Abstract
Pancreatic ductal adenocarcinoma (PDAC), the most prevalent form of pancreatic malignancy, is characterized by its aggressive nature and high mortality rate, with a 5-year survival rate in China of only 9.9%. Polymorphic adenoma gene-like 2 (PLAGL2) has been implicated in the development of various digestive tract tumors, including hepatocellular carcinoma, gastric carcinoma, and colorectal carcinoma, and it influences tumor progression through multiple pathways. However, the specific role and mechanism of PLAGL2 in PDAC requires further investigation. The objective of this study was to evaluate the expression level of PLAGL2 in PDAC and its association with clinical parameters, thereby assessing its prognostic significance for PDAC, and its impact on the malignant process. We first analyzed the expression of PLAGL2 in pancreatic ductal adenocarcinoma (PDAC) via multiple databases, including TCGA and GTEx, to investigate its associated enrichment pathways. We subsequently examined the expression of PLAGL2 via immunohistochemistry and PCR in PDAC tumor tissue samples from our hospital, explored the relationships between PLAGL2 expression and clinical features, and evaluated the predictive value of PLAGL2 for the survival of PDAC patients. we investigated the effect of PLAGL2 on the epithelial‒mesenchymal transition (EMT) process by examining its association with the expression of EMT-related proteins. Finally, We confirmed that PLAGL2 facilitates the proliferation, invasion, and migration of PDAC using conventional cellular assays. The immunohistochemistry and PCR results, combined with the results of GEPIA analysis, indicated that PLAGL2 was significantly overexpressed in patients with pancreatic ductal adenocarcinoma (PDAC). GO analysis revealed that PLAGL2 was intricately linked to protein modification and regulation, as well as nerve projection development. The KEGG and GSEA enrichment analyses indicated that several signaling pathways, including the PI3K-AKT pathway, were significantly enriched, particularly in relation to EMT. The characterization of PLAGL2 expression and its clinicopathological features indicated that high PLAGL2 expression was associated with poor prognosis in PDAC patients and was positively correlated with TNM stge, TNM: T,TNM: N, Tumor size, Nerve infiltration and other clinical characteristics of the tumor. Immunohistochemical analysis suggested that PLAGL2 plays a role in enhancing the epithelial-to-mesenchymal transition (EMT) process in PDAC. By knocking down or overexpressing PLAGL2 in PDAC cell lines and conducting a series of cellular experiments, we ultimately demonstrated that low expression of PLAGL2 inhibits proliferation, migration, and invasion in PDAC, while high expression of PLAGL2 promotes these processes. This study demonstrated that the expression level of PLAGL2 serves as a predictive biomarker for survival in patients with pancreatic ductal adenocarcinoma (PDAC). It is associated with clinical features, including TNM stge, Tumor size, Nerve infiltration, and it also plays a role in promoting epithelial‒mesenchymal transition (EMT) in PDAC, and promotes the proliferation, migration and invasion of PDAC.
Similar content being viewed by others
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most prevalent form of pancreatic malignancy and is characterized by its high lethality and invasiveness1. This disease is characterized by its aggressive nature, rapid progression, and early metastasis, leading to a poor prognosis. In clinical practice, the treatment of PDAC is challenging because of the low rate of radical surgery and the subsequent high incidence of recurrence after surgery2,3. The 5-year survival rate in China is only 9.9%4. At diagnosis, most patients are found to have locally advanced or distant metastases, with only a small fraction (15–20%) eligible for complete surgical resection of their pancreatic tumors4,5.Despite surgical intervention, nearly three-quarters of patients experience recurrence within two years6,7,8. The primary factors contributing to this outcome are aggressive behavior and late diagnosis of PDAC. Currently, there are no reliable biomarkers for predicting or effectively treating PDAC. Thus, elucidating the biological mechanisms underlying the progression of PDAC could lead to improved treatment strategies and improved survival outcomes.
