Abstract
In this work, we proposed a novel inferential procedure assisted by machine learning based adjustment for randomized control trials. The method was developed under the Rosenbaum’s framework of exact tests in randomized experiments with covariate adjustments, replacing the traditional linear model with nonparametric models that capture the complex relationships between covariates and outcomes. Through extensive simulation experiments, we showed the proposed method can robustly control the type I error and can boost the statistical efficiency for a randomized controlled trial (RCT). This advantage was further demonstrated in a real-world example. The simplicity, flexibility, and robustness of the proposed method makes it a competitive candidate as a routine inference procedure for RCTs, especially when nonlinear association or interaction among covariates is expected. Its application may remarkably reduce the required sample size and cost of RCTs, such as phase III clinical trials.
Similar content being viewed by others
Introduction
The randomized controlled trial (RCT) is the gold standard in determining treatment efficacy. RCTs are characterized by their extensive participant involvement, which typically encompass a substantial cohort size, particularly evident in phase III clinical trials. However, this rigorous process comes at a considerable cost. Especially for trials on immunotherapy, the cost of some of the newest therapies can reach $850,000 per patient when including the value of medical support necessary to deliver these treatments1. Given the significant investment required, maximizing the statistical power of RCTs becomes critical.
A common approach to improve the statistical efficiency, or power, is baseline adjustment using linear models, such as analysis of covariance (ANCOVA). Related models and their applications in RCT has been extensively discussed2,3. The method of covariate adjustments is supported by FDA in phase III oncology trials due to the enhanced efficiency and its minimal impact on bias or type I error rate4. Hence, the agency advises sponsors to incorporate adjustments for covariates expected to show the strong association with the outcome of interest. In 2002, Rosenbaum proposed method of exact inference which is free of distributional assumptions5. This method is endorsed by the FDA guidance, which states that “Sponsors can conduct randomization/permutation tests with covariate adjustment (Rosenbaum 2002)”. Although this method was based on ordinary least square (OLS) linear regression, the author raised possibility of using more general forms of covariate adjustment.
Nowadays, due to the ever-decreasing cost of medical testing and sequencing techniques, the number of baseline data available in clinical trials has been growing dramatically. The rich data can provide a much finer picture of individuals and thus capture larger variation in the outcomes. This makes it possible to design inferential procedures with superior efficiency. However, the traditional adjustment methods using OLS method quickly break down when the number of covariates increases to a relatively large scale. Even if penalized regressions6,7,8,9 can accommodate high-dimensional data, such linear models are neither capable of handling nonlinear associations nor complex interactions.
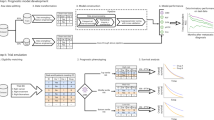
On the other hand, machine learning techniques have been successfully applied in numerous settings during the last decade. Supervised learning methods such as random forests (RFs), boosting machines, neural networks8,10,11,12 are known to be capable of handling high-dimensional data and model complex functional forms. While efforts have been made towards the application of machine learning models for the covariates adjustment in randomized studies, most of the works focused on estimations and asymptotics13,14,15. In this work, we propose a novel approach of machine learning based covariate adjustment under the Rosenbaum’s framework, which makes exact inference possible5. The proposed method focuses on continuous outcomes, and is shown to be an unbiased and flexible adjustment method to boost the statistical efficiency for a RCT. Compared to existing methods, the proposed approach offers greater flexibility and efficiency in controlling Type I error while enhancing statistical power. It enables exact inference without requiring repeated model training, thereby streamlining study planning and simplifying the modeling process. These characteristics make it a novel and powerful inferential procedure for RCTs, especially when nonlinear association or between-covariate interaction is expected. The RF model is used under this framework as a proof of concept, but it can be extended to more general modeling approaches, provided the model avoids overfitting the data. Its application may remarkably reduce the required sample size and cost of RCTs, especially for phase III clinical trials.
