Abstract
Colorectal cancer is one of the common malignant tumors nowadays, with the highest mortality rate among cancers, and most patients have metastasis when diagnosed. Exosomes contain bioactive molecules such as RNA, which can participate in signal transduction between cells. tRF is a new type of non-coding small RNA, which plays an important role in the progression of cancer. The purpose of this study is to explore the value of exosomal tRF as a biological diagnostic marker for colorectal cancer. Through the use of transmission electron microscopy (TEM), qNano, and western blot, exosomes obtained from the serum of both healthy people and patients with colorectal cancer were characterized; the expression of tRNA-Gly-5-0007 was validated by real-time quantitative PCR (qRT-PCR) in the exosome of 172 colorectal cancer patients and 164 healthy people. The diagnostic efficacy of tsRNA-Gly-5-0007 as a diagnostic marker for colorectal cancer was evaluated by receiver operating characteristic curve (ROC) analysis. We further conducted a preliminary verification of how tsRNA-Gly-5-0007 participates in the malignant phenotype of tumors through transwell experiments and cell adhesion experiments. The TCGA database shows that tsRNA-Gly-5-0007 is an RNA fragment derived from tsRNA-Gly-CCC-2-1. The expression of tsRNA-Gly-CCC-2-1 is down-regulated in colorectal cancer tissues and may be involved in Wnt, mTOR and other signaling pathways. tsRNA-Gly-5-0007 can inhibit the adhesion and motor capacity of colorectal cancer cells. Compared with healthy people, the expression of exosomal tsRNA-Gly-5-0007 was significantly down-regulated in patients with colorectal cancer, especially in patients with early colorectal cancer. The diagnostic efficacy of exosomal tsRNA-Gly-5-0007 for the diagnosis of colorectal cancer was 0.7812, and the diagnostic efficacy for the diagnosis of early colorectal cancer was 0.7726. tsRNA-Gly-5-0007 may affect the adhesion and motor capacity of colorectal cancer cells through Wnt signaling pathway, and then participate in the malignant process of colorectal cancer cells. Moreover, the expression of exosomal tsRNA-Gly-5-0007 is down-regulated in colorectal cancer patients and can be used as a promising biomarker for the biological diagnosis of colorectal cancer.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) ranks among the most prevalent malignant neoplasms globally, characterized by high incidence and mortality rates. A substantial proportion of patients present with metastatic disease at initial diagnosis, leading to unfavorable therapeutic outcomes and diminished survival rates1. Early detection during the initial stages of tumorigenesis, followed by timely clinical intervention, could significantly improve prognosis. Although carcinoembryonic antigen (CEA) and carbohydrate antigen 19 − 9 (CA19-9) are among the most widely utilized tumor biomarkers in current clinical practice, their diagnostic utility remains limited by suboptimal specificity and sensitivity, particularly in early-stage CRC2,3,4. Consequently, the identification of more reliable biomarkers for early CRC detection represents a critical unmet need in clinical oncology.
Exosomes are a subclass of extracellular vesicles characterized by a lipid bilayer membrane structure, typically measuring 40–160 nm in diameter. They are ubiquitously distributed in various biological fluids, including blood, urine, and cerebrospinal fluid. Originating from parental cells, exosomes selectively encapsulate bioactive molecules derived from their cells of origin, thereby facilitating intercellular communication and modulating diverse biological processes. As such, they are regarded as pivotal mediators of cell-to-cell signaling. The exosomal surface harbors specific membrane proteins, including CD9, HSP70, and CD81, which serve as canonical markers for exosome identification. Furthermore, exosomes exhibit several advantageous properties for tumor biomarker applications, such as high abundance, exceptional stability, and non-invasive detectability5,6,7,8.
Transfer RNA (tRNA), a class of non-coding RNA (ncRNA) typically comprising 70–90 nucleotides, adopts a characteristic cloverleaf secondary structure. As a fundamental component of protein biosynthesis, tRNA delivers specific amino acids to their corresponding positions on the mRNA-ribosomal complex, thereby facilitating the synthesis of polypeptides with precise amino acid sequences9,10. Emerging evidence has identified tRNA-derived small RNAs (tsRNAs) as functionally important fragments generated through enzymatic cleavage of mature tRNAs or their precursors. These regulatory fragments have been implicated in diverse pathological processes, including immune dysregulation, metabolic disorders, and oncogenesis, positioning them as potential therapeutic targets for various diseases11,12,13,14. Based on their cleavage patterns and genomic origins, tsRNAs are primarily categorized into two subgroups: tRNA-derived fragments (tRFs) and tRNA halves (tiRNAs), which are produced through differential processing by specific endonucleases at distinct tRNA loci15,16,17.
The classification of tRNA-derived fragments (tRFs) is determined by their cleavage positions along the tRNA molecule, resulting in five major subtypes: tRF-5, tRF-3, tRF-2, tRF-1, and tRF-i. Functionally, tRFs exhibit miRNA-like properties by participating in RNA interference pathways. Specifically, tRFs form ribonucleoprotein complexes with Argonaute (AGO) proteins, which subsequently bind to the 3’ untranslated region (3’UTR) of target mRNAs to mediate post-transcriptional gene silencing11,18,19. Beyond their role in gene regulation, emerging evidence indicates that tRFs participate in critical cellular processes, including cell cycle control20,21 and epigenetic regulation22. These multifunctional molecules have thus emerged as important regulators of diverse biological pathways.
tRNA-derived fragments (tRFs) have emerged as significant regulators in tumorigenesis and cancer progression, demonstrating considerable potential as diagnostic biomarkers in oncology. Studies have shown that tRF can be important as a biological diagnostic marker in cancers such as gastric cancer9,23non-small cell lung cancer24 and breast cancer25. And exosomes can protect the RNA inside them from being degraded by RNases26. In recent years, exosome-associated tRFs have gained substantial attention as promising diagnostic biomarkers, with demonstrated potential for cancer screening, diagnosis, and therapeutic monitoring. However, to date, no studies have investigated the role of serum exosomal tsRNA-Gly-5-0007 in the early diagnosis of colorectal cancer, representing a significant gap in current research.
