Abstract
This study aims to investigate the role of MSH6 in the diagnosis and prognosis of bladder cancer and its association with immunity. Various analyses were conducted on The Cancer Genome Atlas (TCGA) data, and the results were validated using the Gene Expression Omnibus (GEO) database. The potential mechanism of MSH6 in bladder cancer was revealed using R tools, Gene Set Enrichment Analysis (GSEA), and Gene Ontology (GO) analysis. Bladder cancer patients admitted to the Affiliated Hospital of Inner Mongolia Medical University from June 2023 to December 2023 were enrolled. Pathological specimens of bladder cancer and adjacent tissues were obtained. Immunohistochemical results showed the expression of MSH6 in bladder cancer tissues was higher than in adjacent tissues. High expression of MSH6 was identified as an independent prognostic factor for bladder cancer. The occurrence of bladder cancer was also influenced by age, pathological T stage, and pathological stage. According to the Kaplan–Meier survival curve, the overall survival rate of the MSH6 high-expression group was lower (P = 0.004). After MSH6 was knocked down in cell experiments, the proliferation, colony formation, and migration abilities of bladder cancer cells were significantly inhibited. MSH6 is significantly associated with the diagnosis and prognosis of bladder cancer, suggesting its potential utility as a biomarker for these purposes. MSH6 is closely related to the tumor immune microenvironment, implying a significant role in immunotherapy.
Similar content being viewed by others
Introduction
Bladder cancer remains the most common malignant tumor of the urinary system. In 2018, 549,393 individuals were diagnosed with bladder cancer, and 199,922 deaths were recorded worldwide1. The most common pathological type of bladder cancer is urothelial carcinoma, accounting for 90% of all cases. Other types, including squamous cell carcinoma and adenocarcinoma, account for approximately 10%2. At diagnosis, 75% of bladder cancers are confined to the mucosal urothelium and are classified as non-muscle-invasive diseases. In the remaining 25–30% of patients, bladder cancer invade the deep layer of the bladder wall or metastasize3. Therefore, more effective biomarkers need to be identified for early diagnosis of bladder cancer. The human MSH6 gene is a mismatch repair (MMR) gene located on chromosome 2. This gene has a length of 23,806 bp and includes 10 exons and 83 untranslated regions. This gene encodes the MSH6 protein, which plays an important role in DNA repair4. Current research indicates that individuals carrying pathogenic MMR variants have a high risk of developing early-stage cancers, particularly colorectal and endometrial cancers5,6. Moreover, abnormal expression of MSH6 is associated with an increased risk of ovarian, gastric, hepatobiliary, small bowel, pancreatic, breast, prostate, and adrenocortical cancers, and glioblastoma7,8,9,10,11,12,13,14. However, the role of MSH6 in bladder cancer remains unclear.
The Cancer Genome Atlas (TCGA) data were used to analyze MSH6 expression in bladder cancer samples, and the Human Protein Atlas (HPA) was used to validate protein expression. The relationship between MSH6 expression and prognostic and clinical parameters was investigated using R (version 4.2.1). To clarify the regulatory mechanism of MSH6 in bladder cancer, GSEA and GO analyses were performed. Additionally, R software was utilized to examine associations between MSH6 and the tumor microenvironment and immune system. A nomogram was developed to predict overall survival (OS) in bladder cancer patients. Calibration curves and receiver operating characteristic (ROC) curves were constructed to evaluate the accuracy of the nomogram. Finally, we explored the genetic alterations in MSH6 in bladder cancer samples.
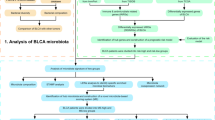
After an in-depth analysis of the association between MSH6 expression and bladder cancer, it was found that elevated levels of MSH6 were associated with poor overall survival. GSEA indicated that enriched pathways were closely related to the immune and central nervous systems. In addition, we found a negative correlation between MSH6 and NK cells, CD56bright T cells, plasmacytoid dendritic cells, immature dendritic cells, mast cells, and T helper cells 17. Thus, potential mechanisms and the role of MSH6 in bladder cancer were partly elucidated. The work design and flow chart of this study are shown in Figure S1.
Materials and methods
Data collection and differential expression analysis
Data on differentially expressed MSH6 from TCGA cancerous tissues were collected and analyzed using the UCSC Xena database (https://xenabrowser.net/datapages/).