Methods
Hypothesis testing under non-parametric adjustment
In this work, we focus on RCTs with continuous outcomes and with an objective to compare two group means. Our method is based on the Rosenbaum’s framework. For details, please see5. As a brief background, suppose there are n individuals, \(j = 1, \dots , n\), and the response of inividual j would be \(r_{T_j}\) or \(r_{C_j}\) if j were assigned to treatment or control, respectively16,17. The treatment effect can be written as \(\tau _i = r_{T_j} - r_{C_j}\), and it can never be observed because individual j can only receive one of the treatments. The treatment effect is additive if the treatment causes the response to change by a fixed amount \(\tau\) for every j. In other words, the treatment effect is the same for all individuals, regardless of their characteristics. The covariate vector \(x_j\) describes the baseline characteristic of j. The variable \(Z_j\) is the treatment indicator, where \(Z_j=1, 0\) indicates j is assigned to the treatment or control group, respectively. Thus, the response of j can be written as \(R_j=Z_jr_{T_j} + (1-Z_j)r_{C_j}\). Following Rosenbaum5, we denote \(\textbf{Z}=(Z_1, Z_2, \dots , Z_n)^T\), \(\textbf{R}=(R_1, R_2, \dots , R_n)^T\), \(\textbf{r}_c=(r_{C_1}, r_{C_2}, \dots , r_{C_n})^T\) and use \(\textbf{X}\) as the covariate matrix whose jth row is \(\textbf{x}_j^T\). The fixed but unobserved vector \(\textbf{r}_C\) are potential outcomes of all participants if assigned to the controlled group, which can be expressed as \(\textbf{r}_C=\textbf{R} - \tau \textbf{Z}\) when the treatment effect is additive. Under this framework, the only stochastic component is the random assignment \(\textbf{Z}\). The test of \(H_0:\tau =\tau _0\) can be performed using the statistic \(t(\textbf{Z}, \textbf{Y})\), where \(\textbf{Y} = \textbf{R} - \tau _0\textbf{Z}\). The response variable \(\textbf{Y}\) is the observed outcome \(\textbf{R}\) adjusted with respect to \(\tau _0\), which will be the variable used in the inference procedure. Thus we have \(\textbf{Y} = \textbf{R}\) when \(\tau _0=0\). Under this framework, the exact inference is the randomization inference derived from the randomization distribution of the statistics. Here, exact inference refers to randomization inference, which is based on the randomization distribution of the statistics5. The randomization distribution is essentially a collection of all possible outcomes one could observe from a study, assuming that any treatment or intervention is assigned randomly. The parametric distributions such as the normal distribution are used as approximations to randomization distributions.
When covariate information \(\textbf{X}\) is available and associated with the response variable \(\textbf{Y}\), we can write \(\textbf{Y}=g(\textbf{X})+\textbf{e}\). The residual \(\textbf{e}\) is expected to have less variation than \(\textbf{Y}\) if the baseline covariates explain part of the variation, and it can be used in place of \(\textbf{Y}\) for inference. Thus the test statistic can be modified to \(t(\textbf{Z}, \textbf{e})\), which can lead to an improvement in efficiency due to smaller variation in \(\textbf{e}\).
Linear models are natural choice of g, but they tend to fail when the association is complex or the data is high dimensional. Here we investigate whether using modern machine learning method will achieve better efficiency for testing \(H_0:\tau =\tau _0\) as compared to the straight two group comparison, while controlling the type I error rate properly. In this work, we select RF for the exact inference approach. The RF model is an ensemble learning method that builds multiple decision trees and merges their outputs to improve prediction accuracy. Each tree is trained on a bootstrapped data set. Further, at each decision tree split, a random subset of p variables (features) is considered rather than using all available predictors. RFs are particularly useful for handling large datasets with many variables, and they can model complex relationships by combining the predictions of multiple trees.
During the training process, for each tree, some data points are left out of the training set due to the bootstrap sampling process. These left-out points are referred to as“out-of-bag.” The Random Forest model can then make predictions for these out-of-bag (OOB) data points using the trees that were not trained on them. The OOB predictions provide an internal validation measure without the need for a separate validation dataset.
The RF approach was selected for our approach due to its minimal requirement of parameter tuning and invariance to monotonic transformation of features10. More importantly, the OOB predictions can be used to attenuate the overfitting, thus avoiding manually splitting the data into training and validation set. Such simplicity makes it ideal for RCTs, which typically requires detailed preplanning.
The proposed procedure for testing \(H_0:\tau =\tau _0\) is straight forward:
-
1.
Use the data from all individuals and build a RF model using \(\textbf{X}\) as the features and \(\mathbf {R - \tau _0\textbf{Z}}\) as the response \(\textbf{Y}\).
-
2.
Obtain the OOB predictions for all individuals and calculate the residuals \(\textbf{e}\).
-
3.
