Abstract
Somatic cell nuclear transfer (SCNT) allows the multiplication of elite livestock and conservation of endangered species. However, restrictions on cow slaughter limit the access to oocytes for SCNT applications in Indian cattle breeds. To overcome this limitation, we utilized transvaginal ovum pick-up (OPU) method to collect oocytes, which were then used for the production of cloned embryos via the handmade cloning (HMC) technique. A total of 98 Sahiwal oocytes were collected, leading to the successful reconstruction of 24 SCNT Gir embryos. Out of these, five developed into blastocysts, which were transferred into five recipient cows. Two pregnancies were confirmed, but one was lost due to hydro-allantois condition. The other pregnancy continued to term, and a healthy Gir female calf weighing 32 kg was born. Microsatellite DNA analysis confirmed the genetic identity of the cloned calf to its donor. Postnatally, the calf was monitored for serum cytokine parameters, telomere length, and reproductive potentials. Cytokine profiling revealed variations between the cloned calf and naturally conceived counterparts; however, the born cloned calf did not exhibit any pathological conditions and has high telomere length compared to age-matched counterparts, and surviving well. Furthermore, to assess cloned cow utility in reproductive biotechnologies, we produced blastocyst stage embryos (35%) through OPU-IVF method and established one pregnancy from five transfers (20%). In conclusion, this study reports the first successful SCNT of Indian cattle breed and demonstrates the feasibility of the cloned cow for the production of OPU-IVF embryos.
Similar content being viewed by others
Introduction
The first successful application of somatic cell nuclear transfer (SCNT) in farm animals was demonstrated in sheep, the birth of famed Dolly, in 19961. This groundbreaking achievement triggered widespread interest in SCNT for the production of elite livestock species and the conservation of endangered animals2. Over the years, the SCNT has been successfully applied to various livestock species, including cattle, pigs, goats, and buffaloes, in different parts of the world3. Cattle, being one of the most important livestock species, have significantly benefited from the application of SCNT technology3. Countries such as the United States, Brazil, China, and Argentina have commercial companies, which enable the production of genetically superior cattle with desirable traits4. These advancements have provided cattle breeders with an effective tool to multiply elite animals, reduce genetic dilution, and sustain high-yielding lineages. Despite these global success, India had not reported any case of cloned cattle until now. Cattle are the backbone of the Indian dairy industry, accounting for 52% (124 million tons) of the total milk production (Basic Animal Husbandry Statistics, 2024, Department of Animal Husbandry, Dairying & Fisheries (DADF) of India, https://dahd.gov.in/schemes/programmes/animal-husbandry-statistics). Indian cattle breeds, including Gir, Sahiwal, Tharparkar, and Red Sindhi, play a crucial role in India’s dairy sector, contributing to both milk yield and genetic diversity. Assisted reproductive technologies (ARTs) such as artificial insemination (AI), multiple ovulation and embryo transfer (MOET), and, in-vitro fertilization (IVF) have been widely employed to improve the reproductive efficiency and genetic merit of Indian cattle population (https://dahd.gov.in/schemes/programmes/rashtriya_gokul_mission). In addition to these technologies, SCNT also offers a unique advantage in multiplying genetically superior animals while maintaining their desired traits5.
Over the past decade, we have successfully implemented handmade cloning (HMC), a simplified and cost-effective SCNT technique, for the production of elite buffalo clones. This technique has resulted in the birth of over 30 cloned buffaloes in India, demonstrating its feasibility and efficiency in large-scale livestock applications6,7,8. One of the major limitations in employing SCNT in cattle is the restricted availability of oocytes due to the ban on cattle slaughter in India9. To overcome this challenge, we utilized the ovum pick-up (OPU) technology to collect oocytes from live animals. These oocytes were subsequently used for the production of cloned embryos. Using this approach, we successfully produced India’s first cloned cattle, belonging to the Gir breed. This success demonstrates the feasibility of SCNT for Indian cattle breeds. Although media coverage of the cloned Gir calf (Ganga) was available in 2023, the current study is the first peer-reviewed scientific report detailing the methods and outcomes of SCNT leading to a live birth in indigenous Indian cattle.
