of 4 spectra. After processing, features were annotated using the Target List utility in MetaboScape, drawing on data from various libraries16. DataAnalysis software was used to visualize the chromatogram peaks.
Results
Culture-based diversity of actinomycetes
Based on cultural, morphological, and 16 S rRNA gene sequencing, 59 actinomycetes strains isolated from 56 soil samples of Nepal belong to 9 genera and 45 species. The dominance of actinomycetes genera was obtained as Streptomyces > Kitasatospora > Nocardia, Nocardiopsis > Nocardioides, Arthrobacter, Amycolatopsis, Promicromonospora, Rhodococcus (Fig. 2).
The 16 S rRNA gene sequences compared on the NCBI database revealed that the highest number of isolates belonged to Streptomyces (n = 44), comprising 31 species, followed by Kitasatospora (n = 5) with 5 different species, Nocardia (n = 3) with 3 different species, and Nocardiopsis (n = 2) with single species. In addition, each single species of Nocardioides, Arthrobacter, Amycolatopsis, Promicromonospora, and Rhodococcus were also identified. Notably, we also isolated and reported the Promicromonospora for the first time from a high-altitude soil sample (2753 m.a.s.l) of Nepal. The strains showed 98.7–100% similarity with the closest known type strains when compared on the NCBI database. All 59 strains have been submitted to NCBI GenBank under the accession numbers PQ104910 to PQ104968 (Table S2).
The culture on AIA agar showed variation in colony color and diffusible pigments (Fig. 3) by different isolates. Most strains exhibited cream-colored colonies, except those identified as Rhodococcus and some Nocardia strains. Red and yellowish diffusing pigments were observed in Nocardia sp. strain 13097 and Streptomyces sp. strain 12932, respectively.
Cultural characteristics of isolated actinomycetes on AIA agar plates showing the variations in morphology and pigments. (A) Amycolatopsis sp. 13008, (B) Arthrobacter sp. 12926, (C) Kitasatospora sp. 13087, (D) Nocardia sp. 12990, (E) Nocardia sp. 13097, (F) Nocardioides sp. 13093, (G) Nocardiopsis sp. 12995, (H) Promicromonospora sp. 12994, (I) Rodococcus sp. 13096, (J) Streptomyces sp. 12929, (K) Streptomyces sp. 12932, (L) Streptomyces sp. 13081.
Figure 4 presents the number of strains isolated from soil samples collected at different altitude ranges in this study. The highest number of actinomycetes was obtained from altitudes > 1000 to 2000 m.a.s.l. However, we could not establish a clear correlation between the actinomycetes diversity and the altitude range due to the unequal numbers of samples collected at different altitude levels. In this study, more samples were collected and processed at altitudes > 1000 to 2000 m.a.s.l, suggesting more isolates.
Evolutionary analysis of the isolated strains
Phylogenetic analysis, based on the 16 S rRNA gene sequence, revealed the positions of the 44 Streptomyces strains, representing the most diverse group comprising 31 species among the isolated actinomycetes in Nepal (Fig. 5). These isolates clustered with different species of Streptomyces. Strains 12935, 13101, 12928, and 13086 constituted the highest number of isolates clustered to a species of Streptomyces (S. lactacystinicus). Among these, strains 12996 and 13097 appear interesting as they showed 98.8% similarity with Streptomyces mangrovi HA11110T and 98.7% similarity with Nocardia pseudovaccinii DSM 43406T, respectively, which might potentially represent a new species of actinomycetes. Further analysis, such as whole genome sequencing, dDDH value, ANI value, phenotypic characterization, and other biochemical tests, is needed to support their unique novelty in describing a new species in the future.
Maximum likelihood phylogenetic tree based on 16 S rRNA gene sequences showing the position of 44 Nepalese Streptomyces strains (in bold) and closely related Streptomyces species with validly published names from the NCBI database. Cellulomonas massiliensis strain JC225T was used as an outgroup. The numbers on the branches indicate the percentage bootstrap value of 1000 replicates. Bar, 0.01 nucleotide substitutions per site.