Perform the permutation test, Wilcoxon rank-sum test or two sample t-test based on \(\textbf{e}\) and \(\textbf{Z}\).
-
4.
Estimate the treatment effect, standard error or confidence interval by treating \(\textbf{e}\) as the outcome variable.
In step 3, the selection of test follows the same principle as in the simple two-group comparisons. In general, Wilcoxon rank-sum test is more robust to extreme values and does not rely on any underlying distribution assumptions. While t-test could be more efficient when the distribution assumption holds. However, as we will show in the simulations studies, the Wilcoxon rank-sum test generally outperforms t-test under the scenarios investigated.
Estimation
Without loss of generality, we focus on the null hypothesis \(H_0: \tau _0=0\) and relax the additivity assumption for the discussion of estimation. When \(H_0: \tau _0=0\), we have \(\textbf{Y} = \textbf{R}\) which is the observed response. Conceptually, the adjustment by \(g(\textbf{X})\) shifts the response at any point in the covariate space by the same amount for both arms. Therefore, the difference in the residuals should be the same as that in the original response, so the treatment effect is estimated as,
where \(n_0\) and \(n_1\) are numbers of individuals in control and treatment groups. The same estimator has been proposed by Opper14 for a different inference procedure. The condition for \(\hat{\tau }\) in Eq. 1 to be unbiased estimator was also given by Theorem 1 in14. In our framework, the estimation of \(g(\textbf{X})\) does not involve treatment assignment, so it is invariant under randomization. Therefore, the only stochastic component is \(Z_i\). We can write \(Y_i = r_{C_i} + Z_i\tau _i, i=1,2,\dots n\), then the expectation of estimated treatment effect is given as,
Under additivity, we have \(\tau _i=\tau , i=1, 2, \dots , n\). Thus, \(E(\hat{\tau }) = \tau + \frac{1}{n}\sum _{i=1}^n(g(X_i)|_{Z_i=1} - g(X_i)|_{Z_i=0})\). Therefore, a sufficient condition for \(\hat{\tau }\) to be an unbiased estimator, i.e., \(E(\hat{\tau }) = \tau\) is
One would assume that this condition is naturally fulfilled since the estimation of g does not involve treatment assignment. However, it can be violated particularly if g overfits the data. Overfitting occurs when \(g(X_i)\) is overly close to the observed response \(Y_i\), meaning that as the flexibility of g increases, we have \(g(X_i)\rightarrow r_{C_i}\) when \(Z_i=0\), and \(g(X_i)\rightarrow r_{C_i} + \tau _i\) when \(Z_i=0\). Consequently, we have \(E(g(X_i)|_{Z_i=1}) - E(g(X_i)|_{Z_i=0})\rightarrow \tau _i\) and \(E(\hat{\tau }) \rightarrow 0\). This suggests when g overfits the data, the estimated treatment effect will be biased towards 0. As long as the machine learning model is adequately flexible, it can always overfit the data even if the sample size is large (e.g., the k Nearest Neighbor with \(k=1\)). Therefore, for models with high complexity, the bias term is O(1) as \(n\rightarrow \infty\), thus simply increasing the sample size does not necessarily mitigate the bias. One way to counter this problem is to use the predictions from models not trained on the observations at question, which can be easily achieved through cross-validations, or OOB predictions.
Next, we derive the variance of the permutation distribution of \(\hat{\tau }\). Note that \(\hat{\tau }\) can be rewritten as,
Recall that in our method, the residuals are fixed after the estimation of g. In addition, the only random components is \(Z_i\), which follows a binomial distribution with \(p=n_1/n\). Therefore, the variance of the estimator is given as,
The equation above suggests the variance will be smallest when \(\sum ^n_{i=1}e_i^2\) is minimized, which corresponds to a model g that is optimized towards minimal mean squared error. This observation bridges the inference problem with classic predictive modeling problems. It is notable that Opper obtained the same form of variance in his work14. However, the author ignored the variability of \(g(\textbf{X})\), and assumed the residuals to be fixed, which does not align with their procedure that requires estimation of \(g(\textbf{X})\) under each randomization. Interestingly, the same expression is correct under our framework, where the residuals are indeed fixed because estimation of \(g(\textbf{X})\) does not depend on \(\textbf{Z}\).