Results and discussion
Reproductive biotechnologies play significant role in the genetic improvement of livestock species. Among these technologies, SCNT has emerged as a tool to produce elite stock of animals and preserve endangered breeds2. Despite the potentials, the cattle SCNT have several challenges, particularly in India, where ethical considerations and logistical issues have hindered the widespread applications9. This study aimed to explore the feasibility of SCNT in Indian cattle breeds using OPU-derived oocytes. A schematic representation of the SCNT methodology used in this study to produce cloned Gir cow is shown in Fig. 1A. A total of 98 oocytes from Sahiwal cows were collected through OPU method (Table 1). These oocytes were then processed via HMC (Fig. 1B), a simplified and efficient method for producing cloned animals10. Previous studies have demonstrated the effectiveness of HMC in producing cloned animals in various species, including cattle11,12,13. In this study, five blastocysts were produced (Table 1), and these were transferred into surrogate crossbred cows (n = 5). Cross-bred surrogate cows were chosen due to the limited availability of Gir cows with us. Two pregnancies were established, of which one was lost to abortion, which is not uncommon in cloned pregnancies and has been reported in previous studies14,15. Second pregnancy resulted in the successful birth of a cloned calf (Fig. 1C), named as GANGA. The birth of this calf represents a significant achievement, as it marks the first known instance of cattle cloning in India. Microsatellite analysis confirmed that the calf’s genetic makeup was identical to that of the donor cow (Table 2). It is important to highlight that the phenotypic similarity between the clone and the donor animal is a critical indicator of successful SCNT. While there have been concerns regarding the abnormal phenotypes in cloned animals16. The cloned calf has no signs of abnormalities in its physical traits, and phenotypically similar to the donor cow (Fig. 1D).
Birth of the India’s first cloned cow. (A) A schematic outline of the SCNT methodology used in this study to produce the cloned Gir cow. (B) Donor cell preparation and SCNT embryo development, which include (a) donor cell outgrowth from primary explant, (b) confluent donor cells before SCNT, (c) OPU-derived oocytes, (d) zona-free oocytes, (e) nucleated part of oocytes stained with DAPI, (f) fused couplets, in which somatic cells are sandwiched between two enucleated oocytes, (g) early-stage cleavage embryos (8–16 cells), and (h) a developed blastocyst. (C) The cloned calf (center) is shown alongside its donor (left) and surrogate mother (right). (D) At 18 months of age, the cloned cow (left) is phenotypically similar with its donor (right).
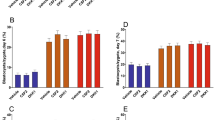
One of the major concerns in SCNT is the health and immune function of cloned animals8,17. It is often believed that the immune system of cloned animals may be weakened or altered due to abnormal reprogramming of somatic cells in SCNT18. To assess the health of the cloned calf, we analyzed 15 cytokines (IFN-γ, IL-1α, IL-1β, IL-4, IL-6, IL-8, IL-10, IL-17, MIP-1α, MIP-1β, IL-36Ra, IP-10, MCP-1, TNFα, and VEGF-A) at 1 day and 10 months of age. Cytokines are important in regulating the immune response, and their levels can help understand the animal’s overall health and stress19. At birth, the cloned calf had higher levels of INF-γ and IL-4 compared to control calves. However, other cytokines such as IL-6, IL-8, and VEGF-A were found to be lowered in the cloned calf (Fig. 2A). This difference in cytokine levels suggests that the cloned pregnancy may have been exposed to more immune stress. The changes in cytokine levels between the cloned calf and the wild-type calves indicate that cloned animals might face immune challenges that need careful monitoring in their early life. When we analyzed the cytokines again at 10 months of age, we found that IL-8 and VEGF-A still had lower levels in the cloned calf compared to the wild-type calves, while INF-γ, IL-4, and IL-6 returned to normal levels similar to control calves (Fig. 2B). This suggests that while some immune markers improved over time, the cloned animal’s immune system—particularly related to IL-8 and VEGF-A—might still be compromised in the long term. More studies focusing on cytokine profiles and long-term immune health of cloned animals are essential to better understand the impact of SCNT on their immune function.