This study systematically investigated the functional role of exosomal tsRNA-Gly-5-0007 in modulating cellular adhesion and motility in colorectal cancer (CRC) cells. Furthermore, we comprehensively evaluated the differential expression patterns of serum exosomal tsRNA-Gly-5-0007 between CRC patients and healthy controls, along with its diagnostic potential for CRC. Our findings demonstrate that serum exosomal tsRNA-Gly-5-0007 exhibits significant potential as a novel diagnostic biomarker for early-stage colorectal cancer detection.
Materials and methods
serum samples from healthy people and colorectal cancer patients
The Affiliated Cancer Hospital of Shandong First Medical University provided plasma from 164 healthy volunteers and serum from 172 patients receiving initial treatment for colorectal cancer for this investigation. The Affiliated Cancer Hospital of Shandong First Medical University’s ethics committee examined and approved studies involving human samples while adhering to the principles of the Declaration of Helsinki (1964). The study’s participants, all of whom were receiving first treatment for colorectal cancer, had no prior anticancer therapy and no other immunological, endocrine, or metabolic disorders.
Serum exosome isolation
First, use the low-temperature centrifuge of Thermo Fisher Scientifc (Shanghai) Instruments Co., Ltd. for centrifugation. Centrifuge the EP tube containing serum at 2000 g at 4℃ for 10 min. After centrifugation, transfer the supernatant to a new EP tube and label the information. Then centrifuge at 10,000 g and 4 ℃ for 30 min to remove small cell fragments. After the centrifugation is completed, transfer the supernatant to the ultracentrifugation tube and mark the sample number and other information on the top of the ultracentrifugation tube wall for distinction. Finally, ultracentrifugation was carried out using the ultracentrifugation centrifuge of Beckman Coulter, Inc. in the United States. The ultracentrifugation tubes were placed into the rotor of the ultracentrifugation centrifuge in sequence according to the serial numbers, and attention was paid to balancing at the same time. In order to facilitate the observation of the position of exosome precipitation in the subsequent work, the ultracentrifugation conditions were set at 100,000 g, 4 ℃, and 2 h. After centrifugation is completed, the supernatant is discarded, and the resulting precipitate is the exosomes of the serum sample.
TEM (transmission electron microscopy)
Firstly, the required exosomes are obtained by ultracentrifugation, using the same method as above; Next, mix the exosomes evenly and centrifuge them instantaneously, and prepare the required copper mesh; Next, first add the filtered PBS to the exosome precipitate, gently pipette to resuspend it and mix well, and then use a pipette to draw 5µL of the exosome sample onto the copper mesh. Next, use filter paper to absorb the remaining exosome samples on the copper mesh. Then, use a pipette to draw the staining solution and carefully drop it onto the prepared copper mesh for staining. Next, use filter paper to absorb the remaining dye solution on the copper mesh. After completion, place the copper mesh in a storage box for later use. Finally, turn on the transmission electron microscope, observe and take photos, and save the pictures well.
qNano analysis
First of all, turn on the instrument (qNano: Izon Sciences Ltd, New Zealand, Model: S/N 601 A 17060506) in advance, prepare the exosome samples to be tested and the standard particle solution cpc100B, and place them on ice for later use. Next, open the computer software, mark the stretching length with a vernier caliper, moisten the nanoporous membrane at the same time, close the air valve, adjust the pressure rod, and apply pressure to 20mBar for one minute. Then, add PBS to the lower slot, install the components, add PBS to the upper slot, adjust the instrument voltage to about 0.5 V, and then adjust the voltage to stabilize the current signal at about 100nA. Determination of standard particles: Aspirate the PBS solution in the upper tank, add the standard particles, apply pressure, and click start when the current stabilizes. stop collecting immediately if there is a fluctuation. When 500 particles have been collected, click stop and record the data. Determination of the sample to be tested: Aspirate the upper bath solution, wash with PBS, add the exosome sample to be tested, apply pressure, and conduct the test under the same conditions using the determination standard particles. Repeat the steps and record the data. Analyze the data After entering the Izon Control Suite v.3.3.2.2001 software, associate the sample results with the standard particle results, analyze the data and generate pictures; Finally, turn off the computer and the instrument, disassemble the components and clean them, then reassemble the instrument and put it back in its original place.
Western blot
Proteins were separated by 10% SDS-PAGE (Dakewe Biotech, Shenzhen, China), transferred to PVDF membrane (Milipore, MA, USA), blocked with 5% nonfat dry milk about 3 h, incubated with anti-GM130 (1:250, Cell Signaling Technology, MA, USA), HSP70 (1: 500, Cell Signaling Technology, MA, USA), CD9 (1: 500, Proteintech, Wuhan, China), primary antibodies overnight at 4 °C. Then, the CD9 band and the HSP70 band were placed in the secondary antibody at a ratio of 1:4000, and the GM130 was placed in the secondary antibody at a ratio of 1:5000, and incubated at room temperature for 1 h. They were left at room temperature for 1 h. Finally the protein bands with ECL blot detection reagents (Thermo Fisher, MA, USA) and flms (FUJIFILM, TKY, Japan) were visualized.
RNA extraction and qRT-PCR
Firstly, exosomes were obtained by ultracentrifugation. Secondly, the total RNA of plasma exosomes was extracted using trizol reagent. Then, the RNA was reverse transcribed into cDNA using the miRNA First Strand Synthesis Kit (AG). Prepare the qPCR reaction system by mixing 2 µl of cDNA with 18 µl of the system, with a volume of 20 µl. The company designed and synthesized the tsRNA-Gly-5-0007 primer sequence as GCGCCGCTGGTGTAG, U6-F as TGGAACGCTTCACGAATTTGCG, and U6-R as GGAACGATACAGAGAAGATTAGC. The LightCycler480 system was used for qPCR analysis. The program was set to pre-denaturation (95 °C, 30 s), denaturation (95 °C, 5 s, 45 cycles), annealing (60 °C, 20 s), and elongation (65 °C, 15 s). Taking U6 as the control, the relative expression level of exosome tRF was calculated as 2^(-ΔΔCt), (∆Ct = CT (tRF) -CT (U6)). Each sample was analyzed repeatedly.