When the treatment effect is not additive, it can vary among patients, which depends on the patients’ covariates. In this case, the treatment effect is defined as \(\tau = \frac{1}{n}\sum _{i=1}^n\tau _i\). Therefore, the above results on unbiasedness still hold. On the other hand, non-additivity implies that there are interactions between treatment and covariates. Thus the function \(g(\textbf{X})\) can be better described as two separate functions for two arms, namely \(g_0(\textbf{X})\) and \(g_1(\textbf{X})\). Although testing the null hypothesis under the non-additivity assumption is not very well defined, a natural question under non-additivity is whether modeling \(g_0(\textbf{X})\) and \(g_1(\textbf{X})\) individually will gain efficiency over estimating a common regression function \(g(\textbf{X})\) by pooling the observations from two arms together. Such a strategy was adopted by Wager13 and Opper14. To investigate this problem, we examined the estimator of treatment effect from the cross-estimation (CE) method by Wager13,
Here, we ignored the \((-i)\) notation, which was used to denote that the estimator of g was not dependent on the ith training ovservation. Then based on their result, the variance of the estimator is given as,
where \(n_z= {n \atopwithdelims ()n_0}\) is the number of all unique randomizations, or possible group assignments, and \(p=n_1/n\). Note that the variance depends on \(g_0\) and \(g_1\) only through a weighted sum \(g(X_i) = (1-p)g_0(X_i)+pg_1(X_i)\), which can be estimated through pooling the observations from two arms and proper weighting. Therefore, even under non-additivity, the knowledge of \(g_0\) and \(g_1\) does not lead to a further gain in efficiency, so the estimation of \(g(\textbf{X})\) across two components is unnecessary. In fact, under a balanced design, the sample size for training the model will be halved to train the models for the control and treatment groups separately, which can result in reduced model performance and consequently lower inference efficiency. Therefore, the proposed method is not only more straightforward for exact inference, but also achieves a better efficiency through pooling observations from both intervention arms.
In practice, the estimation and hypothesis testing can be carried out using the procedure specified in Section 2.1. Additionally, the Random Forest (RF) model for regression can be trained using the default settings, which typically include 500 to 1000 fully-grown trees, and the use of p/3 features for splitting at each node, where p is the number of input features. These default configurations are often sufficient for many practical applications, providing a good balance between model complexity and computational efficiency.
Simulations
To demonstrate the performance of the proposed method, we conducted a set of comprehensive simulation studies under various scenarios for testing a one-sided hypothesis \(H_0: \tau =0\) versus \(H_1: \tau >0\). The results can be readily extended to two-sided tests. In each scenario, we generated samples of size \(N=50, 100, 200\) and 400 independent individuals, who were then randomized into two balanced groups. There were \(p=40\) mutually independent covariates following standard normal distributions. Only the first four covariates are useful predictors. In each setting 10,000 Monte Carlo simulations were performed. The outcome y was generated using the four different models:
-
1.
Model 1 (primarily nonlinear no interaction):
$$y = \tau Z + \beta \sigma (x_1/2) + \beta x_2^2 + \beta cos(x_3) + \beta x_4 + \epsilon ,$$ -
2.
Model 2 (primarily nonlinear with interaction):
$$y = \tau Z + \beta \sigma (x_1/2) + \beta x_2^2 + \beta cos(x_3) + \beta sign[cos(x_3)]x_4 + \epsilon ,$$ -
3.
Model 3 (primarily linear no interaction):
$$y = \tau Z + \beta \sigma (x_1/2) + \beta x_2 + \beta x_3 + \beta x_4 + \epsilon ,$$ -
4.
Model 4 (primarily nonlinear with by treatment interaction):
$$y = \tau Z + \beta [\sigma (x_1/2)-1/2]Z + \beta x^2_2 + \beta cos(x_3) + \beta x_4 + \epsilon ,$$
where Z is the treatment indicator, \(\tau\) is the treatment effect, \(\sigma (x)=\exp (x)(1+\exp (x))^{-1}\) is the sigmoid function, \(x_k\) is the kth covariate, and \(\epsilon\) is the error term. The subscript for individual is omitted. We examined \(\beta =0.2, 0.5, 0.8\) to represent different levels of association between the covariates and the outcome measure. In addition, we utilized error terms from standard normal, log-normal, and Gumbel distributions. For comparing the outcomes between the two treatment arms we utilized the Wilcoxon rank-sum test and the two-sample t-test. We also included t-test and Wilcoxon rank-sum test on the residuals from linear regression on all baseline covariates (t-test-LM and Wilcoxon-LM). Finally, we included the proposed RF adjusted methods, which use the t-test and Wilcoxon rank-rum test on the residuals from RF model (t-test-RF and Wilcoxon-RF). The Wager’s cross-estimation (CE) method was also included for comparison, where a one-sided Z-test was constructed based on the estimate and standard error from the ate.randomForest function in R crossEstimation package13. We examined the type I error control with \(\tau =0\), and the statistical power with \(\tau =0.3\) and 0.6. The results of all simulations are shown in the supplementary materials.