Cancer is a complex process involving multiple transcription factors (TFs), which can be dysregulated to initiate or accelerate various procancer events, such as proliferation, invasion, and metastasis, in a variety of cancers, including hepatocellular carcinoma (HCC)9,10, colorectal cancer (CRC)11,12 and gastric cancer (GC)13,. PLAGL2, a zinc-finger PLAG TF, is overexpressed in gastrointestinal malignancies. In CRC, PLAGL2 binds to the Wnt6 promoter and activates the Wnt/β-catenin pathway, regulating ZEB1-mediated EMT to promote metastasis14,15.In HCC, PLAGL2 enhances cell survival through the regulation of mitochondrial apoptosis and the C-MET/STAT3 signaling axis16.Additionally, PLAGL2 has been shown to promote HCC cell proliferation, migration, and invasion via the EGFR-AKT pathway17. In gastric cancer (GC), PLAGL2 regulates USP37-mediated Snail1 deubiquitination to promote cell proliferation and migration18.
To date, the role of PLAGL2 in pancreatic cancer and its mechanism has not been explored in the literature. Our previous genomics showed that PLAGL2 is differentially expressed among PDAC. Furthermore, literature review and database analysis support its potential role in cancer progression. In this study, we identified PLAGL2 as an oncogene that facilitates the progression of pancreatic ductal adenocarcinoma (PDAC). PLAGL2 overexpression in PDAC tumor tissues is correlated with poorer survival outcomes in PDAC patients. Upregulation of PLAGL2 promotes PDAC progression by triggering epithelial‒mesenchymal transition (EMT) in PDAC.
Materials and methods
Database analysis
GEPIA was used to investigate the differential expression levels of PLAGL2 across multiple organs, including its expression in normal pancreas and pancreatic ductal adenocarcinoma (PDAC), as well as to assess its correlation with clinical stage. Genes related to PLAGL2 were retrieved from the STRING database (https://cn.string-db.org/). A GO enrichment analysis was conducted on PLAGL2 and its related genes, revealing the cellular localization of PLAGL2 and the enriched pathways. The screening of PLAGL2 differential genes in the TCGA and GTEx databases, along with KEGG pathway enrichment analysis and GSEA enrichment analysis, indicated potential related signaling pathways19,20,21.
Patient selection and sample collection
Patient selection and sample collection involved tumor and matched nonneoplastic tissues (surgical samples were immediately flash-frozen in liquid nitrogen and stored at -80 °C for analysis) from patients with a postoperative pathological diagnosis of PDAC at the First Affiliated Hospital of Henan University of Science and Technology between October 2022 and April 2023. Additionally, 73 paraffin-embedded tissue sections (dehydrated paraffin-preserved PDAC tissues sectioned for pathological review) were obtained from 73 patients diagnosed with PDAC between January 2018 and April 2023 from the Department of Pathology at the First Affiliated Hospital of Henan University of Science and Technology. Patients receiving palliative care or those lost to follow-up were excluded. Tumors were staged according to the ninth edition of the TNM classification. The study was approved by the ethics committee (ethical number: NA2024-0537). We confirm that all methods and experiments were conducted in accordance with the relevant guidelines and regulations. Detailed demographic information, clinical data, and the pathology reports are presented in Table 1.
Follow-up
Overall survival is a prognostic measure defined as the time elapsed from the date of surgery to the date of death or the last follow-up examination. Progression-free survival is defined as the time from the date of surgery to the date of death or the date of diagnosis of tumor recurrence.
Immunohistochemical (IHC) staining
Immunohistochemical staining was performed as follows: tissues were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, deparaffinized in xylene, and rehydrated through a graded ethanol series. Antigen retrieval was achieved using sodium citrate/TE buffer. The sections were blocked with 10% sheep serum in phosphate-buffered saline (PBS) to prevent nonspecific binding. The sections were incubated with primary antibodies overnight at 4 °C. After washing, the sections were incubated with secondary antibodies and visualized via diaminobenzidine (DAB). Hematoxylin counterstaining, dehydration, and mounting were then performed. Images were captured via a microscope. Protein expression was quantified via ImageJ software.
Immunohistochemical (IHC) scoring
Scores were calculated based on the sum of the percentage of positive tumor cell staining: 0–5% scored 0, 6-35% scored 1, 36-70% scored 2, and greater than 70% scored 3. Staining intensity: no staining scored 0, weak staining scored 1, moderate staining scored 2, and strong staining scored 3. The final score was determined by multiplying the positivity of cell staining by the staining intensity, and was categorized into low or high expression groups as follows: “-” 0–1, “+” 2–3, “++” 4–6, and “++++” >6. Low expression was defined as a total score < 4, and high expression was defined as a total score ≥ 4.