Based on a BLAST search of the 16 S rRNA gene sequence, strain 13097 showed 98.9% similarity with Nocardia pseudovaccinii DSM 43406T. Phylogenetic analysis of strain 13097 and its closely related Nocardia type species revealed a novel branch in the phylogenetic tree. Figure 6 shows a maximum likelihood (ML) phylogenetic tree based on 16 S rRNA gene sequences with the position of 15 actinomycetes isolates other than Streptomyces strains and closely related type species with validly published names from the NCBI database. The phylogenetic tree was also constructed using the neighbor-joining (NJ) method (Figs. S2, S3) and the minimum evolution (ME) method (Figs. S4, S5). Both of these distance methods (NJ and ME) recovered the same major groupings as ML. For instance, in the Streptomyces trees, all three trees consistently placed isolates 12924/12991/13092 with the S. anulatus and S. fulvissimus lineage and isolates 13082/13003 with S. rhizosphaerihabitans, each supported by bootstrap values > 70%. Similarly, in the non-Streptomyces trees, isolates 12998/125 consistently grouped with Nocardiopsis dassonvillei, and isolate 13097 was placed within the Nocardia vinacea and N. pseudovaccinii clade with strong support, indicating that the taxonomic placements are stable across all ML, NJ, and ME phylogenetic trees. No conflicting placements with strong bootstrap support were observed, suggesting that neither long-branch attraction (addressed by ME) nor model mis-specification (addressed by NJ) biases the inferred relationships. Presenting different trees from distance-based and model-based frameworks meets current best-practice methods for robust microbial taxonomy and highlights the reliability of our phylogenetic inferences.
Maximum likelihood phylogenetic tree based on 16 S rRNA gene sequences showing the position of 15 Nepalese Non-Streptomyces strains (in bold) and closely related species with validly published names from the NCBI database. Myxococcus xanthus ATCC 25232T was used as an outgroup. The numbers on the branches indicate the percentage bootstrap value of 1000 replicates. Bar, 0.05 nucleotide substitutions per site.
Antimicrobial activity of the actinomycetes extracts
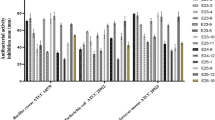
Among 59 isolates, 29 (49.2%) showed a range of antimicrobial activities against tested organisms, indicating the varied antimicrobial potential of actinomycetes strains isolated from the high-altitude soils of Nepal (Table S3). Out of these 29 isolates, 6 exhibited antimicrobial activity against E. coli BW25113, 10 against E. coli JW0451-2, 2 against B. subtilis DSM 10T, 23 against W. anomalus DSM 6766T, and 14 against M. hiemalis DSM 2656T (Fig. 7, Fig. S1). The predominant bioactivity in the crude extracts of Nepalese actinomycetes appears to be antifungal, followed by activity against Gram-negative bacteria. Among the actinomycetes isolates from which secondary metabolites were extracted, Streptomyces olivaceus (13081), and Nocardia thailandica (13105) appeared to be the most potent strains due to their broad range of activity against multiple tested microorganisms. Streptomyces olivaceus 13081 inhibited the growth of both fungal test organisms up to well H (0.78% concentration), inhibited E. coli up to well B (50% concentration) and C (25% concentration), and B. subtilis up to well B (50% concentration). Similarly, Nocardia thailandica (13105) inhibited the growth of fungal test organisms up to well B (50% concentration), inhibited E. coli up to well C (25% concentration) and D (12.50% concentration), and B. subtilis up to well G (0.78% concentration). This reflects the presence of highly potent antifungal and antibacterial compounds capable of maintaining activity even at lower concentrations. Our study presents qualitative screening, which is recognized as a valuable initial step in searching for bioactive compounds and could guide subsequent analysis31.
Compounds annotated from metaboscape
The raw mass data was analyzed using MetaboScape software, where each peak was analyzed, detected, aligned, and annotated. Evaluation of UPLC-MS/MS data of the two active extracts Streptomyces olivaceus (13081) and Nocardia thailandica (13105), revealed several known compounds and compound classes that can support the antimicrobial activity observed from the two active extracts. Antibacterial compounds such as Neoaspergillic acid and Okaramine D, and antifungal compounds such as Antimycin A3, Tricycloalternarene A were annotated and identified from the extracts of Streptomyces olivaceus (13081) (Fig. 8A). Similarly, antibacterial compounds such as Nocardimicin B, and 4-O-methylmelleolide, and antifungal compounds such as Flocculosin and Microcolin A, were annotated and identified from the extracts of Nocardia thailandica (13105) (Fig. 8B). Their identification supports the broad-spectrum antimicrobial activity observed in the initial bioassays and highlights the metabolic richness of the strain.