MSH6 expression levels and associated clinical data were retrieved from the TCGA bladder cancer database. To verify MSH6 mRNA expression in bladder cancer patients, the original gene profile (GSE3167) was obtained from the Gene Expression Omnibus (GEO) database. Visual differential expression analysis was performed using the “ggplot2” R package (version 4.2.1; https://www.r-project.org/). The HPA (http://www.proteinatlas.org) database contains human protein expression and location information for various tissues and organs. Online immunohistochemical staining data from the HPA were used to assess differences in MSH6 protein expression between normal and bladder cancer tissues. A clinicopathological scatter plot was created to visualize the relationship between MSH6 expression and clinicopathological parameters. Logistic regression analysis was conducted with R software to investigate associations between clinical characteristics and MSH6 expression.
Survival analysis
To determine the independent prognostic factors for MSH6, univariate and multivariate Cox regression analyses were performed using the R package survival, and the results were visualized as forest plots using ggplot2. Risk factor maps were drawn based on risk scores and survival status obtained from Cox regression analysis. The predictive power of MSH6 in bladder cancer was evaluated by ROC curve analysis using pROC and visualized with ggplot2. Kaplan–Meier survival analysis was conducted using the R package survminer to assess the relationship between MSH6 expression and patient survival. P < 0.05 was considered to indicate statistical significance.
For overall survival prediction, a Cox regression-based nomogram was constructed using the rms and survival ROC packages in R. The area under the curve (AUC) values were calculated to evaluate the model’s performance. To evaluate the performance of the constructed nomogram, calibration curves were also used.
Functional enrichment analysis
Differential expression analysis between high and low MSH6 expression groups in bladder cancer tissues from TCGA was performed using the DESeq2 R package. P < 0.05 was considered to indicate statistical significance. GO enrichment analysis, including biological processes (BP), molecular functions (MF), and cellular components (CC), was conducted using the clusterProfiler package. P < 0.05 was considered to indicate statistical significance. In addition, GSEA of standardized MSH6 RNA-Seq data from the TCGA cohort was performed with cluster Profiler.
Immune correlation analysis
The correlation between MSH6 expression and immune infiltration was analyzed using the GSVA R package and visualized as a lollipop plot. The TCGA data were divided into high- and low-expression groups based on the median MSH6 expression to investigate differences in immune infiltration between the groups. The TIMER database (https://cistrome.shinyapps.io/timer/) was used to evaluate the relationship of MSH6 with a variety of immune cells.
Genetic alteration analysis through cBioPortal
MSH6 genetic alterations, including alteration frequency, copy number alteration (CNA), structural variations, and mutation types, were explored using the cBioPortal database (http://cbioportal.org). Kaplan–Meier plots were generated to illustrate the relationship between MSH6 gene alterations and survival outcomes in bladder cancer patients.
Protein–protein interaction (PPI), tumor mutational burden (TMB), and microsatellite instability (MSI)
Using the online STRING (https://string-db.org/) database, an analysis of the PPI network was also carried out to find potential relationships between MSH6 and other genes in bladder cancer by querying MSH6 gene in Homo sapiens. In order to explore the relationship between MSH6 gene expression and TMB or MSI, respectively, correlation analyses were conducted by querying MSH6 gene in single-gene pan-cancer analysis tool of Sangerbox.
Immunohistochemical analysis
Paraffin-embedded sections from patients admitted to the Affiliated Hospital of Inner Mongolia Medical University from June 2023 to December 2023, who underwent surgical treatment and were pathologically confirmed to have bladder cancer after surgery, were collected. These sections included both cancer tissues and adjacent normal tissues. This study strictly adhered to the principles of medical ethics and was authorized and supervised by the Ethics Committee of Inner Mongolia Medical University (No. YKD202401093).
Immunohistochemical staining was conducted, and the percentage of positive cells was calculated. Immunohistochemical staining was performed according to the manufacturer’s instructions, and the MaxVision two-step immunohistochemical staining was also used in this study. The staining scores were determined by evaluating membrane staining in tumor cells. According to the 2018 ASCO/CAP criteria, the IHC scores were classified as negative (score of 0 or 1 +), equivocal (score of 2 +), or positive (score of 3 +)15.
Results were observed under a laser scanning confocal microscope, and images were acquired using an image analysis system. Tissue staining data were obtained using ImageJ 1.8 software, and average optical density (AOD) was calculated. Data analysis was performed with GraphPad Prism 10.3.1, and results were presented as the mean of each group. A paired T test was applied for comparisons between groups, and differences were considered statistically significant at P < 0.05.