Cell culture
Human pancreatic cancer cell lines PANC-1 (CL-0184-Wuhan Pricella Biotechnology Co.,Ltd), MIA PaCa-2 (CL-0627-Wuhan Pricella Biotechnology Co.,Ltd), BxPC-3 (CL-0042-Wuhan Pricella Biotechnology Co.,Ltd) and pancreatic ductal cell line HTERT HPNE (KC4033-Guangzhou Kinlogix Biotech Co.,Ltd) were used. Cells were grown at 37 °C in a 5% CO2 humidified environment and cultured in DMEM supplemented with 10% fetal bovine serum, 2.5% horse serum, and 1% penicillin-streptomycin.
Western blot analysis
total protein was extracted from pancreatic ductal adenocarcinoma (PDAC) cell lines. The protein concentration was determined via a BCA protein assay kit (Merck). Samples of 40 µg of lysate were electrophoresed in a 10% polyacrylamide gel and then transferred to a polyvinylidene difluoride (PVDF) membrane. The protein was detected via a rabbit anti-plagl2 polyclonal antibody via Western blot analysis. A mouse monoclonal β-actin antibody was used in conjunction with a goat anti-mouse IgG antibody for the detection of β-actin. Finally, the immunoblotting signals were analyzed via an image station (4000R PRO scanner).
Transient transfection
Small interfering RNA (siRNA), siPLAGL2 and pcDNA-PLAGL2 plasmid were synthesized by Zhengzhou Baiogi Biotechnology Co. The oligonucleotide sequence of the synthesized oligonucleotide siPLAGL2 was forward 5’-AAAGCAGGAGGAGGAAGTGG-3’ and reverse 5’-TTCTGGGGCTGAGTGGGGTGG-3’. One day prior to transfection, Cells were cultured at 30–50% confluency into 6-well plates. Cells were transfected using Lipofectamine 8000 (Invitrogen, Carlsbad, CA) as directed.
RNA extraction and qRT‒PCR analysis
Total RNA was extracted from pancreatic ductal cells and various PDAC cell lines via an RNA extraction and reverse transcription kit. Subsequently, quantitative real-time PCR (qRT‒PCR) was conducted on a real-time PCR system. The expression data were normalized to that of GAPDH. The fold change in PLAGL2 expression between PDAC and normal tissue pairs was analyzed by calculating the 2-ΔΔCt values. An identical approach was adopted to compare PDAC cells with HPNE cells. Each sample was subjected to three replicate runs.
CCK-8 assay
Cells were inoculated into 96-well culture plates at a density of 3*103 cells/well. Cell Counting Kit-8 (CCK-8) solution (10 µL, Analysis Quiz, China) was added to the culture medium. Cell proliferation was assessed by measuring the absorbance at 450 nm using an enzyme marker at the indicated time points (0, 24 and 48 h).
Wound healing assay
Twenty-four hours after infection or transfection, cells in the confluent state were treated with 1 µg/mL mitomycin C (Sigma, USA) in serum-free medium for 1 h. Subsequently, cells were scraped in a straight line along the center of the well with a 200 µL pipette tip, then rinsed with PBS and incubated in serum-free medium for 24 h. The wound gap was measured under a microscope at the beginning (0 h) and end (24 h) of the experiment.
Transwell assay
The upper surface of the Transwell chamber (Corning, USA) was pre-coated with Matrigel gel. A total volume of 200 µL of serum-free RPMI 1640 medium containing 1 × * 105 cells (3 × 104 cells/well) was added to the upper chamber. The lower chamber was filled with DMEM containing 10% FBS as an inducer. After 24 h of incubation at 37 °C and 5% CO2, cells invading the lower chamber were fixed with 4% paraformaldehyde for 20 min and stained with 0.5% crystal violet for 5 min. Finally, the number of cells was recorded under a microscope.
Statistical analysis
Each experiment was conducted in triplicate. The data were analyzed via SPSS (version 26.0) software. Appropriate statistical tests, including t tests, one-way ANOVA, chi-square tests, and Cox regression analysis for clinical data to identify prognostic associations, were employed.