In addition to cytokine analysis, we also examined telomere length, which is an important indicator of cellular aging and overall health. There have been concerns about premature aging in cloned animals16. Our results showed that the cloned calf had longer telomeres (Fig. 3A), suggesting that Gir cloned cow may experience normal aging. Previous studies on telomere length in cloned animals have reported varying results. Some studies found shorter telomeres20while others reported comparable lengths8,21,22,23and some even observed longer telomeres24,25 compared to age-matched control calves. These differences in telomere length between cloned animals and their controls are likely influenced by several factors, such as the type of donor cells used, the nuclear transfer method employed, species variations, and the overall health of the animals8,16,23.
To fully capitalize on the potential of SCNT for genetic trait propagation, it is crucial to assess the reproductive performance of animals produced through this process26. In this study, we observed that the cloned heifer reached puberty, and her estrus cycle length was similar to that of control animals. Most previous studies have focused on comparing cloned animals to age-matched controls27,28,29 rather than to their genetic donors. Additionally, reproductive performance in previous studies has typically been assessed after artificial insemination (AI) or natural mating. Fewer studies have compared the reproductive outcomes of cloned animals to those of their genetic donors, especially in terms of producing OPU-IVF embryos and calves26,28. After the cloned heifer reached puberty, we used her to produce embryos via OPU-IVF (Fig. 3C). Following FSH stimulation, we recorded follicular counts, which were found to be similar to those of the donor cow (Fig. 3B). This indicates that cloned animals have reproductive potential comparable to that of their genetic donors. The blastocyst production rate was 35%, which is typical for bovine species26,28. These embryos were transferred into surrogate animals (n = 5), resulting in one successful birth. The results aligns with findings from previous studies26,28suggesting that cloned animals can have similar reproductive success to their genetic donors.
In conclusion, SCNT is a reproductive technique that allows breeders to replicate the best female animals and enhance the spread of their desirable genetics. This study is the first to report the successful cattle SCNT in India. We also found that the cloned cattle showed normal health, aging, and reproductive performance. Therefore, the combination of SCNT and OPU-IVF, as shown in this study, offers a valuable tool for the faster spread of superior genetics and conservation of endangered Indian cattle breeds.
Methods
Ethics statements
All experiments were performed according to the ethical standards of the institute. Animal procedures including biopsy collection were approved by the Institute Animal Ethics Committee, ICAR-National Dairy Research Institute (NDRI), Karnal, India. The study is reported in accordance with ARRIVE guidelines. Cattle used in this study were available at the Livestock Research Centre, ICAR-NDRI, Karnal, and managed as per standard farm practices.