Cell experiments
Human bladder cancer cell lines (5637, J82, and SV-HUC-1) were purchased from Haixing Biology (Suzhou, China). Cells were cultured in specialized medium provided by Haixing at 37 °C with 5% CO2. MSH6 expression was silenced using small interfering RNA (siRNA) transfection technology. RNA extraction, reverse transcription, and real-time quantitative PCR were performed on the three cell lines. Subsequently, cell scratch assays, CCK-8 cell proliferation assays, and colony formation assays were conducted. Data were analyzed using Graphpad Prism 10.3.1 and expressed as \(\overline{x }\) ± s for each group. One-way ANOVA was used for multiple comparisons among groups, and T tests were performed for comparisons between two normally distributed groups.
Results
MSH6 expression analysis
We analyzed pancancer data from the TCGA and GTEx data to assess MSH6 expression. MSH6 expression was elevated in 22 cancer types and reduced in 5 compared to normal tissues (Fig. 1a). Based on TCGA-BLCA data, MSH6 expression levels in 19 paired samples, 19 normal tissues, and 412 bladder cancer samples were plotted (Fig. 1b, d). The analysis showed significantly increased MSH6 expression in bladder cancer tissues (P < 0.05). These findings were validated using microarray data from GEO database GSE3167, confirming higher MSH6 expression in bladder cancer compared to normal tissues (Fig. 1c). Immunohistochemistry data from the HPA database further confirmed these results (Fig. 1e–h).
MSH6 expression analysis. (a) MSH6 expression in normal and tumor tissues in the TCGA and GTEx pancancer datasets (***P < 0.001; **P < 0.01; *P < 0.05). (b) Expression of MSH6 in an unpaired bladder cancer sample. c. MSH6 expression in the GSE3167 database. (d) Expression of MSH6 in paired bladder cancer samples. (e, f) Representative image of an immunohistochemical image showing the expression of MSH6 in bladder cancer tissues. (g, h) Representative image of the immunohistochemical analysis showing MSH6 expression in normal bladder tissue.
Relationships between MSH6 expression and clinicopathological parameters
Various clinicopathological parameters of bladder cancer patients were compared with MSH6 expression. Results showed that MSH6 expression was significantly associated with race (P < 0.01; Fig. 2a), subtype (P < 0.001; Fig. 2b), and histological grade (P < 0.001; Fig. 2c). Logistic regression analysis indicated that MSH6 expression was significantly correlated with primary therapy outcomes (PR&CR vs. PD&SD, P = 0.013) and histologic grade (low grade vs. high grade, P = 0.003) (Table 1).
Survival analysis
As shown in Table 2, we explored the relationship between MSH6 expression and overall survival in patients with bladder cancer using Cox regression analysis. Univariate Cox regression analysis showed that age (HR = 1.424, P = 0.018), pathological T stage (HR = 2.157, P < 0.001), pathological stage (HR = 1.677, P = 0.016), and MSH6 expression (HR = 1.551, P = 0.004) were significantly associated with OS. Multivariate Cox regression analysis (Fig. 3a, forest plot) revealed MSH6 expression (P = 0.012) as an independent prognostic factor. The distribution of MSH6 expression, patient survival status, and predicted risk scores based on the MSH6 Cox model are shown in Fig. 3b. In addition, according to the K‒M survival curve, patients in the MSH6 high-expression group had a lower overall survival rate (P = 0.004; Fig. 3c). The ROC curve indicated that MSH6 had good prognostic capability (AUC = 0.615) (Fig. 3d).
Survival analysis of patients stratified by MSH6 expression. (a) Multivariate Cox regression analysis of MSH6 expression and other clinicopathological variables. (b) MSH6 expression distribution and survival status (0 = survival; 1 = death). c. MSH6 mRNA expression levels and overall survival. d. ROC curve for MSH6.
Using this nomogram, survival rates for bladder cancer patients at 1, 3, and 5 years could be predicted with increased accuracy (Fig. 4a). The AUC values of the nomogram were 0.583, 0.555, and 0.516, indicating moderate prediction accuracy (Fig. 4b). Our nomogram calibration curves were plotted at 1, 3, and 5 years (Fig. 4c–e).