However, a significant portion of the detected features, specifically, 143 chromatographic peaks from the extracts of Streptomyces olivaceus (13081) and 244 chromatographic peaks from Nocardia thailandica (13105), remained unannotated when cross-referenced against the MetaboScape spectral library. This lack of annotation suggests the presence of previously uncharacterized or novel compounds within these extracts. A list of all the compounds annotated and identified from these two actinomycetes’ crude extracts, along with their m/z value, retention time, and formula, is given in Tables S4 and S5.
Discussion
Nepal has multiple ecological regions with increased elevations from south to north within approximately 200 km, which could be hidden treasures of microbial biodiversity. The full potential of useful actinomycetes in this region is underexplored, although they could be a source of new natural products. This study demonstrates the culture-based diversity of soil actinomycetes collected at different altitudes and locations in Nepal. We obtained 45 species belonging to 9 genera of actinomycetes, which indicates intra-generic and intra-species diversity of soil actinomycetes from high altitudes. However, a culture-independent metagenomic approach could explain the diversity in depth. In this study, Streptomyces appeared to be the most dominant and diverse genus, comprising 31 species. The previous studies corroborating these findings showed Streptomyces as a dominant genus, comprising 60–70.7% of the total isolates4,18. In India, a country with nearly a similar topography as Nepal, Streptomyces has also been isolated from soils32. Besides Streptomyces, different species of Kitasatospora, Nocardia, Nocardiopsis, Nocardioides, Arthrobacter, Amycolatopsis, Promicromonospora, and Rhodococcus were also isolated in our study. Similarly, other researchers also discovered and isolated various species of actinomycetes belonging to genera such as Nocardiopsis33,34, Nocardia35,36, Micromonospora37, Kitasatospora3,38, Rhodococcus29,36 and Arthrobacter36 suggesting their greater diversity in Nepalese soils. To our knowledge, this is the first report of Promicromonospora iranensis from the high-altitude soils of Nepal. However, it was isolated previously only from low-altitude soil habitats39. Promicromonospora is known to produce bioactive compounds with cytotoxic activities against ACC-MESO-1 and HeLa cells40.
Although many studies have mainly focused on terrestrial soils7,32,41,42,43, mangrove forests6,44,45,46, marine sediments17,47 and desert dunes48,49 for the isolation of actinomycetes, our soil samples were collected from diverse lands, including forest areas, cultivated lands, bare lands, and trekking routes at different altitudes (957 to 5550 m.a.s.l).
We found the majority of isolates (n = 34, 57.6%) were isolated at comparatively lower altitudes (1000 to 2000 m.a.s.l.), followed by 15 isolates (25.4%) from 2001 to 3000 m.a.s.l. This indicates that the abundance and diversity of actinomycetes is shrinking towards higher altitudes. This might be because there is a reduction in nutrient availability and harsher environmental conditions at higher altitudes, such as lower temperatures and higher UV radiation50. However, the occurrence of actinomycetes at higher altitudes acquire adaptations to cold due to the production of antifreeze proteins51 and cold-active enzymes52. Additionally, species richness declined diversity at higher altitudes. Nine genera of actinomycetes were isolated below the 3000 m.a.s.l. while only strains belonging to Kitasatospora and Streptomyces were isolated between 3000 and 5500 m.a.s.l., no actinomycetes could be isolated at altitudes above 5500 m.a.s.l. In support of this, previous studies reported that Streptomyces was isolated from altitudes ranging from > 2100 to < 5400 m.a.s.l. and Kitasatospora from 3114 to 5000 m.a.s.l., suggesting that these microorganisms are capable of surviving in high altitude environments3,53,54,55,56. In our study, Streptomyces thermocarboxydus was recovered from an altitude of 5356 m.a.s.l., which is known for antibacterial and antifungal properties due to the presence of compounds like alkyl halides, alkenes, sulfoxides, carboxylic acids, and alkanes57. Additionally, Streptomyces thermocarboxydus can produce chitinase, an enzyme with broad applications in medicine and agriculture due to its ability to degrade chitin58.
Therefore, our study showed the occurrences of diverse actinomycetes having diverse functional potential in high-altitude soils. However, in our study, some soil samples did not yield any actinomycetes despite following the standard isolation procedure on the AIA medium. Considering the variable properties of different soils, the coverage of culture-based diversity of actinomycetes could be increased by applying different isolation media and culture conditions. Although Actinomycetes Isolation Agar (AIA) was used exclusively in this study due to its effectiveness and selectivity, it is essential to note that other media such as Starch Casein Agar, Glycerol Asparagine Agar, and ISP media are also commonly employed for the isolation of actinomycetes and may help in recovering a broader diversity of strains. Further research is warranted to confirm that certain strains have developed specific adaptations to the low temperatures, reduced atmospheric pressure, or increased UV radiation typically associated with higher altitude environments. In addition, the stress tolerance capacity of these actinomycetes isolates may explain distinctive mechanisms, making them well-adapted to high altitudes.