Production of cloned embryos
To produce cloned embryos, primary fibroblast cells were established from tail biopsies of new born Indian Gir cattle as previously described by us30. Briefly, tail tissue samples were collected aseptically from a 2-month-old donor female Gir calf. The tissues were minced into ~ 1 mm³ explants and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. After 5th day of culture, the fibroblast outgrowths were observed, then monitored for any kind of contaminations and passaged at ~ 80% confluence using trypsin-EDTA. Cells at passage 5–6 were allowed to grow for three days to achieve total confluence before SCNT experiments. For oocyte collection, the Sahiwal cows were treated with GnRH, and performed transvaginal OPU after 35–40 h as previously described26. Follicles, ≥ 5 mm in diameter, were aspirated using an 18-gauge disposable needle under vacuum pressure. The contents of all follicles were collected into a 50 mL tube containing Dulbecco’s Phosphate-Buffered Saline (DPBS) supplemented with heparin (10,000 IU/L). Recovered cumulus–oocyte complexes (COCs) were washed in TCM-199 supplemented with 10% FBS, and matured in in-vitro maturation (IVM) medium (TCM-199, 10% FBS, 1 µg⁄mL estradiol 17β, 5 µg⁄mL porcine FSH, 0.81 mM sodium pyruvate, 0.68 mM L-glutamine, 50 µg/mL gentamicin sulphate) in a 5% CO₂ incubator at 38.5 °C for 24 h. In-vitro matured oocytes were passed through the steps of HMC using Gir fibroblast cells as the nuclear donor31. Briefly, post-IVM, oocytes were denuded and their zona pellucida removed using enzymatic treatment, followed by enucleation with a microblade under a stereomicroscope. Enucleated oocytes were individually paired with Gir fibroblasts using phytohemagglutinin, and fusion was carried out via electrofusion. Reconstructed embryos were activated using 5 µM ionomycin for 7 min, followed by incubation in 2 mM 6-DMAP for 4 h in T10 medium. Activated embryos were cultured in RVCL medium (Cook, Queensland, Australia) for 8 days in a 5% CO₂ incubator. Cleavage and blastocyst development rates were recorded on day 8.
Embryo transfer, birth of cloned calf, and genotype verification
Recipient cross-bred cows with a functional corpus luteum (CL) were synchronized using the prostaglandin F2α analogue (cloprostenol sodium, 500 µg). Cows that exhibited estrus within 72 h post-treatment were selected for embryo transfer. On day 8 of embryonic development, cloned blastocysts were non-surgically transferred into the uterine horn ipsilateral to the CL after confirming its presence. Each recipient received a single embryo. Pregnancy was diagnosed via trans-rectal ultrasonography on day 30 post-transfer, and subsequent calving outcomes were recorded to assess the efficiency of the SCNT approach. To confirm the genetic identity of a born cloned calf, microsatellite analysis was conducted using genomic DNA from the blood of cloned calf, donor, and the surrogate mother. Twelve microsatellite markers were amplified using specific primer sets to verify genetic fidelity. The parentage genetic tests were conducted by the CALF Lab, National Dairy Development Board, Anand, Gujarat, India (https://nddbcalf.com/).
Serum cytokine analysis
Blood samples were collected from the cloned calf and age-matched AI-born calves (n = 2) at both day 1 and 10 months of age. Blood samples were drawn using vacutainer tubes containing a serum activator and incubated for 3 h to allow clot formation. The clots were then removed by centrifugation at 1500 g for 10 min. The resulting serum samples were stored at − 80 °C, ensuring minimal freeze-thaw cycles to prevent degradation of cytokine components. Cytokine levels, including IFN-γ, IL-1α, IL-1β, IL-4, IL-6, IL-8, IL-10, IL-17, MIP-1α, MIP-1β, IL-36Ra, IP-10, MCP-1, TNFα, and VEGF-A, were quantified using a bovine cytokine magnetic bead panel multiplex assay kit (BCYT1-33 K-PX15, Merck Millipore, Darmstadt, Germany) according to the manufacturer’s instructions. Data acquisition was performed using a Bio-Plex MAGPIX multiplex reader, with a digital processor handling output analysis. The Bio-Plex Manager 6.0 software provided results in terms of median fluorescence intensity (MFI) and cytokine concentrations (pg/mL).