Results
Analysis of publicly available data indicates that PLAGL2 affects the progression of PDAC
In the GEPIA dataset (Includes TCGA and GTEx databases), PLAGL2 expression was elevated in several cancer types (Fig. 1A), whereas PLAGL2 was overexpressed in PDAC compared with normal pancreatic tissues (Fig. 1B) and was positively correlated with the grading stage of PDAC (Fig. 1C). GO analysis (Metascape) of the gene data associated with PLAGL2 in the String database (Fig. 1D ) indicated that PLAGL2 is primarily localized in the perinuclear region of the cytoplasm and plays a crucial role in various critical biological processes, including the modification and regulation of proteins and the development of nerve projections (Fig. 1E), KEGG and GSEA enrichment analyses further investigated the relevant signaling pathways, indicating that the differential expression of PLALG2 is closely associated with several pathways(Fig. 1F), including PI3K-Akt(Fig. 1G), MAPK, TNF, and TGFβ(Fig. 1H), and that some of these pathways are linked to the EMT process. This information indicates that PLAGL2 may be differentially expressed in PDAC and may influence tumor progression.
Database analysis of PLAGL2 expression and role in PDAC. (A) GEPIA was used to analyze the differential expression of PLAGL2 in various tumors and corresponding normal tissues in the TCGA and GTEx database; (B) PDAC differential expression with normal pancreatic tissues; (C) PLAGL2 expression in PDAC with different stages;. (D) PLAGL2 related genes in the String database; (E) GO enrichment analysis of PLAGL2-related genes; (F) KEGG pathway enrichment analysis of PLAGL2-related genes; G and H: GSEA pathway enrichment analysis of PLAGL2-related genes;
IHC and WB revealed that PLAGL2 protein expression was higher in PDAC tissues than in paraneoplastic tissues
Immunohistochemistry was conducted on paraffin-embedded, archival samples of pancreatic ductal adenocarcinoma (PDAC) tumors and their corresponding pancreatic paracarcinoma tissues from our hospital. PLAGL2 was predominantly detected in the nuclei of cells, with some expression also observed in the cytoplasm (Fig. 2A). PLAGL2 expression is higher in PDAC tumors than in their corresponding pancreatic tissues (Fig. 2A). Notably, the protein levels of PLAGL2 were found to be greater in PDAC tissues than in paracarcinoma tissues (Fig. 2B).
PLAGL2 expression was higher in PDAC than pancreatic tissues. (A) PLAGL2 expression in PDAC was higher than the corresponding paracancerous tissues expression as shown by arrows, which was mainly located in the nucleus for expression; (B) Western blot (WB) analysis of six pairs of PDAC and their corresponding paracancerous tissue; (C) mRNA levels of PLAGL2 were analyzed by qPCR in PDAC cell lines (PANC-1, MIA-2, BxPC-3) and normal pancreatic cells (HPNE). (D) Western blot (WB) analysis of the protein levels of PLAGL2 in PDAC cell lines (PANC-1, MIA-2) and normal pancreatic cells (HPNE) in the protein level of PLAGL2.
PCR and WB to detect PLAGL2 expression in normal pancreatic cell lines and PDAC cell lines
To verify that PLAGL2 was significantly upregulated in PDAC, the mRNA expression of PLAGL2 in pancreatic ductal cells and two pancreatic cancer cell lines was assessed via quantitative RT‒PCR (qRT‒PCR). The analysis revealed that PLAGL2 was elevated in tumor cells compared with pancreatic ductal cells (Fig. 2C). Western blotting further demonstrated that the protein expression of PLAGL2 in pancreatic cancer cell lines was also greater than that in pancreatic ductal cells (Fig. 2D).
Relationships between PLAGL2 expression and the clinicopathologic features of PDAC patients
High expression of PLAGL2 was significantly correlated with Gender, TNM stge, TNM: T,TNM: N, Tumor size, Nerve infiltration.Conversely, there was no significant association between PLAGL2 expression and patient age, Total bilirubin increased, Elevated aminotransferase, CA199 (Table 1).