MSH6 enrichment analysis
GO and GSEA were used to explore the potential biological functions of MSH6. As shown in Fig. 5a, GO terms related to BP, MF, and CC closely associated with MSH6 included axon development, cell fate, neurons, intermediate filaments, keratin fibers, serine hydrolase activity, serine-type peptidase activity, and serine-type endopeptidase activity. As shown in Fig. 5b, GSEA results indicated that genes within KEGG and Reactome pathways were downregulated16,17,18.
Functional and pathway enrichment analysis of MSH6 in bladder cancer. (a) Important Gene Ontology terms associated with MSH6, including biological process (BP), cellular component (CC), and molecular function (MF)(www.kegg.jp/kegg/kegg1.html). (b) Significant GSEA results associated with MSH6, including KEGG pathway and REACTOME pathway enrichment analyses (www.kegg.jp/kegg/kegg1.html).
The relationship between MSH6 expression and immune system and tumor microenvironment
We investigated the different immune microenvironment in patients with bladder cancer with different tumor MSH6 levels. As shown in Fig. 6a, compared to the low-expression group, the proportions of T helper 2 cells, helper T cells, central memory T cells, T helper 1 cells, aDCs, macrophages (P < 0.001), Tgd (P = 0.0131), regulatory T cells (P = 0.0003), effector memory T cells (P = 0.0022), NK cells (CD56dim subsets) (P = 0.0032), T cells(P = 0.00213), and T follicular helper cells (P = 0.00213) were increased in the high-expression group. As shown in Fig. 6b, MSH6 expression levels correlated with the activation of dendritic cells, macrophages, helper T cells, central memory T cells, TGDS cells, T helper 1 cells, T helper 2 cells (P < 0.001), NK cells, the CD56dim subset (P = 0.0048), effector memory T cells (P = 0.0011), regulatory T cells (P = 0.0006), NK cells, the CD56bright subset, plasmacytoid dendritic cells (P < 0.001), immature dendritic cells (P = 0.0118), mast cells (P = 0.0016) and T helper cell 17 (P = 0.0206). Based on these results, MSH6 may play a key role in the immune infiltration of bladder cancer. To investigate the association between MSH6 expression and immune cell infiltration in bladder cancer, correlation analyses were performed using the TIMER database. We found that the expression of MSH6 was positively correlated with the infiltration of CD4+ T cells, CD8+ T cells, neutrophils, macrophages, dendritic cells, and B cells (P < 0.001) (Fig. 6c).
MSH6 expression is correlated with the immune system and tumor microenvironment. (a) The varied proportions of 24 subtypes of immune cells in high and low MSH6 expression groups in tumor samples. (b) Correlation between MSH6 expression and the level of immune infiltration. (c) Correlation between the expression of MSH6 and immune cell infiltration.
Genetic variant analysis
Mutation characteristics of MSH6 in bladder cancer patients from TCGA were explored using the cBioPortal database, showing a genetic alteration frequency of 6.08% (Fig. 7a). As shown in Fig. 7b, alterations in the MSH6 gene were not significantly associated with patient survival outcomes. Figure 7c illustrates the mutation sites of MSH6 in bladder cancer.
Mutational signature of MSH6 in patients with bladder cancer in the TCGA cohort determined using the cBioPortal tool. (a) Type of MSH6 in patients with bladder cancer from the TCGA cohort and the frequency of mutation changes. (b) K‒M survival analysis of OS with or without MSH6 alterations. (c) MSH6 mutation sites in bladder cancer.
Relationships between MSH6 and PPI, MSI and TMB in bladder cancer
With the help of the online STRING database, MSH6 was analyzed to find possible relationships with other genes in bladder cancer using PPI network analysis (Fig. 8a). According to Fig. 8b,c, our results revealed that MSH6 expression was not related to MSI and TMB.
Detection of MSH6 expression level in paired tissues by immunohistochemical staining
Bladder cancer tissues collected at the Affiliated Hospital of Inner Mongolia Medical University were further verified by detecting MSH6 expression level through immunohistochemical staining. A total of 8 pairs of bladder cancer tissues and adjacent tissues were collected (Fig. 9a). Quantitative analysis (Fig. 9b) showed that the expression level of MSH6 in bladder cancer tissues was significantly higher than in adjacent tissues (0.36 ± 0.03 vs. 0.49 ± 0.1, P < 0.01).