Figure 1 shows that all tests robustly control the type I error rates when \(\tau =0\) under non-Gaussian error terms. In models with primary nonlinear associations, the RF adjusted tests (t-test-RF and Wilcoxon-RF) show higher power than the other methods. In the right panel of Fig. 2, we see that the RF tests attains over 80% power at around \(N=170\), while the other tests require approximately 80 more individuals to achieve the same power. It is notable that, when the error term is non-Gaussian, the Wilcoxon-RF further outperforms t-test-RF. In models with primary linear associations, the performance of RF tests is mostly comparable with that of the linear adjusted methods. Only when the association is very strong (\(\beta =0.8\)), the linear model methods will have an advantage of around 7% (Figures S3 & S7). This is because linear model is sufficient to capture the relationship between response and covariates. It should be noted that under some scenarios (e.g. small sample sizes), the linear model approach tends to hurt the efficiency, having power lower than the straight t-test and Wilcoxon test (Figs. 1 and 2, N=40, 100), even when the true association is linear (Figures S3, S7 and S11, N=40, 100). This is due to the instability of ordinary least square estimators when p is close to N. On the other hand, the RF tests have similar or better performance than the unadjusted tests in most cases. One exception is the setting with weak associations and log-normal errors (Fig. 3). However, as the association becomes stronger, the RF test still outperforms other methods. In addition, although the Rosenbaum’s framework assumes an additive treatment effect, the proposed method shows equally well type I error control and efficiency improvement when there are interactions between treatment and covariates (Figures S4, S8, S12). It is also notable that our proposed method consistently outperforms the CE method13, even when the covariate by treatment interaction is present. This demonstrates that the explicit modeling of g separately for two arms using nonparametric methods reduces statistical efficiency due to the smaller training sample sizes. The results are similar when the randomization is unbalanced (data not shown). Overall, our simulations suggest in most cases the Wilcoxon-RF test as the best option among the methods tested. All simulation results are also provided in the supplementary materials.
The t-test-RF and Wilcoxon-RF rely on the same assumptions of the residuals as the straight t-test and Wilcoxon rank-sum test. It is also notable that occasionally a simple linear model based test can outperform the RF tests. In practice, when preliminary data is available, it can be used to inform the selection of the most appropriate modeling methods, e.g. linear versus RF, by examining the relationship between the response variable and covariates. Additionally, as shown in Eq. 4, a model that minimizes prediction error also maximizes statistical power. Consequently, when sufficient preliminary data is available, the choice of testing methods can be viewed more broadly as a model selection problem in a more general sense.
While the focus of this work is on the hypothesis testing procedures, we also performed a simulation study on the coverage of the 95% confidence intervals and bias of the estimator under Wilcoxon-RF test. The exact confidence interval was obtained by the algorithm described in Bauer18. For the simulations, we focused on Model 1 with a treatment effect of 0.3. The results are shown in the supplementary table. Under all scenarios, the coverage probability of the confidence intervals are well controlled at 0.95 and the bias was close to 0.