PLAGL2 overexpression is associated with poor prognosis in patients with PDAC
To investigate the impact of PLAGL2 overexpression on progression-free survival and overall survival (OS) in patients with pancreatic ductal adenocarcinoma (PDAC), we conducted a univariate survival analysis. As indicated in the table, TNM stge, lymphatic metastasis, and PLAGL2 expression were significantly associated with poor prognosis in patients with PDAC. The findings of the multivariate Cox regression analysis indicated that PLAGL2 expression is independent prognostic factors influencing OS in PDAC patients (Table 2).
We present representative images of high and low PLAGL2 expression levels in IHC (Fig. 3A). Kaplan‒Meier survival curves were generated to elucidate the effects of PLAGL2 expression on progression-free survival and overall survival in PDAC patients. As illustrated in KM, progression-free survival and overall survival were significantly shorter in patients with high PLAGL2 expression than in those with low PLAGL2 expression (Fig. 3B).
PLAGL2 overexpression is associated with poor prognosis in PDAC patients. (A) Representative images of high and low PLAGL2 expression levels in immunohistochemistry of some PDACs are listed. (B) KM plots of overall survival as well as progression-free survival according to PLAGL2 expression, with the high expression group suggestive of a poor prognosis.
IHC and WB revealed a positive correlation between PLAGL2 expression and the levels of EMT-associated factors
A correlation analysis via GEPIA indicated that PLAGL2 expression was positively associated with β-catenin, N-calmodulin and adhesin but negatively associated with E-calmodulin (Fig. 4B). IHC was employed to assess the expression of EMT-related proteins, including β-catenin, N-calmodulin, Adhesin, and E-calmodulin, in patients with PDAC(Fig. 4A), which was in agreement with the GEPIA results. Furthermore, WB was used to compare the expression of associated factors in PANC-1 cells that down- or up-regulated PLAGL2 (Fig. 4C). Taken together, these findings suggest that PLAGL2 can act as a EMT-promoting factor.
IHC and WB shows that PLAGL2 is positively correlated with the expression of EMT-associated factors. (A) IHC indicates β-catenin, N-calmodulin, adhesin, and E-calmodulin expression under high and low PLAGL2 expression subgroups. (B) GEPIA indicates that PLAGL2 expression is associated with β-catenin, N-calmodulin, adhesin, and E-calmodulin expression. (C) WB shows reduced expression of β-catenin, N-calmodulin, adhesin, and E-calmodulin in PANC-1 cells that down- or up-regulated PLAGL2.
PLAGL2 promotes proliferation, invasion and migration of PDAC cells
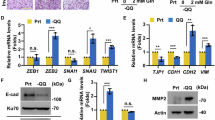
To further investigate the role of PLAGL2 in pancreatic ductal adenocarcinoma (PDAC), the PANC-1 and MIA PaCa-2 cell lines were transfected with specific PLAGL2 shRNA. Western blot analysis confirmed successful knockdown of PLAGL2 protein expression(Fig. 5A). Cell viability assessed by the CCK-8 assay demonstrated that PLAGL2 downregulation significantly inhibited cellular proliferation in both cell lines (Fig. 5B). Subsequent Wound healing and Transwell assays revealed that PLAGL2 silencing markedly suppressed migration (Fig. 5C) and invasion (Fig. 5D) capacities of PDAC cells.
To further validate the above findings, PLAGL2 was overexpressed in the BxPC-3 cell line and its overexpression efficiency was verified by WB (Fig. 6A). Cell viability assessed by the CCK-8 assay demonstrated that PLAGL2 overexpression significantly promoted cell proliferation in cell lines (Fig. 6B). Subsequent Transwell experiments showed that PLAGL2 overexpression significantly promoted the migration (Fig. 6C) and invasion (Fig. 6D) abilities of PDAC cells.These findings collectively indicate that PLAGL2 promotes proliferation, migration, and invasion in PDAC pathogenesis.
Low expression of PLAGL2 inhibited PDAC cell proliferation, invasion and migration. (A) WB validation of PLAGL2 knockdown effect in cells (B) Cell viability was determined using the CCK-8 assay (0/24/48/72/96 h time points). (C) Cell migration was evaluated by wound healing assay (scale bar: 200 μm; 24 h culture). (D) Cell invasion was detected by Transwell assay. *P < 0.05.