The biological function of MSH6 in bladder cancer cells
We determined MSH6 expression levels in cells using real-time fluorescence quantitative PCR.As shown in Fig. 10a, MSH6 expression in bladder cancer cell lines was significantly higher than in normal urothelial cells. After knockdown of MSH6 using siRNA, proliferation, colony formation, and migration of bladder cancer cells were significantly inhibited. Figure 10b,c further confirmed the crucial role of MSH6 in bladder cancer progression. As shown in Fig. 10d–g, Cell proliferation and colony formation abilities decreased significantly following MSH6 knockdown. Scratch assay results (Fig. 10d–k) demonstrated a significant reduction in the migration ability of bladder cancer cells after MSH6 knockdown.
The biological function of MSH6 in bladder cancer cells. (a) The expression level of MSH6 mRNA in normal bladder epithelial cell lines, J82 and 5637 cell lines (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (b) qRT-PCR detection of MSH6 mRNA expression in 5637 cell lines after knockdown (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (c) qRT-PCR detection of MSH6 mRNA expression in J82 cell lines after knockdown (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (d) CCK-8 assay to detect the proliferation ability of 5637 cells after si-NC and si-MSH6 treatment. (e) CCK-8 assay to detect the proliferation ability of J82 cells after si-NC and si-MSH6 treatment. (f–g) Clonogenic assay of 5637 cells. (h–i) Scratch assay to detect the migration ability of 5637 cells after si-NC and si-MSH6 treatment (*P < 0.05; **P < 0.01). (j–k) Scratch assay to detect the migration ability of J82 cells after si-NC and si-MSH6 treatment (*P < 0.05; **P < 0.01).
Discussion
MSH6 is a very attractive biomarker in cancer diagnosis; for example, the MSH6 protein exhibits low expression in glioblastoma19 and endometrial cancer20 but high expression in prostate cancer21. According to our results, MSH6 expression was lower in five types of cancer tissues and higher in 22 types, including bladder cancer, compared to normal tissues.
In this article, we analyzed the differential expression of MSH6 in bladder cancer using multiple public databases. To further verify MSH6 expression in bladder cancer, we collected eight pathological specimens, including adjacent tissues. Immunohistochemical analysis revealed high expression of MSH6 in bladder cancer tissues. MSH6 expression is associated with various tumor characteristics, such as race, subtype, and histological grade. Nomograms have been widely used to aid in clinical decision-making. GO enrichment and GSEA analyses indicated that MSH6 upregulation was closely associated with signaling pathways, the immune system, and the central nervous system, particularly the Wnt signaling pathway. The Wnt signaling pathway is activated in various cancers and contributes to tumorigenesis22.
We further explored the potential association between MSH6 and the immune system, primarily regarding immune infiltration and the tumor microenvironment. The tumor microenvironment is a highly complex system23,24. The tumor microenvironment can facilitate tumor growth, metastasis, and resistance to chemotherapy and immunotherapy25,26. Th1, Th2 and Th17 cells are mostly differentiated from naive CD4+ T cells27. Th1 and Th2 cells participate in cellular immunity and humoral immunity, respectively, by secreting various specific cytokines28. CD8+ T cells mediate cytotoxic activity mainly by inducing tumor cell apoptosis29.
Human NK cells are typically classified into CD56brightCD16- and CD56dimCD16+ subsets30, with CD56dimCD16+ NK cells exhibiting strong cytotoxic activity upon stimulation by target cells31. In the TCGA cohort, the frequency of genetic alterations in MSH6 was 6.08%. However, no association was observed between genetic alterations of MSH6 and bladder cancer prognosis. We utilized the online STRING database to conduct PPI network analysis on MSH6 in the context of bladder cancer. This approach allowed exploration of potential relationships between MSH6 and other genes. These mutations can hinder tumor evolution by triggering an immune response to the tumor32. Cancer immunotherapy involves using drugs to activate or enhance the immune system’s natural mechanisms to attack cancer cells33,34. Therefore, immunotherapy is considered a promising strategy for treating or even curing certain types of cancer.
In our MSH6 expression analysis, significant differences were identified between bladder cancer cell lines (5637 and J82) and normal urothelial cells (SV-HUC-1). These findings suggest that abnormal MSH6 expression may be closely related to bladder cancer occurrence. Biological function experiments further confirmed the critical role of MSH6 in bladder cancer. The CCK-8 proliferation assay, colony formation assay, and scratch assay showed significant inhibition of proliferation, colony formation, migration, and invasion abilities in bladder cancer cells (5637 and J82) following MSH6 knockdown by siRNA. These findings indicate that MSH6 plays a crucial role in promoting the proliferation and self-renewal of bladder cancer cells.