The second important aspect of this study was the screening of the antimicrobial potential of isolated actinomycetes. Among the 59 isolated actinomycetes, nearly half (49.2%) exhibited antimicrobial activity. The antifungal activity showed the highest efficacy, comprising over 86.0% against tested strains such as Wickerhamomyces anomalis DSM 6766 and Mucor hiemalis DSM 2656. In one study59, 52.3% of the actinomycetes isolated from ants showed inhibitory properties against phytopathogenic fungi. Two species of Streptomyces exhibited a selective antifungal effect against Xylaria60. The antifungal activity in actinomycetes was also described in several more studies involving co-cultivations61 and some crude extractions62. Some peptides produced by Streptomyces misionensis V16R3Y1 showed antifungal activity against Fusarium, Aspergillus, Penicillium, and Candida63.
Only two strains (6.9%), 13081 (Streptomyces olivaceus) and 13105 (Nocardia thailandica), in our study, showed antimicrobial properties against Gram-positive bacteria (B. subtilis DSM 10T). Streptomyces isolated from agricultural soil in India showed good activity against Gram-positive Staphylococcus aureus32. In a similar study, 51 isolates (66.0%) exhibited antimicrobial activity against Staphylococcus aureus MTCC 96, and 49 isolates (64.0%) showed antimicrobial activity against methicillin-resistant Staphylococcus aureus (MRSA)64. Micromonospora sp. and Streptomyces sp. were also found to be active against Bacillus subtilis29.
Twelve out of 29 active strains (41.4%) show antimicrobial properties against Gram-negative bacteria (E. coli BW25113, E. coli JW0451-2). In a study64, 19 isolates inhibited the growth of E. coli MTCC 40. Among 29 active actinomycetes against human pathogens isolated from the Qinghai-Tibetan Plateau, 2 isolates (6.9%) demonstrated antimicrobial activity against E. coli21. Micromonospora sp. IB 2015I12-1 and Streptomyces sp. IB2015I9-1, isolated by29 from Baikal endemic algae, which were active against E. coli. Antimicrobial activity against E. coli BW25113, being the parental strain for the Keio collection, and E. coli JW0451-2, which carries specific gene deletion, also shows the importance of actinomycetes strains genes related to antibiotic resistance and efflux systems65. These findings suggest that crude extracts from actinomycetes isolated from high altitudes in Nepal possess a broad spectrum of antimicrobial activities with antifungal activities being most prevalent. This is highly significant since there is an increasing trend of antifungal resistance in both clinical and agricultural settings. Overall, nearly 50% of the strains showing antimicrobial activities in our study show the potential of Nepalese actinomycetes as a rich source of novel antimicrobial agents.
The crude extract of Streptomyces olivaceus (13081) and Nocardia thailandica (13105) showed broad-spectrum antimicrobial activity against all the tested gram-positive and gram-negative bacteria and fungi. This activity is likely attributed to the presence of antibacterial compounds such as Neoaspergillic acid66,67 and Okaramine D68,69 and antifungal compounds such as Antimycin A370 and Tricycloalternarene A71 annotated and identified from the extracts of Streptomyces olivaceus (13081). Similarly, antibacterial compounds such as Nocardimicin B72 and 4-O-methylmelleolide73 and antifungal compounds such as Flocculosin74 and Microcolin A75 were annotated and identified from the extracts of Nocardia thailandica (13105). The presence of these compounds supports the antimicrobial activity observed in the crude extracts of Streptomyces olivaceus (13081) and Nocardia thailandica (13105) and highlights the potential of actinomycetes as valuable sources for novel antimicrobial agents to combat multidrug-resistant pathogens. Identifying unknown compounds not annotated in existing libraries presents a valuable opportunity to discover novel antimicrobial metabolites with potential efficacy against resistant pathogens. Utilizing untargeted metabolomic approaches, we can detect and characterize these unrecognized bioactive molecules, expanding the range of antimicrobial agents available for therapeutic development.
In this study, we assessed the production of secondary metabolites and their antimicrobial activities by cultivating isolated actinomycetes strains in a single culture medium, ISP-2 broth, under standardized conditions. However, cultivating actinomycetes in different broth media under optimal conditions could allow the expression of different secondary metabolite biosynthesis pathways, activating specific silent gene clusters76. Therefore, future in-depth studies should include varied media and maximum possible optimized conditions to harness the hidden bioactivity of actinomycetes.