High expression of PLAGL2 promotes PDAC cell proliferation, invasion and migration. (A) WB validation of PLAGL2 knockdown effect in cellsWB validation of PLAGL2 overexpression in cells (B) Cell viability was determined using the CCK-8 assay (0/24/48/72/96 h time points). (C) Cell migration was evaluated by wound healing assay (scale bar: 200 μm; 24 h culture). (D) Cell invasion was detected by Transwell assay. *P < 0.05.
Discussion
The high lethality of pancreatic ductal adenocarcinoma (PDAC) is a significant concern in clinical practice, primarily due to the difficulty in early detection, which often leads to its poor prognosis. Currently, there are no reliable biomarkers for predicting the late-stage prognosis in patients with PDAC. Researchers have explored various approaches in this direction, including the use of clinically relevant data such as hemoglobin and erythrocyte distribution width22, glutamyltransferase (GGT), lactate dehydrogenase (LDH)23, and different genes, such as lncRNAs24, miRNAs25, IgG26, EGFR, HER227, and the expression of various marker proteins, to predict PDAC prognosis. However, no specific markers have been recognized as accurate guides for clinical treatment, highlighting the importance of identifying potential prognostic markers for PDAC.
PLAGL2 has been shown to exhibit oncogenic and oncostatic activities in various tissues, and its role in digestive tract tumors, such as colon cancer11,12, hepatocellular carcinoma9,10, and gastric cancer13, has been of particular interest. We are also interested in the role of PLAGL2 in digestive system tumors, including colon cancer, hepatocellular carcinoma, and gastric cancer, where the procarcinogenic effect of PLAGL2 has been confirmed. Additionally, high PLAGL2 expression is associated with gliomas28, prostate cancer29, and colorectal cancer30. However, no study has reported the exact role of PLAGL2 expression in PDAC, and we aimed to determine whether PLAGL2 is abnormally expressed in PDAC and whether it can predict the prognosis in patients with PDAC.
In this study, Analysis of publicly available data revealed a difference in PLAGL2 expression between PDAC and normal pancreatic tissues. we compared multiple groups of PDAC and paracancerous samples via IHC and WB, which revealed that PLAGL2 was significantly overexpressed in PDAC patients compared with normal pancreas and paracancerous tissues, which is consistent with previously reported changes in PLAGL2 expression in certain tumors.
To further validate the conclusion, Additionally, we detected the expression level of PLAGL2 mRNA and protein in pancreatic cell lines and pancreatic cancer cell lines via qRT‒PCR and WB, which revealed that the expression of PLAGL2 mRNA and protein was significantly elevated in PDAC cell lines. Overall, these data strongly support the notion that PLAGL2 is overexpressed in PDAC, which is consistent with its procarcinogenic effects observed in gastrointestinal tumors. These results suggest that PLAGL2 expression is increased in PDAC and may play a role in promoting tumor progression.
To further explore the impact, we combined the clinical data of selected pathological samples for further statistical analysis. We found that high PLAGL2 expression was significantly correlated with Gender, TNM stge, TNM: T,TNM: N, Tumor size, Nerve infiltration. These findings suggest that PLAGL2 may be associated with these clinical conditions, potentially influencing the common mechanisms of these clinical factors. Additionally, we observed a significant correlation between PLAGL2 expression and patient survival, indicating that it is an independent factor of poor prognosis in PDAC patients. These findings suggest that PLAGL2 may serve as a biomarker for predicting prognosis in the clinic.
We hypothesize that it may influence PDAC through several biological processes, possibly by promoting tumor metastasis in colon cancer through the epithelial‒mesenchymal transition (EMT) process14,15. The EMT process is also known to contribute to PDAC metastasis, invasion, neural infiltration31, drug resistance32, and immune escape33. To investigate whether PLAGL2 affects the EMT process, We conducted KEGG and GSEA pathway enrichment analyses using the GEPIA database and identified associations with several signaling pathways, including PI3K-Akt, MAPK, TNF, and TGFβ, most of which are involved in the EMT process, and analyzed its correlation via GEPIA and found that PLAGL2 expression was consistent with the expression of EMT-related proteins, positively correlated with N-calmodulin and adhesin, and negatively correlated with the expression of E-calmodulin. Histochemical results further verified this correlation.