Cell adhesion assay
The HCT116, HT29 and SW480 cells involved in this study were derived from Cell Resource Center (http://crcpumc.com/login.jsp? url=%2Fmcart.jsp&errno = 11). Spread Matrigel into 96-well plates after diluting it with serum-free 1640 medium. Let it air dry in the incubator overnight. 5000 transfected cells were added to 96-well plates, which were then incubated at 37 °C with 5% CO2. The media was taken away at the correct time interval, and the non-adherent cells were removed by washing the cells three times in PBS. To test absorbance, add CCK8 reagent and incubate for 1 h.
Cell migration assay
Following 48 h of transfection, cells were digested into a cell suspension and counted. Then, 800ul of 1640 medium containing 15% serum was added to the transwell’s lower chamber, 200ul of serum-free cell suspension containing 80,000 cells was added to the transwell’s upper chamber, and the transwell was placed in an incubator for culture. The upper chamber was rinsed, dried, and positioned under the microscope for inspection after 30 min in the fixing solution and crystal violet solution to fix the staining.
Cell invasion assay
Spread Matrigel into the transwell’s upper chamber after diluting it with serum-free 1640 medium, and let it in the incubator overnight to air-dry. After being digested to create a cell suspension and being counted, the cells were transfected for 48 h. Following this, 800ul of 1640 medium containing 15% serum and 200ul of serum-free cell suspension containing 80,000 cells were added to the transwell, which was then placed in an incubator for incubation. After incubation, the upper chamber was cleaned, dried, and submerged under a microscope for 30 min in each of the fixing solution and crystal violet solution to fix the staining.
tsRNA-Gly-5-0007 information in the database
We obtained transcriptomic data and clinicopathological data containing survival information of the TCGA colorectal cancer cohort from the tRic database. Colorectal cancer plasma exosomal RNA data (GSE71008) were downloaded from the GEO database. In order to clarify the difference between tsRNA-Gly-CCC-2-1 expression in cancer and paracancer, we used the R software package “Limma” to conduct differential analysis. We then combined TCGA transcriptome data with clinical data based on sample names to explore the correlation between tsRNA-Gly-CCC-2-1 expression and various stages using willcoxon tests. In addition, we extracted tsRNA-Gly-CCC-2-1 expression from transcriptome data, integrated it with survival time and survival state, and performed survival analysis and visualization using R software package “Survival”.
GO and KEGG analysis
In order to clarify the potential molecular mechanisms and biological functions of tsRNA-Gly-CCC-2-1 involved in colorectal cancer progression, we first performed differential expression analysis in a colorectal cancer cohort using the R-package “Limma”, and those differences meeting P < 0.05 and logFC > 2 were identified as differentially expressed genes. Subsequently, we used the SVM-GA algorithm to obtain the potential target genes of tsRNA-Gly-CCC-2-1 on the website (http://trftar.cmuzhenninglab.org:3838/tar/#). Then we intersected the differentially expressed genes and potential target genes, and used the R package “clusterprofiler” to enrich the intersected genes by GO and KEGG.
Statistical analysis
Prism 9.0 was used to statistically evaluate the qRT-PCR experiment results. When the conditions for a normal distribution were met, the mean and standard deviation were expressed using the t-test, and the median and interquartile range were expressed using the Mann-Whitney test. The clinical diagnostic efficacy of tRF was evaluated by calculating ROC curves using Prism 9.0. The difference was considered statistically significant at P < 0.05. The tests were two-tailed.
Results
tsRNA-Gly-5-0007 is involved in the malignant progression of colorectal cancer
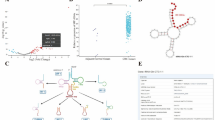
tsRNA-Gly-5-0007 is an RNA fragment derived from tsRNA-Gly-CCC-2-1. As shown in Fig. 1C, the tsRFun database (http://rna.edu.cn/tRFfinder/) of tsRNA-Gly-5-0007 at the tsRNA-Gly-CCC-2-1 cleavage site on the secondary structure was demonstrated. The TCGA database showed that the expression of tsRNA-Gly-CCC-2-1 was down-regulated in colorectal cancer tissues (Fig. 1A) and plasma exosomes of colorectal cancer (Fig. 1B). The tsRFun database showed that the expression of tsRNA-Gly-5-0007 was also down-regulated in colorectal cancer tissues (Fig. 1D), which was consistent with the TCGA database results showed consistency.
tsRNA-Gly-5-0007 may be involved in the malignant progression of colorectal cancer. (A) The expression of tsRNA-Gly-CCC-2-1 in colorectal cancer tissues and normal tissues in the TCGA database. (B) The expression of tsRNA-Gly-CCC-2-1 in colorectal cancer cells and colorectal extracellular vesicles in the TCGA database. (C) The secondary structure of tsRNA-Gly-5-0007. (D) The expression of tsRNA-Gly-5-0007 in colorectal cancer tissues and normal tissues in the tsRFun database. (E) The relationship between tsRNA-Gly-CCC-2-1 and S stage in the database. (F) The relationship between tsRNA-Gly-CCC-2-1 and T stage in the database. (G) The expression of tsRNA-Gly-CCC-2-1 in colorectal cancer tissues with distant metastasis and non-distant metastasis. (H) The difference of tsRNA-Gly-CCC-2-1 in lymph node metastasis. (I) The relationship between tsRNA-Gly-CCC-2-1 and survival.
To further verify whether tsRNA-Gly-5-0007 was involved in the malignant process of colorectal cancer, we looked again at the effect of tsRNA-Gly-5-0007 on colorectal cancer invasion and metastasis. First, we analyzed the combined colon and rectal cancer datasets in the TCGA database. clinical data analysis showed that tsRNA-Gly-CCC-2-1 was associated with S-staging (Fig. 1E), T-staging (Fig. 1F), and distant metastasis (Fig. 1G). Compared with SI SII, the expression of tsRNA-Gly-CCC-2-1 was significantly decreased in SIII SIV (Fig. 1E). tsRNA-Gly-CCC-2-1 expression was lower in T3 T4 than in T1 T2 (Fig. 1F). tsRNA-Gly-CCC-2-1 expression in colorectal cancer patients with distant metastases was lower than that in colorectal cancer patients with non-distant metastases (Fig. 1G), possibly due to the reason of individual differences in the samples, the database information showed that tsRNA-Gly-CCC-2-1 was not associated with lymph node metastasis (Fig. 1H). And the higher expression of tsRNA-Gly-CCC-2-1 was associated with better overall survival (Fig. 1I). In conclusion, tsRNA-Gly-5-0007 could be involved in colorectal cancer invasion and metastasis.