By elucidating the bioactive compounds produced by these microbial strains, researchers can uncover new leads for drug development, particularly in addressing health challenges prevalent in Nepal and similar regions. Some of the strains isolated in this study represent an exciting prospect for further studies to characterize its phenotypic and genotypic properties and determine its taxonomic status as a new actinomycetes species.
Conclusions
Diverse indigenous species belonging to nine genera of actinomycetes were isolated from soils at different altitudes and locations in Nepal. Streptomyces accounted for the most frequent isolates with the highest species richness, and previously unreported Promicromonospora was identified in the high-altitude soil. Screening of the crude extracts revealed the significance of actinomycetes strains as a source of antimicrobial bioactive compounds, as almost half of the isolates exhibited lower to higher antimicrobial potential. Notably, Streptomyces olivaceus (13081) and Nocardia thailandica (13105) exhibited broad-spectrum antimicrobial activity, underscoring their potential as promising candidates for further exploration and development. The activities demonstrated here highlight the biotechnological importance of continued isolation of environmental strains for compound discovery, as they serve as the basis for natural product drug discovery. Expanding the natural product research area and optimizing the downstream cultivations could be future steps leading to the discovery of antimicrobial compounds.
Data availability
The nucleotide sequences for the bacteria sequenced and assembled in this study are available on NCBI GenBank under the following accession numbers: PQ104910-PQ104968.
References
Cheng, M. J., Chen, J. J., Wu, M., Der, Leu, J. Y. & Tseng, M. Antifungal activities of compounds produced by newly isolated Acrocarpospora strains. Antibiotics 12, 95 (2023).
Ait Assou, S., Anissi, J., Sendide, K. & El Hassouni, M. Diversity and antimicrobial activities of actinobacteria isolated from mining soils in midelt region, Morocco. Sci. World J. (2023).
Singh, L. S., Sharma, H. & Sahoo, D. Actinomycetes from soil of lachung, a pristine high altitude region of Sikkim himalaya, their antimicrobial potentiality and production of industrially important enzymes. Adv. Microbiol. 09, 750–773 (2019).
Sapkota, A. et al. Isolation, characterization, and screening of antimicrobial-producing actinomycetes from soil samples. Int. J. Microbiol. 2020, 2716584 (2020).
Shivabai, C. & Gutte, S. Isolation of actinomycetes from soil sample using different pretreatment methods and its comparative study. Int. J. Res. Anal. Rev. 6, 697–702 (2019).
Sengupta, S., Pramanik, A., Ghosh, A. & Bhattacharyya, M. Antimicrobial activities of actinomycetes isolated from unexplored regions of sundarbans Mangrove ecosystem. BMC Microbiol. 15, 170 (2015).
Elbendary, A. A. et al. Isolation of antimicrobial producing Actinobacteria from soil samples. Saudi J. Biol. Sci. 25, 44–46 (2018).
Khadayat, K. et al. Molecular identification and Antimicrobial Potential of Streptomyces Species from Nepalese Soil. Int J Microbiol (2020). (2020).
Sebak, M. et al. Bioassay- and metabolomics-guided screening of bioactive soil actinomycetes from the ancient City of Ihnasia, Egypt. PLoS One. 14, 1–29 (2019).
Adam, D. et al. Isolation, characterization, and antibacterial activity of hard-to-culture actinobacteria from cave moonmilk deposits. Antibiotics 7, 28 (2018).
Aryal, S., Neupane, L., Adhikari, R., Regmi, B. & Joshi, D. R. Nocardia nepalensis sp. nov., a novel actinobacterium isolated from forest soil in pokhara, Nepal. Microbe 6, 100282 (2025).
Thapa, B. B. et al. Metabolic comparison and molecular networking of antimicrobials in Streptomyces species. Int. J. Mol. Sci. 25, 4193 (2024).
Das, P. et al. An antibacterial compound pyrimidomycin produced by Streptomyces sp. PSAA01 isolated from soil of Eastern Himalayan foothill. Sci. Rep. 12, 10176 (2022).
Iwu, C. D., Korsten, L. & Okoh, A. I. The incidence of antibiotic resistance within and beyond the agricultural ecosystem: a concern for public health. Microbiologyopen 9, 1–28 (2020).