Telomere length analysis
Genomic DNA was extracted from blood samples of cloned cow’s and age-matched controls (n = 3) using the genomic DNA purification kit (Promega Wizard®, Madison, USA). Telomere length was determined by quantitative PCR (qPCR), employing telomere-specific primers (TEL-F-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT, TEL-R-GGCTTGCCTTACCCTTACCCTTACCCTTACCTTACCCT) and a single-copy reference gene primers (HBG-F-GCTTCTGACACAACTGTGTTCACTAGC, HBG-R-CACCAACTTCATCCACGTTCACC) as mentioned previously (Joglekar et al., 2020). Each qPCR reaction was performed in a total volume of 10 µL, which included 100 ng of genomic DNA (1µL), SYBR green master mix (5µL), 0.5 µM of each primer (0.5µL), and nuclease-free water (3µL). The thermal cycling conditions for telomere amplification began with an initial step of denaturation at 95 °C for 3 min, followed by 40 cycles consisting of 15 s at 95 °C, 1 min at 58 °C, and 1 min at 72 °C, concluding with a melt curve stage of 15 s at 95 °C, 1 min at 65 °C, and 15 s at 95 °C. All tests were performed in triplicate using a real-time PCR system (CFX 96 cycler, Bio-Rad, Hercules, CA, USA). The relative telomere length was determined by calculating the T/S ratio (Telomere/single copy reference) using the ΔΔCt method as described previously32.
Production of OPU-IVF embryos of the cloned cow
To produce in-vitro embryos, the cloned cow and its donor were undergone the FSH stimulation protocol as described in the previous section. The numbers of visible follicles were counted, and oocytes were aspirated using transvaginal OPU method. Aspirated COCs were washed in Medium-199 containing 10% FBS and then kept in IVM medium for 24 h. After IVM, oocytes were subjected to in-vitro fertilization (IVF) as per the standard method of the Vitrogen media manufacture (Vitrogen–Biotecnologia em Reprodução Animal, Cravinhos, SP, Brazil). Matured oocytes were washed in 100 µL droplets of IVF medium (#BOV03), then, 15–20 oocytes were placed in 70 µL droplets of IVF medium in which 50 µL of sperm suspension (106 sperm/mL) was added to each droplet, and incubated for 18 h at 38.5 °C in a 5% CO2 incubator. After completion of IVF duration, fertilized oocytes were treated with hyaluronidase to remove cumulus cells, washed in IVC medium (#BOV04), and cultured in 100 µL droplets (15–20 zygotes per drop) of IVC medium for 7 days. Cleavage rates were assessed on the second day of IVC, and the blastocyst production rates were recorded on the seventh day. Some of the blastocyst stage embryos were transferred into surrogate mothers to determine in-vivo developmental competence of produced embryos.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 5 software. The datasets (telomere length) underwent analysis through one-way analysis of variance (ANOVA), followed by the Tukey test for post hoc comparisons. Statistical significance was considered at a threshold of P < 0.05. The results are presented as mean ± standard error of the mean (SEM).
Data availability
The data during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J. & Campbell, K. H. Viable offspring derived from fetal and adult mammalian cells. Nature 385 (6619), 810–813. https://doi.org/10.1038/385810a0 (1997).
Keefer, C. L. Artificial cloning of domestic animals. Proceedings of the National Academy of Sciences (PNAS). 112 (29), 8874–8878. (2015). https://doi.org/10.1073/pnas.1501718112
Klinger, B. & Schnieke, A. 25th anniversary of cloning by somatic-cell nuclear transfer twenty-five years after dolly: how Far have we come? Reproduction 162 (1). https://doi.org/10.1530/REP-20-0652 (2021). F1-F10.
Galli, C. & Lazzari, G. 25th anniversary of cloning by somatic-cell nuclear transfer: current applications of SCNT in advanced breeding and genome editing. Livest. Reproduction. 162 (1), F23–32. https://doi.org/10.1530/rep-21-0006 (2021).
Selokar, N. L., Singh, M. K., Kumar, D., Yadav, P. S. & Chauhan, M. S. Milestones and Recent Developments in Farm Animal Cloning. Frontier technologies in bovine reproduction. (eds Kumaresan, A., Srivastava, A.K.) Springer. (2022). https://doi.org/10.1007/978-981-19-3072-0_12 (233–254).