Based on the information in the database, we speculated whether tsRNA-Gly-5-0007 could affect the malignant process by influencing cell adhesion ability and cell motor capacity. First, we first examined the background expression level of tsRNA-Gly-5-0007 in colorectal cancer cells SW480, HCT116, and HT29, and found that the expression of tsRNA-Gly-5-0007 was relatively low in SW480 cells and relatively high in HCT116 cells (Fig. 2A). Therefore, We chose to overexpress the expression of tsRNA-Gly-5-0007 in SW480 and HT29 cells, and knock down the expression of tsRNA-Gly-5-0007 in HCT116 and HT29 cells. Next, to verify the effect of tsRNA-Gly-5-0007 on the adhesion ability of colorectal cancer cells, we did Matrigel adhesion experiment. We found that after knocking down tsRNA-Gly-5-0007 in colorectal cancer cells HCT116 (Fig. 2B) HT29 (Fig. 2C), the adhesion ability of colorectal cancer cells was reduced. After overexpressing tsRNA-Gly-5-0007 in colorectal cancer cells SW480 (Fig. 2D) HT29 (Fig. 2E), the adhesion ability of colorectal cancer cells was enhanced. Subsequently, we verified the effect of tsRNA-Gly-5-0007 on the motor capacity of colorectal cancer cells by transwell migration and invasion assays. We found that knockdown of tsRNA-Gly-5-0007 in colorectal cancer cells HCT116, HT29 promoted cell migration (Fig. 3A, B) and invasion (Fig. 3E, F). After overexpression of tsRNA-Gly-5-0007 in colorectal cancer cells SW480, HT29, cell migration (Fig. 3C, D) and invasion were inhibited (Fig. 3G, H). In other words, tsRNA-Gly-5-0007 could promote malignant progression of colorectal cancer by affecting intercellular adhesion ability and cell motor capacity.
tsRNA-Gly-5-0007 inhibits the adhesion ability between colorectal cancer cells. (A) To observe the background expression level of tsRNA-Gly-5-0007 in colorectal cancer cells. The effect of knockdown of tsRNA-Gly-5-0007 on cell adhesion ability in colorectal cancer cells HCT116 (B) HT29 (C). The effect of tsRNA-Gly-5-0007 overexpression on cell adhesion ability in colorectal cancer cells SW480 (D) HT29 (E).
tsRNA-Gly-5-0007 inhibits the migration and invasion of colorectal cancer cells. Knockdown of tsRNA-Gly-5-0007 promoted the migration of colorectal cancer cells (A, B), while overexpression of tsRNA-Gly-5-0007 inhibited the migration of colorectal cancer cells (C, D). Knockdown of tsRNA-Gly-5-0007 promoted the invasion ability of colorectal cancer cells (E, F), and overexpression of tsRNA-Gly-5-0007 inhibited the invasion ability of colorectal cancer cells (G, H).
To further clarify the biological processes involved in tsRNA-Gly-5-0007 in colorectal cancer, we performed pathway enrichment analysis. Firstly, the intestinal cancer samples were divided into high and low expression groups by tsRNA-Gly-CCC-2-1 expression in the TCGA database, and mRNAs of protein-coding genes that had a regulatory relationship with tsRNA-Gly-CCC-2-1 were obtained by differential analysis. 480 + 846 target genes were screened by log2 fold change (FC) > 1, P < 0.05, and subsequently we obtained 26 common target genes (Fig. 4A). The KEGG database serves as a pivotal resource for the integration and analysis of genomic, proteomic, metabolic pathway, disease, and pharmaceutical data, playing a crucial role as a core database for biological pathway annotation and gene functional analysis27,28. Consequently, we performed KEGG pathway analysis on these 26 candidate genes. Our results demonstrated that tsRNA-Gly-CCC-2-1 primarily modulates several key signaling pathways, including the Wnt signaling pathway, mTOR signaling pathway, and Rap1 signaling pathway (Fig. 4B). Furthermore, Gene Ontology (GO) analysis revealed that tsRNA-Gly-CCC-2-1 is potentially involved in biological processes such as cell adhesion, insulin response, and regulation of JUN kinase activity (Fig. 4C). tsRNA-Gly-5-0007 may affect the malignant process by influencing cell adhesion, insulin response and other processes. In other words, tsRNA-Gly-5-0007 may inhibit the motor capacity of colorectal cancer cells through the Wnt signaling pathway and may inhibit the adhesion of colorectal cancer cells through the Wnt signaling pathway.
tsRNA-Gly-5-0007 may affect the malignant progression of colorectal cancer through wnt signaling pathway. (A) The intersection of tsRNA-Gly-CCC-2-1 target genes in SVM-GA and DEG datasets. (B) Enrichment analysis of pathways involved in tsRNA-Gly-CCC-2-1 target genes. (C) The process of tsRNA-Gly-CCC-2-1 target genes participating in biological activities.
In conclusion, we suggest that tsRNA-Gly-5-0007 may be involved in the malignant progression of colorectal cancer by inhibiting the adhesion ability and motor capacity of colorectal cancer cells.
Identification of serum exosomes and the detection of tsRNA-Gly-5-0007 in serum exosomes
We extracted serum exosomes by ultracentrifugation and then validated the extracted exosomes using transmission electron microscopy, qNano and Western blot. We observed that exosomes were oval vesicles under electron microscopy (Fig. 5A), and qNano showed that the size of exosomes was distributed at 40–150 nm (Fig. 5B). Western blot showed that exosome marker proteins CD9 and HSP70 were expressed in exosomes and not expressed in cells. GM130 (negative control) was expressed in cells and not expressed in extracted exosomes (Fig. 5C).