Ahmed, R. N., Daniel, F., Gbala, I. D. & Sanni, A. Potentials of actinomycetes from reserved environments as antibacterial agents against Drug-Resistant clinical bacterial strains. Ethiop. J. Health Sci. 30, 251–258 (2020).
Aryal, S., Adhikari, R., Regmi, B. & Joshi, D. R. Antibacterial compounds of actinomycetes isolated from altitude soils. J. Nepal. Health Res. Counc. 22, 784–791 (2025).
Ibrahimi, M. et al. Marine actinobacteria: screening for predation leads to the discovery of potential new drugs against multidrug-resistant bacteria. Antibiotics 9, 1456 (2020).
Rai, K., Khadka, S., Shrestha, B. & Actinomycetes Isolation, characterization and screening for antimicrobial activity from different sites of chitwan, Nepal. Int. J. Microbiol. Biotechnol. 3, 25–30 (2018).
Mondal, H. & Thomas, J. Isolation and characterization of a novel actinomycete isolated from marine sediments and its antibacterial activity against fish pathogens. Antibiotics 11, 1546 (2022).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species 1. Int. J. Syst. Evol. Microbiol. 16, 313–340 (1966).
Ma, A. et al. Phylogenetic and physiological diversity of cultivable actinomycetes isolated from alpine habitats on the Qinghai-Tibetan plateau. Front. Microbiol. 11, 555351 (2020).
Kumar, S. et al. MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 41, 1423 (2024).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10, 512–526 (1993).
Saitou, N. & Nei, M. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Rzhetsky, A. & Nei, M. A. Simple method for estimating and testing Minimum-Evolution trees. Mol. Biol. Evol. 9, 945 (1992).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evol. (N Y) 39, 783 (1985).
Ribeiro, I. et al. Diversity and bioactive potential of actinobacteria isolated from a coastal marine sediment in Northern Portugal. Microorganisms 8, 1691 (2020).
Axenov-Gribanov, D. V. et al. Cultivable actinobacteria first found in Baikal endemic algae is a new source of natural products with antibiotic activity. Int. J. Microbiol. 2020, 5359816 (2020).
Hoffmann, T., Krug, D., Hüttel, S. & Müller, R. Improving natural products identification through targeted LC-MS/MS in an untargeted secondary metabolomics workflow. Anal. Chem. 86, 10780–10788 (2014).
Bérdy, J. Bioactive microbial metabolites. J. Antibiot. (Tokyo). 58, 1–26 (2005).
Singh, V. et al. Isolation and purification of antibacterial compound from Streptomyces levis collected from soil sample of North India. PLoS One. 13, e0200500 (2018).
Lekhak, B., Singh, A. & Bhatta, D. R. Antibacterial and antifungal property of actinomycetes isolates from soil and water of Nepal. J. Nepal. Health Res. Counc. 16, 136–139 (2018).
Gamaleldin, N. M. et al. Exploration of chemical diversity and antitrypanosomal activity of some red sea-derived actinomycetes using the OSMAC approach supported by LC-MS-based metabolomics and molecular modelling. Antibiotics 9, 1–16 (2020).
Borah, A. & Thakur, D. Phylogenetic and functional characterization of culturable endophytic Actinobacteria associated with Camellia spp. For growth promotion in commercial tea cultivars. Front. Microbiol. 11, 318 (2020).
Silva, L. J. et al. Actinobacteria from Antarctica as a source for anticancer discovery. Sci Rep 10, 4125(2020).
Pansomsuay, R. et al. Micromonospora thermarum sp. nov., an actinobacterium isolated from hot spring soil. Arch. Microbiol. 205, 123 (2023).
Srivastava, N., Nandi, I., Ibeyaima, A., Gupta, S. & Sarethy, I. P. Microbial diversity of a Himalayan forest and characterization of rare actinomycetes for antimicrobial compounds. 3 Biotech. 9, 27 (2019).
Mohammadipanah, F. et al. Promicromonospora iranensis sp. nov., an actinobacterium isolated from rhizospheric soil. Int. J. Syst. Evol. Microbiol. 64, 3314–3319 (2014).
Izumikawa, M., Takagi, M. & Shin-Ya, K. Isolation of a novel macrocyclic dilactone-JBIR-101- from Promicromonospora sp. RL26. J. Antibiot. 64, 689–691 (2011).
Al-Dhabi, N. A., Esmail, G. A., Ghilan, A. K. M. & Arasu, M. V. Isolation and screening of Streptomyces sp. Al-Dhabi-49 from the environment of Saudi Arabia with concomitant production of lipase and protease in submerged fermentation. Saudi J. Biol. Sci. 27, 474–479 (2020).