There are several limitations to this study. First, the sample size was small, and the samples were collected from a single location, suggesting the need for a randomized study with an expanded sample of PDAC patients to strengthen the conclusions and avoid confounding bias. Furthermore, owing to the limited experimental conditions, we were unable to conduct animal experiments to explore the mechanism of PLAGL2, and its mechanism of inducing PDAC progression needs further verification.
To the best of our knowledge, this paper shows, for the first time, that PLAGL2 expression is upregulated in human PDAC tissues compared with paracancerous tissues. Furthermore, increased PLAGL2 expression was strongly associated with several adverse clinicopathologic features of PDAC. Our study also provides clinical evidence that PLAGL2 is an independent prognostic factor for progression-free survival and overall survival in patients with PDAC. Additionally, cellular experiments demonstrated that high expression of PLAGL2 promotes cell proliferation, migration, and invasion in PDAC. In addition, these findings suggest the possible existence of EMT-influenced pathways. In summary, PLAGL2 may function as a potential prognostic indicator for PDAC and may facilitate the malignant progression of this disease.
Data availability
The attachment includes the clinical data referenced in this article, while the section pertaining to patient privacy has been omitted.Individuals seeking to obtain data from this study are encouraged to contact the corresponding author, Hua Fan:fanhua19851229@126.com.
References
Grimaud, J. A., Peyrol, S., Paliard, P. & Vachon, A. [Experimental hepato-biliary toxicity of lithocholic acid: early ultrastructural changes of hepatocytes (author’s transl)]. Gastroenterol. Clin. Biol. 1 (4), 353–360 (1977).
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74 (1), 12–49 (2024).
Siegel, R.L., Miller, K.D., Fuchs, H.E. & Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 71 (1), 7–33 (2021).
Sun, D., Cao, M., Li, H., He, S. & Chen, W. Cancer burden and trends in china: A review and comparison with Japan and South Korea. Chin. J. Cancer Res. 32 (2), 129–139 (2020).
Hessmann, E. et al. Microenvironmental determinants of pancreatic Cancer. Physiol. Rev. 100 (4), 1707–1751 (2020).
Wood, L. D., Canto, M. I., Jaffee, E. M. & Simeone, D. M. Pancreatic cancer: pathogenesis, screening, diagnosis, and treatment. Gastroenterology 163 (2), 386–402e1 (2022).
Halbrook, C. J., Lyssiotis, C. A., Pasca di Magliano, M. & Maitra, A. Pancreatic cancer: advances and challenges. Cell 186 (8), 1729–1754 (2023).
Kolbeinsson, H. M., Chandana, S., Wright, G. P. & Chung, M. Pancreatic cancer: A review of current treatment and novel therapies. J. Invest. Surg. 36 (1), 2129884 (2023).
Dai, D. L. et al. AXIN1 boosts antiviral response through IRF3 stabilization and induced phase separation. Signal. Transduct. Target. Ther. 9 (1), 281 (2024).
Xin, X. et al. Hepatocyte-specific Smad4 deficiency inhibits hepatocarcinogenesis by promoting CXCL10/CXCR3-dependent CD8(+)- T cell-mediated anti-tumor immunity. Theranostics 14 (15), 5853–5868 (2024).
Lei, Z. et al. Reciprocal interactions between LncRNAs and MYC in colorectal cancer: partners in crime. Cell. Death Dis. 15 (7), 539 (2024).
Sun, L., Xing, J., Zhou, X., Song, X. & Gao, S. Wnt/β-catenin signalling, epithelial-mesenchymal transition and crosslink signalling in colorectal cancer cells. Biomed. Pharmacother. 175, 116685 (2024).
Jin, C. et al. Crucial role of the transcription factors family activator protein 2 in cancer: current clue and views. J. Transl Med. 21 (1), 371 (2023).
Li, N. et al. Overexpressed PLAGL2 transcriptionally activates Wnt6 and promotes cancer development in colorectal cancer. Oncol. Rep. 41 (2), 875–884 (2019).
Wu, L. et al. PLAGL2 promotes epithelial-mesenchymal transition and mediates colorectal cancer metastasis via β-catenin-dependent regulation of ZEB1. Br. J. Cancer. 122 (4), 578–589 (2020).