Identification of serum exosomes and stability detection of serum exosomal tsRNA-Gly-5-0007.
The extracted serum exosomes were detected by (A) transmission electron microscopy (B) qNano analysis (C) Western blotting. The expression of tsRNA-Gly-5-0007 in (D) exosome-depleted supernatant (EDS) and exosomes (EXO) (E) expression after RNaseA treatment (F) expression at different times after incubation at room temperature.
We performed a characteristic analysis of tsRNA-Gly-5-0007. Firstly, we compared the expression of tsRNA-Gly-5-0007 between exosomes (EXO) and exosome-depleted supernatant (EDS). The results showed that the content of tsRNA-Gly-5-0007 in exosomes was higher than that in the exosome-depleted supernatant (Fig. 5D), so we carried out subsequent experiments on exosomal tsRNA-Gly-5-0007. Next, we used RNase A to verify the stability of exosomal tsRNA-Gly-5-0007. It was found that there was no difference in the expression of exosomal tsRNA-Gly-5-0007 after RNase A treatment (Fig. 5E). Finally, we placed the extracted exosomes at room temperature for 0, 6, 12, 18, and 24 h. The results showed that the expression level of tsRNA-Gly-5-0007 did not change significantly (Fig. 5F). In summary, tsRNA-Gly-5-0007 is relatively high in exosomes, and exosomes are stable, which can protect tsRNA-Gly-5-0007 from external factors.
tsRNA-Gly-5-0007 may be used as a biomarker for diagnosis of early colorectal cancer
Previous experiments have shown that tsRNA-Gly-5-0007 is more enriched in exosomes, and because exosomes have a protective effect, the expression of exosomal tsRNA-Gly-5-0007 is more stable. Therefore, we verified the expression of serum exosomal tsRNA-Gly-5-0007 of colorectal cancer patients and healthy people, and observed whether tsRNA-Gly-5-0007 could be used as a new tumor marker for colorectal cancer. First, we used serum exosomes from 24 patients with colorectal cancer and serum exosomes from 24 healthy people to verify tsRNA-Gly-5-0007 by qRT-PCR. It was found that the expression of tsRNA-Gly-5-0007 was different in patients with colorectal cancer and healthy people. Then, the expanded sample of tsRNA-Gly-5-0007 was verified by qRT-PCR in 148 patients with colorectal cancer and 140 healthy people, and it was found that its expression was down-regulated in patients with colorectal cancer (Fig. 6A). Next, we analyzed the expression in 79 patients with early colorectal cancer and found that it was also down-regulated (Fig. 6A). And compared with healthy people, S1-S4 (Fig. 6B) and T1-T4 (Fig. 6C) colorectal cancer patients were significantly reduced.
tsRNA-Gly-5-0007 may be used as a diagnostic marker for early colorectal cancer. (A) The expression of tsRNA-Gly-5-0007 in healthy people, early colorectal cancer patients and colorectal cancer patients. (B) Relationship with S stage. (C) Relationship with T staging. (D) The diagnostic efficacy of tsRNA-Gly-5-0007 for CRC. (E) The diagnostic efficacy of CEA for CRC. (F) The diagnostic efficacy of CA199 for CRC. (G) The diagnostic efficacy of tsRNA-Gly-5-0007 combined with CEA for CRC. (H) The diagnostic efficacy of tsRNA-Gly-5-0007 combined with CA199 for CRC. (I) The diagnostic efficacy of tsRNA-Gly-5-0007, CEA and CA199 for CRC. (J) The diagnostic efficacy of tsRNA-Gly-5-0007 for early CRC. (K) The diagnostic efficacy of CEA for early CRC. (L) The diagnostic efficacy of CA199 for early CRC. (M) The diagnostic efficacy of tsRNA-Gly-5-0007 combined with CEA for early CRC. (N) The diagnostic efficacy of tsRNA-Gly-5-0007 combined with CA199 for early CRC. (O) The diagnostic efficacy of tsRNA-Gly-5-0007, CEA and CA199 for early CRC.
In order to evaluate the diagnostic efficacy of serum exosomal tsRNA-Gly-5-0007 in colorectal cancer, we calculated the receiver operating characteristic curve (ROC). The AUC of serum exosomal tsRNA-Gly-5-0007 for the diagnosis of colorectal cancer was 0.7812, the sensitivity was 72.67%, the specificity was 73.65% (95% CI, 73.05–83.19%), and the cut off was 1.4532 (Fig. 6D). In this population sample, the AUC of the commonly used tumor marker CEA for the diagnosis of colorectal cancer was 0.7328, the sensitivity was 94.4%, and the specificity was 45.93% (95% CI, 67.74–78.82%) (Fig. 6E). The AUC of CA199 in the diagnosis of colorectal cancer was 0.7397, the sensitivity was 84.62%, and the specificity was 58.72% (95% CI, 62.54–85.40%) (Fig. 6F). The diagnostic efficacy of serum exosomal tsRNA-Gly-5-0007 for colorectal cancer was higher than that of CEA and CA199.
The AUC of serum exosomal tsRNA-Gly-5-0007 combined with CEA in the diagnosis of colorectal cancer was 0.8628, the sensitivity was 77.33%, and the specificity was 81.97% (95% CI, 82.16–90.41%) (Fig. 6G). The AUC of serum exosomal tsRNA-Gly-5-0007 combined with CA199 in the diagnosis of colorectal cancer was 0.8272, the sensitivity was 69.19%, and the specificity was 81.82% (95% CI, 71.81–93.62%) (Fig. 6H). The AUC of serum exosomal tsRNA-Gly-5-0007 combined with CEA and CA199 in the diagnosis of colorectal cancer was 0.9302, the sensitivity was 78.49%, and the specificity was 100% (95% CI, 87.70-98.35%) (Fig. 6I). The combination of serum exosomal tsRNA-Gly-5-0007, CEA and CA199 improved the diagnostic efficacy of CEA and CA199. This indicates that serum exosomal tsRNA-Gly-5-0007 may be used as a biological diagnostic marker in the diagnosis of colorectal cancer.