Selokar, N. L. et al. Cloning of buffalo, a highly valued livestock species of South and Southeast asia: any achievements. Cell. Reprogram. 20 (2), 89–98. https://doi.org/10.1089/cell.2017.0051 (2018).
Palta, P., Selokar, N. L. & Chauhan, M. S. Production of water Buffalo SCNT embryos by handmade cloning. Methods Mol. Biol. 2647, 245–258. https://doi.org/10.1007/978-1-0716-3064-8_13 (2023).
Yadav, P. S. et al. Evaluation of postnatal growth, hematology, telomere length and semen attributes of multiple clones and re-clone of superior Buffalo breeding bulls. Theriogenology 213, 24–33. https://doi.org/10.1016/j.theriogenology.2023.09.024 (2024).
Chigateri, S. Negotiating the ‘sacred’ cow. cow.Slaughter and the regulation of difference in India. Democracy, Religious Pluralism and the Liberal Dilemma of Accommodation (ed Mookherjee)) https://doi.org/10.1007/978-90-481-9017-1_82011. (137–159). Springer (2011).
Vajta, G., Lewis, I. M., Hyttel, P., Thouas, G. A. & Trounson, A. O. Somatic Cell. Cloning Without Micromanipulators Cloning, 3(2), 89–95. DOI: https://doi.org/10.1089/15204550152475590 (2001).
Tecirlioglu, R. T. et al. Birth of a cloned calf derived from a vitrified hand-made cloned embryo. Reprod. Fertil. Dev. 15 (7–8), 361–366. https://doi.org/10.1071/RD03052 (2003).
Du, Y. et al. Piglets born from handmade cloning, an innovative cloning method without micromanipulation. Theriogenology 68 (8), 1104–1110. https://doi.org/10.1016/j.theriogenology.2007.07.021 (2007).
Shah, R. A. et al. Pregnancies established from handmade cloned blastocysts reconstructed using skin fibroblasts in Buffalo (Bubalus bubalis). Theriogenology 71 (8), 1215–1219. https://doi.org/10.1016/j.theriogenology.2008.10.004 (2009).
Hill, J. R. et al. Evidence for placental abnormality as the major cause of mortality in first-trimester somatic cell cloned bovine fetuses. Biol. Reprod. 63 (6), 1787–1794. https://doi.org/10.1095/biolreprod63.6.1787 (2000).
Edwards, J. L. et al. Cloning adult farm animals: a review of the possibilities and problems associated with somatic cell nuclear transfer. Am. J. Reprod. Immunol. 50 (2), 113–123. https://doi.org/10.1034/j.1600-0897.2003.00064.x (2003).
Lanza, R. P. et al. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science 288 (5466), 665–669. https://doi.org/10.1126/science.288.5466.665 (2000).
Chavatte-Palmer, P. et al. Health status of cloned cattle at different ages. Cloning Stem Cells. 6 (2), 94–100. https://doi.org/10.1089/1536230041372274 (2004).
Wells, D. N., Forsyth, J. T., McMillan, V. & Oback, B. The health of somatic cell cloned cattle and their offspring. Cloning Stem Cells. 6 (2), 101–110. https://doi.org/10.1089/1536230041372300 (2004).
Vlasova, A. N. & Saif, L. J. Bovine immunology: implications for dairy cattle. Front. Immunol. 12, 643206. https://doi.org/10.3389/fimmu.2021.643206 (2021).
Betts, D. H. et al. Telomere length analysis in goat clones and their offspring. Mol. Reprod. Dev. 72 (4), 461–470. https://doi.org/10.1002/mrd.20371 (2005).
Tian, X. C., Xu, J. & Yang, X. Normal telomere lengths found in cloned cattle. Nat. Genet. 26 (3), 272–273. https://doi.org/10.1038/81559 (2000).