Hamid, M. E. et al. Diversity and geographic distribution of soil streptomycetes with antagonistic potential against actinomycetoma-causing Streptomyces sudanensis in Sudan and South Sudan. BMC Microbiol. 20, 33 (2020).
Singh, V. et al. Isolation, purification, and characterization of heparinase from Streptomyces variabilis MTCC 12266. Sci. Rep. 9, 6482 (2019).
Elias, F., Muddada, S., Muleta, D. & Tefera, B. Antimicrobial Potential of Streptomyces spp. Isolated from the Rift Valley Regions of Ethiopia. Adv. Pharmacol. Pharm. Sci. 2022, 1724906 (2022).
Huang, H. et al. Streptomyces caeni sp. Nov., isolated from Mangrove mud. Int. J. Syst. Evol. Microbiol. 68, 3080–3083 (2018).
Sharifi, M. & Bipinraj, N. K. Isolation and identification of actinomycetes with anticandida activity from Mangrove soil. Biosci. Biotechnol. Res. Asia. 16, 611–615 (2019).
Li, Z. et al. Engineering Bafilomycin high-producers by manipulating regulatory and biosynthetic genes in the marine bacterium Streptomyces lohii. Mar. Drugs. 19, 29 (2021).
Sivakala, K. K. et al. Desert environments facilitate unique evolution of biosynthetic potential in Streptomyces. Molecules 26, 588 (2021).
Hamid, M. E. et al. Isolation and identification of Streptomyces spp. From desert and savanna soils in Sudan. Int. J. Environ. Res. Public. Health. 17, 1–10 (2020).
Kumar, S., Suyal, D. C., Yadav, A., Shouche, Y. & Goel, R. Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS One. 14, e0213844 (2019).
Baskaran, A. et al. Screening and production of antifreeze proteins from Actinobacteria. In Methods in Actinobacteriology (ed. Dharumadurai, D.) 467–470 (Springer US, 2022). https://doi.org/10.1007/978-1-0716-1728-1_67.
Sivasankar, P. et al. Exogenous production of cold-active cellulase from Polar Nocardiopsis sp. with increased cellulose hydrolysis efficiency. Arch. Microbiol. 204, 218 (2022).
Bhat, A. M., Hussain, A., Hassan, Q. P. & Bhat, A. Culturable Streptomyces spp. From high-altitude, oligotrophic North Western himalaya: a comprehensive study on the diversity, bioactivity and insights into the proteome of potential species. FEMS Microbiol. Ecol. 100, fiae026 (2024).
Jiang, K., Chen, X., Zhang, W., Guo, Y. & Liu, G. Nonribosomal antibacterial peptides isolated from Streptomyces agglomeratus 5-1-3 in the Qinghai-Tibet plateau. Microb. Cell. Fact. 22, 5 (2023).
Yadav, J., Shrestha, U. T., Tiwari, K. B., Sahukhal, G. S. & Agrawal, V. P. Streptomycin - Like antibiotic from Streptomyces spp. Isolated from Mount everest base camp. Nepal. J. Sci. Technol. 9, 73–77 (2008).
Zhu, H. et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiol. (Reading). 160, 1714–1726 (2014).
Nayaka, S. et al. A potential bioactive secondary metabolites and antimicrobial efficacy of Streptomyces thermocarboxydus strain KSA-2, isolated from Kali river, Karwar. Curr. Res. Microbiol. Infect. 1, 5–13 (2020).
Tran, T. N., Doan, C. T., Nguyen, V. B., Nguyen, A. D. & Wang, S. L. The isolation of Chitinase from Streptomyces thermocarboxydus and its application in the preparation of Chitin oligomers. Res. Chem. Intermed. 45, 727–742 (2019).
Wang, Z. et al. Community composition, antifungal activity and chemical analyses of ant-derived actinobacteria. Front. Microbiol. 11, 201 (2020).
Yin, C., Jin, L., Li, S., Xu, X. & Zhang, Y. Diversity and antagonistic potential of Actinobacteria from the fungus-growing termite Odontotermes formosanus. 3 Biotech. 9, 45 (2019).
Shamikh, Y. I. et al. Actinomycetes from the red sea sponge Coscinoderma mathewsi: Isolation, diversity, and potential for bioactive compounds discovery. Microorganisms 8, 783 (2020).