Next, we evaluated the diagnostic ability of serum exosomal tsRNA-Gly-5-0007 for patients with early colorectal cancer. Compared with healthy people, the AUC of serum exosomal tsRNA-Gly-5-0007 in the diagnosis of early colorectal cancer was 0.7726, the sensitivity was 69.62%, and the specificity was 77.25% (95% CI, 71.35–83.16%) (Fig. 6J). The AUC of CEA in the diagnosis of early colorectal cancer was 0.6661, the sensitivity was 39.24%, and the specificity was 94.4% (95% CI, 58.76–74.45%) (Fig. 6K). The AUC of CA199 in the diagnosis of early colorectal cancer was 0.6913. The sensitivity was 83.54%, and the specificity was 54.27% (95% CI, 54.69–83.58%) (Fig. 6L). The diagnostic efficacy of serum exosomal tsRNA-Gly-5-0007 in early colorectal cancer was also significantly higher than that of CEA and CA199.
The AUC of serum exosomal tsRNA-Gly-5-0007 combined with CEA in the diagnosis of early colorectal cancer was 0.8371, the sensitivity was 73.42%, and the specificity was 81.15% (95% CI, 78.29–89.13%) (Fig. 6M). The AUC of serum exosomal tsRNA-Gly-5-0007 combined with CA199 in the diagnosis of early colorectal cancer was 0.7934, the sensitivity was 72.15%, and the specificity was 72.73% (95% CI, 65.83–92.86%) (Fig. 6N). The AUC of serum exosomal tsRNA-Gly-5-0007 combined with CEA and CA199 in the diagnosis of early colorectal cancer was 0.9013, the sensitivity was 68.35%, and the specificity was 100% (95% CI, 82.24–98.02%) (Fig. 6O). In early colorectal cancer, the combination of serum exosomal tsRNA-Gly-5-0007, CEA and CA199 improved the diagnostic efficacy of CEA and CA199. Therefore, we believe that serum exosomal tsRNA-Gly-5-0007 may be used as an early diagnostic marker for colorectal cancer.
Discussion
Colorectal cancer (CRC) represents a significant global health burden, characterized by high incidence and mortality rates. Current diagnostic approaches are limited by their invasive nature and suboptimal sensitivity and specificity, resulting in only 30–40% of cases being detected at early stages29,30. While established tumor biomarkers exist, their clinical utility is constrained by inadequate diagnostic performance. These limitations underscore the critical need for novel non-invasive biomarkers with improved specificity and sensitivity to guide clinical decision-making. Our findings demonstrate that serum exosomal tsRNA-Gly-5-0007 exhibits promising potential as a diagnostic biomarker for early-stage CRC detection.
Exosomes, a subclass of extracellular vesicles characterized by a lipid bilayer membrane structure, are ubiquitously present in biological fluids. Their inherent stability and low immunogenicity enable them to evade immune surveillance while facilitating precise drug delivery to target cells, positioning them as promising natural nanocarriers for therapeutic applications31,32. Notably, cancer-associated fibroblasts (CAFs) - highly plastic cellular components of the tumor microenvironment - represent a significant source of exosomes. These CAF-derived vesicles participate in extracellular matrix (ECM) remodeling and contribute to tumor stroma formation, thereby establishing the structural framework for their targeted delivery capacity33. Exosomes encapsulate tumor-specific molecular cargo including proteins, nucleic acids (RNA/DNA), and metabolic products that faithfully reflect the pathophysiological state of parent cancer cells. Functioning as intercellular messengers, they mediate the transfer of these bioactive molecules to recipient cells, thereby facilitating critical oncogenic processes such as tumor progression and metastasis. Longitudinal monitoring of exosomal molecular profiles offers valuable insights into cancer dynamics, including disease progression, therapeutic response, prognostic evaluation, and early detection of recurrence5,34.
tRNA-derived fragments (tRFs) represent a novel class of small non-coding RNAs generated through enzymatic cleavage at specific positions of precursor or mature tRNAs. In this study, we focus on tsRNA-Gly-5-0007, a 5’-end derived fragment of tsRNA-Gly-CCC-2-1. Emerging evidence indicates that exosomes are enriched with diverse tRF populations, which exert regulatory functions through interactions with RNA-binding proteins. A seminal study by Goodarzi et al. demonstrated that specific tRFs (including those derived from tRNA (Glu), tRNA (Asp), tRNA (Gly), and tRNA (Tyr)) compete with oncogenic transcripts for binding to Y-box binding protein 1 (YBX1) in breast cancer35. Remarkably, under cellular stress conditions, malignant cells upregulate tRF production to preferentially sequester YBX1 from pro-tumorigenic transcripts. These findings highlight the crucial role of tRFs in modulating key cancer hallmarks including proliferation, cell cycle progression, apoptosis, and metastatic potential36. Clinical investigations have identified several tRFs with diagnostic potential. Notably, tRF-Gly-CCC-046, tRF-Tyr-GTA-010, and tRF-Pro-TGG-001 show significant downregulation in both tissue specimens and circulation of breast cancer patients, including early-stage cases, establishing their utility as minimally invasive diagnostic biomarkers25. Similarly, in gastric cancer, tRF-31-U5YKFN8DYDZDD expression correlates strongly with clinicopathological parameters including TNM stage, invasion depth, lymph node metastasis, and vascular involvement across tissue, serum, and cellular compartments23. The remarkable stability and abundance of tRFs in biological fluids, coupled with their disease-specific expression patterns, position them as promising candidates for diagnostic, prognostic, and therapeutic monitoring applications across various pathological conditions. This growing body of evidence underscores the translational potential of tRFs in precision medicine.