However, this study has limitations. First, animal model experiments were not conducted to further verify biological effects in vivo. Second, immune-related experiments were limited, and the relationship between MSH6 and the immune microenvironment will be explored in future studies. Additionally, only selected bladder cancer cell lines were analyzed; therefore, the expression and functional differences of MSH6 in other cell lines across different clinical stages and pathological types require further investigation.
Conclusions
In summary, our findings indicate that MSH6 may have significant roles in the immune system and bladder cancer prognosis. MSH6 significantly promotes the proliferation, self-renewal, migration, and invasion of bladder cancer cells, suggesting its potential as a therapeutic target. Ultimately, we anticipate that our results will provide new insights into bladder cancer immunotherapy for future research.
Data availability
Data is provided within the manuscript or supplementary information files.
Abbreviations
- TCGA:
-
The Cancer Genome Atlas
- BLCA:
-
Bladder cancer
- GEO:
-
Gene Expression Omnibus
- GSEA:
-
Gene Set Enrichment Analysis
- GO:
-
Gene Ontology
- HPA:
-
Human Protein Atlas
- OS:
-
Overall survival
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- AOD:
-
Average optical density
- BP:
-
Biological process
- MF:
-
Molecular function
- CC:
-
Cellular component
- BP:
-
Biological process
- MF:
-
Molecular function
- CAN:
-
Copy number alteration
- K‒M:
-
Kaplan–Meier survival curve
- siRNA:
-
Small interfering RNA
- Treg:
-
Regulatory T cells
- Th1 cells:
-
Type 1 Th cells
- aDC:
-
Activated dendritic cells
- Th2 cells:
-
Type 2 Th cells
- Tfh:
-
T follicular helper
- Tem:
-
T effector memory
- pDC:
-
Plasmacytoid dendritic cell
- NK cells:
-
Natural killer cells
- Th17 cells:
-
Type 17 Th cells
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Iwata, T. et al. The role of adjuvant radiotherapy after surgery for upper and lower urinary tract urothelial carcinoma: A systematic review. Urol. Oncol. 37, 659–671 (2019).
Teoh, J. Y. et al. Global trends of bladder cancer incidence and mortality, and their associations with tobacco use and gross domestic product per capita. Eur. Urol. 78, 893–906 (2020).
Szadkowski, M. & Jiricny, J. Identification and functional characterization of the promoter region of the human MSH6 gene. Genes Chromosomes Cancer 33, 36–46 (2002).
Møller, P. The prospective lynch syndrome database reports enable evidence-based personal precision health care. Hered Cancer Clin. Pract. 18, 6 (2020).
Wang, C., Wang, Y., Hughes, K. S., Parmigiani, G. & Braun, D. Penetrance of colorectal cancer among mismatch repair gene mutation carriers: A meta-analysis. JNCI Cancer Spectr. 4, 27 (2020).
Win, A. K. et al. Risks of primary extracolonic cancers following colorectal cancer in lynch syndrome. J. Natl. Cancer Inst. 104, 1363–1372 (2012).
Braun, M. M., Overbeek-Wager, E. A. & Grumbo, R. J. Diagnosis and management of endometrial cancer. Am. Fam. Physician 93, 468–474 (2016).
Watson, P. et al. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int. J. Cancer 123, 444–449 (2008).
Kastrinos, F. et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA 302, 1790–1795 (2009).
Raymond, V. M. et al. Elevated risk of prostate cancer among men with Lynch syndrome. J. Clin. Oncol. 31, 1713–1718 (2013).
Ryan, S., Jenkins, M. A. & Win, A. K. Risk of prostate cancer in Lynch syndrome: A systematic review and meta-analysis. Cancer Epidemiol. Biomark. Prev. 23, 437–449 (2014).
Senter, L. et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 135, 419–428 (2008).
Giardiello, F. M. et al. Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology 147, 502–526 (2014).
Gordian-Arroyo, A. M., Zynger, D. L. & Tozbikian, G. H. Impact of the 2018 ASCO/CAP HER2 guideline focused update. Am. J. Clin. Pathol. 152, 17–26 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Felsberg, J. et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int. J. Cancer 129, 659–670 (2011).