Ouchari, L., Boukeskasse, A., Bouizgarne, B. & Ouhdouch, Y. Antimicrobial potential of actinomycetes isolated from the unexplored hot Merzouga desert and their taxonomic diversity. Biol. Open. 8, bio035410 (2019).
Saadouli, I. et al. Isolation, characterization and chemical synthesis of large spectrum antimicrobial cyclic dipeptide (l-leu-l-pro) from Streptomyces misionensis V16R3Y1 bacteria extracts. A novel 1H NMR metabolomic approach. Antibiotics 9, 270 (2020).
Sharma, P. & Thakur, D. Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Sci. Rep. 10, 4104 (2020).
Tamae, C. et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190, 5981–5988 (2008).
Morales-Sánchez, V. et al. Bioactive metabolites from the endophytic fungus Aspergillus sp. SPH2. J. Fungi. 7, 109 (2021).
Yuan, B. et al. Identification of the Neoaspergillic acid biosynthesis gene cluster by establishing an in vitro CRISPR-Ribonucleoprotein genetic system in Aspergillus melleus. ACS Omega. 8, 16713–16721 (2023).
Petersen, L. M., Hoeck, C., Frisvad, J. C., Gotfredsen, C. H. & Larsen, T. O. Dereplication guided discovery of secondary metabolites of mixed biosynthetic origin from Aspergillus aculeatus. Molecules 19, 10898–10921 (2014).
Labuda, R. et al. Chemical composition of anti-microbially active fractions derived from extract of filamentous fungus Keratinophyton lemmensii including three novel bioactive compounds. Sci. Rep. 14, 1423 (2024).
Hosotani, N. et al. A10∼A16, seven new antimycin antibiotics produced by Streptomyces spp. SPA-10191 and SPA-8893. J. Antibiot. (Tokyo) 58, 460–467 (2005).
Zhang, H., Zhao, Z., Chen, J., Bai, X. & Wang, H. Tricycloalternarene analogs from a symbiotic fungus Aspergillus sp. D and their antimicrobial and cytotoxic effects. Molecules 23, 1423 (2018).
Ikeda, Y. et al. Siderophores with muscarinic M3 receptor inhibiting activity from Nocardia sp. TP-A0674. J. Nat. Prod. 68, 1061–1065 (2005).
Donnelly, D. M. et al. Antibacterial sesquiterpene Aryl esters from Armillaria mellea. J. Nat. Prod. 48, 10–16 (1985).
Mimee, B., Pelletier, R. & Bélanger, R. R. In vitro antibacterial activity and antifungal mode of action of flocculosin, a membrane-active cellobiose lipid. J. Appl. Microbiol. 107, 989–996 (2009).
Meickle, T., Matthew, S., Ross, C., Luesch, H. & Paul, V. Bioassay-guided isolation and identification of Desacetylmicrocolin B from Lyngbya cf. polychroa. Planta Med. 75, 1427–1430 (2009).
Aryal, S. et al. Novel Streptomyces sp. reported in 2018: A meta-analysis. Antiinfect Agents 19, e030921181212 (2020).
Acknowledgements
We express our gratitude to the Department of Microbial Natural Products (MINS), Helmholtz Institute for Pharmaceutical Research Saarland (HIPS) for providing the laboratory facilities, space, and partial scholarship. We thank Prof. Dr. Rolf Müller and Dr. Ronald Garcia from MINS for the helpful advice and guidance during the research stay of S.A. at HIPS. The Ph.D. research work of S.A. was partially funded by the University Grants Commission- Nepal (Award no.: PhD/74_75/S&T-12) and the German Academic Exchange Service (Bi-nationally Supervised Doctoral Degrees, 2019/20).
Funding
The Ph.D. research work of Sagar Aryal was partially funded by the University Grants Commission- Nepal (Award no.: PhD/74_75/S&T-12) and the German Academic Exchange Service (Bi-nationally Supervised Doctoral Degrees, 2019/20).
Author information
Authors and Affiliations
Contributions
S.A.: Conceptualization, Methodology, Investigation, Data Curation, Writing - Original Draft. R.A.: Writing - Review & Editing, Supervision. B.R.: Writing - Review & Editing, Supervision. D.R.J.: Conceptualization, Writing - Review & Editing, Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aryal, S., Adhikari, R., Regmi, B. et al. Antimicrobial potential of actinomycetes from high altitude Nepalese soils. Sci Rep 15, 33915 (2025). https://doi.org/10.1038/s41598-025-09357-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-09357-5