In this study, we first found that the expression level of tsRNA-Gly-5-0007 in tissues or extra-tissue vesicles was correlated with S-stage, T-stage, and distant metastasis. The Kaplan-Meier analysis revealed that the low expression group had a worse overall survival rate. These results indicate that the clinical biomarker tsRNA-Gly-5-0007 is highly promising for the diagnosis and prognosis of colorectal cancer. In subsequent experiments on the biological function of tsRNA-Gly-5-0007 on colorectal cancer cells, we discovered that inhibiting the expression of tsRNA-Gly-5-0007 improved colorectal cancer cell adhesion and motor capacity, which provided evidence that tsRNA-Gly-5-0007 may play an oncogenic role in colorectal cancer. We made predictions in our database and found that tsRNA-Gly-5-0007 could act on Wnt, mTOR and other signaling pathways. The Wnt signaling pathway is known to have a significant impact on cell adhesion and cell motor capacity, and it has been reported that the exosome tRF-19-PNR8YPJZ produced by pancreatic stellate cells targets AXIN2 in pancreatic cancer cells and reduces its expression, thereby activating the Wnt pathway and encouraging metastasis37,38,39,40. Therefore, we preliminarily speculated that tsRNA-Gly-5-0007 may affect the adhesion and motor capacity of colorectal cancer cells through the Wnt signaling pathway, thus affecting the malignant progression of colorectal cancer.
In order to explore the diagnostic value of serum exosomal tsRNA-Gly-5-0007 for colorectal cancer and whether it can be used as a new tumor marker for colorectal cancer, we conducted a large sample verification. The results showed that the expression of serum exosomal tsRNA-Gly-5-0007 was lower in colorectal cancer than in healthy people. Importantly, when we compared serum exosomal tsRNA-Gly-5-0007 with clinical colorectal cancer biomarkers CEA and CA199, we found that serum exosomal tsRNA-Gly-5-0007 had higher diagnostic ability than other biomarkers and further improved the diagnostic efficacy after combination. It is worth noting that many of the current biomarkers are not easy to detect in the early stage, and are usually detected in the late stage of colorectal cancer. Therefore, we found that the expression level of serum exosomal tsRNA-Gly-5-0007 in patients with different stages was different from that in healthy people, especially in the early stage of colorectal cancer (stage I and stage II). Its diagnostic efficacy is higher than the commonly used clinical biomarkers CEA and CA199. The above results suggest that tsRNA-Gly-5-0007 is more promising for the diagnosis of early colorectal cancer, and may be used not only for diagnosis, but also as a potential therapeutic target for colorectal cancer.
However, there are some limitations in our study. Firstly, this study included samples from our hospital, which is geographically limited due to limited conditions, small sample size, and did not cover specimens from more than one region. The results of this experiment may not be representative and the sample size still needs to be supplemented. Secondly, using the database, we projected the pathways in which tsRNA-Gly-5-0007 may primarily act and the activities in which it might be engaged, and we initially hypothesized that it might affect the adhesion and motor capacity of colorectal cancer cells through the Wnt signaling pathway, and we have only explored initially how tsRNA-Gly-5-0007 is involved in the malignant process of colorectal cancer cells, and we need to further in-depth validation.
In summary, our data show that the expression of exosomal tsRNA-Gly-5-0007 is down-regulated in colorectal cancer, and its diagnostic efficiency is higher than that of CEA and CA199 even in the early stage. Therefore, we speculate that tsRNA-Gly-5-0007 may be involved in the malignant process of colorectal cancer by affecting the Wnt signaling pathway to inhibit the adhesion and motor capacity of colorectal cancer cells. Therefore, exosomal tsRNA-Gly-5-0007 can be used as a valuable diagnostic marker for early colorectal cancer.
Data availability
These data are derived from the following available resources in the public domain: TCGA (https://portal.gdc.cancer.gov/); tsRFun database (http://rna.sysu.edu.cn/tRFfinder/). Corresponding author can be directed for further inquirements.
References
Yu, H. et al. The mRNA level of MLH1 in peripheral blood is a biomarker for the diagnosis of hereditary nonpolyposis colorectal cancer. Am. J. cancer Res. 6, 1135–1140 (2016).
Nikolaou, S. et al. Systematic review of blood diagnostic markers in colorectal cancer. Tech. Coloproctol. 22, 481–498. https://doi.org/10.1007/s10151-018-1820-3 (2018).
Wise, J. New bowel cancer screening test is recommended for England. BMJ 352, i273. https://doi.org/10.1136/bmj.i273 (2016).
Young, G. P. et al. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 5, 2763–2772. https://doi.org/10.1002/cam4.868 (2016).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367 https://doi.org/10.1126/science.aau6977 (2020).
Raposo, G. & Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. https://doi.org/10.1083/jcb.201211138 (2013).
Zhang, L. & Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Et Biophys. Acta Reviews cancer. 1871, 455–468. https://doi.org/10.1016/j.bbcan.2019.04.004 (2019).
Doyle, L. M. & Wang, M. Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8 https://doi.org/10.3390/cells8070727 (2019).
Weng, Q. et al. Extracellular vesicles-associated tRNA-derived fragments (tRFs): Biogenesis, biological functions, and their role as potential biomarkers in human diseases. J. Mol. Med. 100, 679–695. https://doi.org/10.1007/s00109-022-02189-0 (2022).
Gong, M., Deng, Y., Xiang, Y. & Ye, D. The role and mechanism of action of tRNA-derived fragments in the diagnosis and treatment of malignant tumors. Cell. Communication Signaling: CCS. 21, 62. https://doi.org/10.1186/s12964-023-01079-3 (2023).
Liu, B. et al. Deciphering the tRNA-derived small rnas: origin, development, and future. Cell Death Dis. 13 https://doi.org/10.1038/s41419-021-04472-3 (2021).
Hu, F. et al. tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Translational Lung cancer Res. 10, 3957–3972. https://doi.org/10.21037/tlcr-21-829 (2021).
Qi Chen, M. Y. et al. Sperm TsRNAs contribute to intergenerational inheritance of an acquired. Science (2016).
Jin, F. et al. A novel class of TsRNA signatures as biomarkers for diagnosis and prognosis of pancreatic cancer. Mol. Cancer. 20, 95. https://doi.org/10.1186/s12943-021-01389-5 (2021).
Xie, Y. et al. Action mechanisms and research methods of tRNA-derived small RNAs. Signal. Transduct. Target. Therapy. 5 https://doi.org/10.1038/s41392-020-00217-4 (2020).
Akiyama, Y. et al. Selective cleavage at CCA ends and anticodon loops of tRNAs by Stress-Induced RNases. Front. Mol. Biosci. 9, 791094. https://doi.org/10.3389/fmolb.2022.791094 (2022).