Yang, T. et al. Selenium sulfide disrupts the PLAGL2/C-MET/STAT3-induced resistance against mitochondrial apoptosis in hepatocellular carcinoma. Clin. Transl Med. 11 (9), e536 (2021).
Hu, W. et al. PLAGL2-EGFR-HIF-1/2α signaling loop promotes HCC progression and erlotinib insensitivity. Hepatology 73 (2), 674–691 (2021).
Wu, L. et al. PLAGL2 promotes the proliferation and migration of gastric cancer cells via USP37-mediated deubiquitination of Snail1. Theranostics. 11(2): 700–714. (2021).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53 (D1), D672–D677 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28 (11), 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Zhou, G., Yang, L., Lu, Y. & Lu, G. Prognostic value of hemoglobin to red blood cell distribution width ratio in pancreatic ductal adenocarcinoma: a retrospective study. BMC Gastroenterol. 24 (1), 288 (2024).
Del Campo-Pedrosa, R., Martín-Carnicero, A., González-Marcos, A. & Martínez, A. New model to predict survival in advanced pancreatic ductal adenocarcinoma patients by measuring GGT and LDH levels and monocyte count. Front. Oncol. 14, 1411096 (2024).
Wu, G. et al. LncRNA BCAN-AS1 stabilizes c-Myc via N(6)-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer. Cell. Death Differ. 30 (10), 2213–2230 (2023).
Jafari, S. et al. The roles of LncRNAs and MiRNAs in pancreatic cancer: a focus on cancer development and progression and their roles as potential biomarkers. Front. Oncol. 14, 1355064 (2024).
Cui, M. et al. Cancer-cell-derived sialylated IgG as a novel biomarker for predicting poor pathological response to neoadjuvant therapy and prognosis in pancreatic cancer. Int. J. Surg. 109 (2), 99–106 (2023).
Long, A. W. et al. Heterodimerization of T cell engaging bispecific antibodies to enhance specificity against pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 17 (1), 20 (2024).
Wang, G. et al. High expression of PLAGL2 is associated with poor prognosis in High-Grade glioma. Front. Genet. 12, 787746 (2021).
Guo, J., Wang, M., Wang, Z. & Liu, X. Overexpression of pleomorphic adenoma Gene-Like 2 is a novel poor prognostic marker of prostate Cancer. PLoS One. 11 (8), e0158667 (2016).
Liu, X. et al. DNA-methylation-mediated Silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell. Death Dis. 9 (10), 1037 (2018).
Fujii-Nishimura, Y. et al. Mesenchymal-epithelial transition of pancreatic cancer cells at perineural invasion sites is induced by Schwann cells. Pathol. Int. 68 (4), 214–223 (2018).
Deng, D. et al. NFAT5 governs cellular plasticity-driven resistance to KRAS-targeted therapy in pancreatic cancer. J. Exp. Med. 221 (11), e20240766 (2024). [pii].
Wu, R. et al. Gasdermin C promotes stemness and immune evasion in pancreatic Cancer via Pyroptosis-Independent mechanism. Adv. Sci. (Weinh) : e2308990. (2024).
Author information
Authors and Affiliations
Contributions
Yang.Y.H and Wang.H wrote the content of the article.Wang.H and Xing.Z.H completed the experiment shown in Figs. 2, 3, 4, 5 and 6.Bai.Q.Q、Zhang.C and Liu.Y.B completed specimen collection and bioinformatics analysis.Liu.W.F and Li.D.H collected clinical data of patients and analyzed them in Table 1.Zhang.S.M and Liu.F.F completed immunohistochemical scores and data analysis in Table 2.Fan.H guides subject selection and coordinates the project as a whole.All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Human and animal rights
The clinical information and pathological materials used in this study have been approved by the relevant ethics committee. Animal experiments are not involved in this study.
Informed consent
Informed consent was obtained from patients and their families.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, YH., Wang, H., Xing, ZH. et al. PLAGL2 as a prognostic biomarker and an EMT-promoting factor in PDAC. Sci Rep 15, 25425 (2025). https://doi.org/10.1038/s41598-025-09591-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-09591-x