Kim, J. et al. DNA mismatch repair protein immunohistochemistry and MLH1 promotor methylation testing for practical molecular classification and the prediction of prognosis in endometrial cancer. Cancers (Basel) 10, 279 (2018).
Albero-González, R. et al. Immunohistochemical expression of mismatch repair proteins (MSH2, MSH6, MLH1, and PMS2) in prostate cancer: Correlation with grade groups (WHO 2016) and ERG and PTEN status. Virchows Arch. 475, 223–231 (2019).
Fan, G. et al. RTL1 promotes melanoma proliferation by regulating Wnt/β-catenin signalling. Oncotarget 8, 106026–106037 (2017).
Giraldo, N. A. et al. The clinical role of the TME in solid cancer. Br. J. Cancer 120, 45–53 (2019).
Hinshaw, D. C. & Shevde, L. A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 79, 4557–4566 (2019).
Wu, H. W. et al. Anti-CD105 antibody eliminates tumor microenvironment cells and enhances anti-GD2 antibody immunotherapy of neuroblastoma with activated natural killer cells. Clin. Cancer Res. 25, 4761–4774 (2019).
Liu, H. et al. Global, regional, and national mortality trends of female breast cancer by risk factor, 1990–2017. BMC Cancer 21, 459 (2021).
Zhu, J. T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb. Perspect. Biol. 10, a030338 (2018).
Zhang, Y., Zhang, Y., Gu, W., He, L. & Sun, B. Th1/Th2 cell’s function in immune system. Adv. Exp. Med. Biol. 841, 45–65 (2014).
Farhood, B., Najafi, M. & Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell Physiol. 234, 8509–8521 (2019).
Cooper, M. A. et al. Human natural killer cells: A unique innate immunoregulatory role for the CD56(bright) subset. Blood 97, 3146–3151 (2001).
Luetke-Eversloh, M. et al. NK cells gain higher IFN-γ competence during terminal differentiation. Eur. J. Immunol. 44, 2074–2084 (2014).
Lakatos, E. et al. Evolutionary dynamics of neoantigens in growing tumors. Nat. Genet. 52, 1057–1066 (2020).
Rosenberg, S. A. IL-2: the first effective immunotherapy for human cancer. J. Immunol. 192, 5451–5458 (2014).
Ahmed, S. & Rai, K. R. Interferon in the treatment of hairy-cell leukemia. Best Pract. Res. Clin. Haematol. 16, 69–81 (2003).
Funding
This work was supported by the General Project of Inner Mongolia Natural Science Foundation (No. 2022MS08063), the Key Research and Development and Achievement Transformation Project in the Social Welfare Field of the 14th Five-Year Plan in the Inner Mongolia Autonomous Region (No. 2022YFSH0087), Science and Technology Program of the Joint Fund of Scientific Research for the Public Hospitals of Inner Mongolia Academy of Medical Sciences (2024GLLH0337), Inner Mongolia Medical University Affiliated Hospital Talent Training Project—Sailing Series, the Inner Mongolia Health Science and Technology Project in 2022 (No. 202201293), the National Natural Science Foundation of China (No. 81960143), the Inner Mongolia Grassland Talents Program Young Innovative Talent Project (No. Q2022082), the Shanghai Key Laboratory of Kidney and Blood Purification (No. 14DZ226022, 20DZ2271600), the Science and Technology Commission of Shanghai, and the Trinity College Students Innovation and Entrepreneurship Cultivation Project of Inner Mongolia Medical University (No.SWYT2020008).
Author information
Authors and Affiliations
Contributions
Ning He analyzed the data and wrote the manuscript. Yan Meng designed the experiments. Lu-Ri Bao was responsible for data curation. Peng-cheng Ma conducted the cell experiments and immunohistochemical experiments. Yan Meng reviewed and approved the manuscript. Zhen-Li Fu, Li-Gang Chen, Jing-Yuan Wang, Wu-niri Gao were responsible for the project administration. Yan Meng supervised the experiments.
Corresponding author
Ethics declarations
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Inner Mongolia Medical University (No. YKD202401093). The patients/participants provided their written informed consent to participate in this study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, N., Bao, Lr., Ma, Pc. et al. MSH6 as a prognostic biomarker in bladder cancer and its correlation with immunity. Sci Rep 15, 24837 (2025). https://doi.org/10.1038/s41598-025-09644-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-09644-1