Slack, F. J. & Chinnaiyan, A. M. The role of Non-coding RNAs in oncology. Cell 179, 1033–1055. https://doi.org/10.1016/j.cell.2019.10.017 (2019).
Cernilogar, F. M. et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480, 391–395. https://doi.org/10.1038/nature10492 (2011).
Okamura, K. & Lai, E. C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 9, 673–678. https://doi.org/10.1038/nrm2479 (2008).
Veneziano, D. et al. Noncoding RNA: Current deep sequencing data analysis approaches and challenges. Hum. Mutat. 37, 1283–1298. https://doi.org/10.1002/humu.23066 (2016).
Shao, Y. et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem. Biol. Drug Des. 90, 730–738. https://doi.org/10.1111/cbdd.12994 (2017).
Gupta, T., Malkin, M. G. & Huang, S. tRNA function and dysregulation in Cancer. Front. Cell. Dev. Biol. 10, 886642. https://doi.org/10.3389/fcell.2022.886642 (2022).
Huang, Y. et al. Elucidating the role of serum tRF-31-U5YKFN8DYDZDD as a novel diagnostic biomarker in gastric Cancer (GC). Front. Oncol. 11, 723753. https://doi.org/10.3389/fonc.2021.723753 (2021).
Zheng, B. et al. Plasma Exosomal tRNA-derived fragments as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 12, 1037523. https://doi.org/10.3389/fonc.2022.1037523 (2022).
Zhang, Y. et al. tRNA-derived fragments: tRF-Gly-CCC-046, tRF-Tyr-GTA-010 and tRF-Pro-TGG-001 as novel diagnostic biomarkers for breast cancer. Thorac. cancer. 12, 2314–2323. https://doi.org/10.1111/1759-7714.14072 (2021).
Fan, Q. et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 414, 107–115. https://doi.org/10.1016/j.canlet.2017.10.040 (2018).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462. https://doi.org/10.1093/nar/gkv1070 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Schönfeld Vorsorge des Kolorektalen karzinoms: Koloskopie Oder Immunologischer stuhltest. Z. Gastroenterol. 51, 299–300. https://doi.org/10.1055/s-0032-1330299 (2013).
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet 394, 1467–1480. https://doi.org/10.1016/s0140-6736(19)32319-0 (2019).
Nedaeinia, R. et al. The role of cancer-associated fibroblasts and Exosomal miRNAs-mediated intercellular communication in the tumor microenvironment and the biology of carcinogenesis: a systematic review. Cell. Death Discovery. 10, 380. https://doi.org/10.1038/s41420-024-02146-5 (2024).
Hasannejad, F. et al. A thermosensitive hydrogel for the sustained delivery of exosomes extracted from menstrual blood mesenchymal stem cells and frizzled antibody on triple-negative breast cancer cells in vitro. J. Drug Deliv. Sci. Technol. 101, 106144. https://doi.org/10.1016/j.jddst.2024.106144 (2024).
Jafarpour, S. et al. MSC-derived exosomes enhance the anticancer activity of drugs in 3D spheroid of breast cancer cells. J. Drug Deliv. Sci. Technol. 92, 105375. https://doi.org/10.1016/j.jddst.2024.105375 (2024).
Pegtel, D. M., Gould, S. J. & Exosomes Annu. Rev. Biochem. 88, 487–514, doi:https://doi.org/10.1146/annurev-biochem-013118-111902 (2019).
Goodarzi, H. et al. Endogenous tRNA-Derived fragments suppress breast Cancer progression via YBX1 displacement. Cell 161, 790–802. https://doi.org/10.1016/j.cell.2015.02.053 (2015).
Sun, C. et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 414, 16–25. https://doi.org/10.1016/j.canlet.2017.10.031 (2018).
Cao, W. et al. Pancreatic stellate cell-derived Exosomal tRF-19-PNR8YPJZ promotes proliferation and mobility of pancreatic cancer through AXIN2. J. Cell. Mol. Med. 27, 2533–2546. https://doi.org/10.1111/jcmm.17852 (2023).
Brunt, L. & Scholpp, S. The function of endocytosis in Wnt signaling. Cell. Mol. Life Sci. 75, 785–795. https://doi.org/10.1007/s00018-017-2654-2 (2018).
Dale, T. C. Signal transduction by the Wnt family of ligands. Biochem. J. 329 (Pt 2), 209–223. https://doi.org/10.1042/bj3290209 (1998).
Akira, K., Shosei, K. & Hideki, Y. Regulation of Wnt signaling by protein-protein interaction and post-translational modifications. Exp Mol Med. 38, 1-10. https://doi.org/10.1038/emm.2006.1(2006).
Acknowledgements
First of all, we would like to thank TCGA, tsRFun database, for their valuable resources, in addition, we would like to thank the editors and reviewers for their valuable comments and suggestions to us.
Funding
This work was supported by Collaborative Academic Innovation Project of Shandong cancer hospital (FC005), the Taishan Scholars Program (NO. tstp20240857), Shandong Provincial Natural Science Foundation(ZR2024LZL001), China Postdoctoral Science Foundation (2023M732126), University-Industry Collaborative Education Program (231007010030301).
Author information
Authors and Affiliations
Contributions
The experiment was designed and supervised by LX and GHL. ZZ and LL wrote the first draft and conducted the data analysis. WXJ, SWW and NZ conducted data collection and compilation. XZ, FFL, JZ were used for material preparation and sample collection. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Ethics Committee of Shandong Cancer Hospital and Institute (No. 20171008). The patients/participants provided written informed consent for the participant’s reference.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Z., Li, G., Li, L. et al. Exosomal tsRNA-Gly-5-0007 may be used as a diagnostic marker for colorectal cancer. Sci Rep 15, 26751 (2025). https://doi.org/10.1038/s41598-025-09830-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09830-1
Keywords
This article is cited by
-
BMSC-derived extracellular vesicles affect gluconeogenesis and lipogenesis by releasing 5'-tRF-GlyCCC to improve MAFLD insulin sensitivity
Cell Biology and Toxicology (2026)