Diet intervention data example
In this example, the investigators studied whether a brief diet intervention can reduce the symptoms of depression in young adults19. There were 76 participants randomly allocated to a brief 3-week diet intervention (Diet Group) or a habitual diet control group (Control Group), with \(n=38\) per group. The primary and secondary outcome measures were assessed at both baseline and after the intervention, which included symptoms of depression (Centre for Epidemiological Studies Depression Scale, CESD-R; and Depression Anxiety and Stress Scale-21 depression subscale, DASS-21-D), current mood (Profile of Mood States, POMS), self-efficacy (New General Self-Efficacy Scale, GSES) and memory (Hopkins Verbal Learning Test). There are a total of 23 baseline variables that can be used as covariates. The GSES score were log-transformed as specified in the article. The ANCOVA was used for the analysis of both primary and secondary outcomes with the baseline scores adjusted as covariates. The results showed that the Diet group had significantly lower self-reported depression symptoms than the Control Group on the CESD-R and DASS-21 depression subscale. However, none of the POMS scores showed significant difference. Here we used the Wilcoxon-RF method to re-analyze the difference in POMS fatigue scores between two groups. Two-sided tests were used at \(\alpha =0.05\). The results in Table 1 show that only the Wilcoxon-RF shows a significant difference in the post-intervention fatigue scores between two groups (\(p=0.0451\)). This demonstrates the higher statistical power of the proposed method. Based on the residuals, the final estimated treatment effect is -1.57 (Diet vs. Control), with 95% confidence interval of (-3.01, -0.05). The variable importance from RF model suggests baseline depression is the most important predictor. The partial dependence plot (Figure 4) suggests the post-intervention fatigue does not change with the baseline depression within the range of \(0 \sim 5\), but starts to show a linear trend in the range of \(6\sim 12\), and then plateaus. The sigmoid shape of what may not be well described by a linear model, but can be better captured by a nonlinear model like RF, which explains why our method achieves better efficiency.
In medical research, a phase II trial is typically conducted to preliminarily assess the efficacy before conducting a large scale phase III RCT. The data collected from the phase II trial provides important parameters that can guide the design of the phase III trial. One can simply use the two-sample t-test formula as an approximation for the exact test, and estimate the proportion of variance that will be reduced by including the covariates based on assumptions or data available, e.g. from a phase II trial. Approximately, if variance explained by the model is \(\gamma\), then one would expect a reduction of \(N\gamma\) in sample size requirement, where N is the sample size needed using an unadjusted two-sample test.
In this part, we use an imaginary scenario of the study above to show how the proposed method can be used to reduce the sample size at the planning stage. In this hypothetical scenario, the study above failed to meet the endpoint but a promising trend was observed, and the data is used to plan for a large scale confirmatory trial. Based on the data, the within-group standard deviation of the response is 4. The RF model explained 19% variance in the response, and the standard deviation of the residual is 3.6. Suppose the minimal clinically meaningful difference we plan to detect is 1, then at a significance level of 0.05, the sample size required to achieve 80% power based on the proposed method is \(N=100\) per group. Without adjustment, the sample size needed is \(N=126\) per group. This corresponds to 20% reduction in required sample size.
Discussion
In this work, we presented a novel inferential procedure for RCTs assisted by a machine learning adjustment. Through a set of simulation experiments and a real-world example, we showed the proposed method remarkably boosts the statistical power and can reduce 1/3 sample size requirement as compared with other approaches. Such improvement is grounded in the capability of machine learning in capturing nonlinear relationships and higher order interactions, which cannot be fully exploited by linear models. We also demonstrated that the proposed method has proper type I error control. Meanwhile, as long as the outcome has a moderate level of dependency with the baseline covariates, the Wilcoxon-RF method never performs worse than the standard two-sample tests. Hence, the risk of using the proposed method is minimal, while the potential gain is substantial. In contrary, the methods using adjustment by linear regression can sometimes harm the efficiency, which can be attributed to its instability when p is large relatively to N. Shrinkage methods such as Ridge6, Lasso7 or Elastic net9 may be used, but they all require careful tuning of hyperparameters, and they do not address the difficulty brought by non-linearities and interactions.
The RF model is often considered an “off-the-shelf” supervised learning method due to its simplicity of usage. Typically, with the recommended settings one can obtain a model with good performance11. The usage of OOB observations further makes explicit training, validation set splitting unnecessary. Therefore, in an ideal scenario, the inference procedure can be completed in a single run of RF model training. This feature makes it particularly advantageous because the FDA has clear expectations that the analyses of primary and secondary endpoints in clinical trials need to be pre-specified. The post-hoc analyses are prone to data-driven manipulations which can lead to biased interpretation of the results. Therefore, any modeling method that is difficult to preplan and requires extensive tuning of hyperparameters can negatively affect the weight of evidence from an RCT for the regulatory decision-making. Another motivation for using RF is that it usually shows better or comparable prediction performance than neural networks in the range of sample sizes typically used in a randomized study. Deep learning models usually require larger sample size to gain advantage12. However, no model can be optimal under all scenarios. Although RF is used as a proof of concept in our study, other modeling methods can be superior to RF in many settings. Therefore, investigation of the proposed approach to incorporate other machine learning techniques is important.