Wani, N. A., Praveen, K. K., Hong, S. & Umer, M. A. Telomere length in dromedary camels (Camelus dromedarius) produced by somatic cell nuclear transfer (SCNT) and their age-matched naturally produced counterparts. Theriogenology 177, 151–156. https://doi.org/10.1016/j.theriogenology.2021.10.012 (2022).
Bao, L. et al. Impact of telomere length and mitochondrial DNA copy number variants on survival of newborn cloned calves. Theriogenology 225, 1–8. https://doi.org/10.1016/j.theriogenology.2024.05.019 (2024).
Wakayama, T. et al. Cloning of mice to six generations. Nature 407 (6802), 318–319. https://doi.org/10.1038/35030301 (2000).
Williams, N. E. et al. A comparison of reproductive characteristics of boars generated by somatic cell nuclear transfer to highly related conventionally produced boars. Cloning Stem Cells. 8 (3), 130–139. https://doi.org/10.1089/clo.2006.8.130 (2006).
Polejaeva, I. A. et al. Longitudinal study of reproductive performance of female cattle produced by somatic cell nuclear transfer. Plos One. 8 (12), e84283. https://doi.org/10.1371/journal.pone.0084283 (2013).
Enright, B. P. et al. Reproductive characteristics of cloned heifers derived from adult somatic cells. Biol. Reprod. 66 (2), 291–296. https://doi.org/10.1095/biolreprod66.2.291 (2002).
Heyman, Y. et al. Zootechnical performance of cloned cattle and offspring: preliminary results. Cloning Stem Cells. 6 (2), 111–120. https://doi.org/10.1089/1536230041372364 (2004).
Saini, M. et al. Semen parameters and fertility potency of a cloned water Buffalo (Bubalus bubalis) bull produced from a semen-derived epithelial cell. Plos One. 15 (8), e0237766 (2020).
Selokar, N. L. et al. Establishment of a somatic cell bank for Indian Buffalo breeds and assessing the suitability of the cryopreserved cells for somatic cell nuclear transfer. Cell. Reprogram. 20 (3), 157–163. https://doi.org/10.1089/cell.2017.0066 (2018).
Selokar, N. L. et al. Effect of post-fusion holding time, orientation and position of somatic cell-cytoplasts during electrofusion on the development of handmade cloned embryos in Buffalo (Bubalus bubalis). Theriogenology 78 (4), 930–936. https://doi.org/10.1016/j.theriogenology.2012.03.018 (2012).
Joglekar, M. V. et al. An optimised Step-by-Step protocol for measuring relative telomere length. Methods Protocols. 3 (2), 27. https://doi.org/10.3390/mps3020027 (2020).
Acknowledgements
This work was supported by a grant, ICAR-LBS award/801/1014521, from the Indian Council of Agricultural Research, New Delhi, India. We are grateful to Dr. Rahul Meena for assisting in initial OPU experiments and Ms. Anushka for telomere length analysis.
Author information
Authors and Affiliations
Contributions
NLS and MKS were involved in ovum pick-up (OPU), somatic cell nuclear transfer (SCNT), embryo transfer (ET) experiments, and data collection. GT contributed to SCNT procedures and cytokine analysis. KP and RV were responsible for the preparation of OPU-donor and embryo-recipient animals, embryo transfer, and the care and management of the resulting calf. PS performed in-vitro fertilization (IVF) procedures. AS assisted in initial OPU experiments. NLS and MSC provided project supervision, secured funding, and were responsible for manuscript review and editing. All authors reviewed and approved the final version of the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Selokar, N.L., Singh, M.K., Tripathi, G. et al. Birth of India’s first cloned cattle and analysis of its health and reproduction status: A case study. Sci Rep 15, 26957 (2025). https://doi.org/10.1038/s41598-025-10225-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-10225-5