A practical challenge is how to translate the gain in efficiency into a reduction of the sample size. This requires the knowledge of the variation that can be explained by the machine learning model at the planning stage. As shown in the example, such information can typically be obtained from an early phase trial or other preliminary data in medical research. In such case, if the variance explained by the model is \(\gamma\), then sample size requirement after adjustment will be \(N(1-\gamma )\). The parameters may also be used to run simulations to estimate the sample sizes required for the machine learning adjusted tests. Another option is an interim analysis on the efficiency gain and adjust the sample size accordingly. Such strategy may require further methodological investigation.
Data availability
The code for implementing the method is available at https://github.com/hyu-ub/ML_RCT No datasets were generated or analysed during the current study. The data used for the real world example is publicly available from supplement data file (S3) of the article: https://doi.org/10.1371/journal.pone.0222768
References
Schaft, N. et al. The future of affordable cancer immunotherapy. Frontiers in Immunology 14, 1248867 (2023).
Ye, T., Shao, J., Yi, Y. & Zhao, Q. Toward better practice of covariate adjustment in analyzing randomized clinical trials. Journal of the American Statistical Association 118(544), 2370–2382 (2023).
Van Lancker, K., Bretz, F. & Dukes, O. Covariate adjustment in randomized controlled trials: General concepts and practical considerations. Clinical Trials, page 17407745241251568, (2024).
Center for Drug Evaluation, Research, Center for Biologics Evaluation, Food Research, U.S. Department of Health Drug Administration (FDA), and Human Services. Adjusting for covariates in randomized clinical trials for drugs and biological products, draft guidance for industry. (2021).
Rosenbaum, P. R. Covariance adjustment in randomized experiments and observational studies. Statistical Science 17(3), 286–327 (2002).
Hoerl, A. E. & Kennard, R. W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 12(1), 55–67 (1970).
Tibshirani, R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B: Statistical Methodology 58(1), 267–288 (1996).
Chen, T. & Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, 785–794, (2016).
Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society Series B: Statistical Methodology 67(2), 301–320 (2005).
Breiman, L. Random forests. Machine learning 45, 5–32 (2001).
Hastie, T., Tibshirani, R., Friedman, J. H. & Friedman, J. H. The elements of statistical learning: data mining, inference, and prediction, volume 2. Springer, (2009).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. nature 521(7553), 436–444 (2015).
Wager, S., Wenfei, D., Taylor, J. & Tibshirani, R. J. High-dimensional regression adjustments in randomized experiments. Proceedings of the National Academy of Sciences 113(45), 12673–12678 (2016).
Opper, I. M. Improving average treatment effect estimates in small-scale randomized controlled trials. In EdWorkingPaper: 21–344. (2021).
Williams, N., Rosenblum, M. & Díaz, I. Optimising precision and power by machine learning in randomised trials with ordinal and time-to-event outcomes with an application to covid-19. Journal of the Royal Statistical Society Series A: Statistics in Society 185(4), 2156–2178 (2022).
Splawa-Neyman, J., Dabrowska, D. M. & Speed, T. P. On the application of probability theory to agricultural experiments. essay on principles. section 9. Statistical Science, pages 465–472, (1990).
Rubin, D. B. Estimating causal effects of treatments in randomized and nonrandomized studies. Journal of educational Psychology 66(5), 688 (1974).
Bauer, D. F. Constructing confidence sets using rank statistics. Journal of the American Statistical Association 67(339), 687–690 (1972).
Francis, H. M. et al. A brief diet intervention can reduce symptoms of depression in young adults-a randomised controlled trial. PloS one 14(10), e0222768 (2019).
Funding
This work was supported by Roswell Park Cancer Institute and National Cancer Institute (NCI) grant P30CA016056, NRG Oncology Statistical and Data Management Center grant U10CA180822 and IOTN Moonshot grant U24CA232979-01, ARTNet Moonshot grant U24CA274159-01, CAP-IT grant U24CA274159-02.
Author information
Authors and Affiliations
Contributions
H.Y. and A.H. conceived of the presented idea. H.Y. developed the method. H.Y. and X.M. performed the computations. All authors wrote and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, H., Hutson, A. & Ma, X. Machine learning assisted adjustment boosts efficiency of exact inference in randomized controlled trials. Sci Rep 15, 24454 (2025). https://doi.org/10.1038/s41598-025-10566-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10566-1