Abstract
Rice suffers from drought stress due to its shallow roots, hindering water uptake from deeper soil layers. Transfer of deeper rooting QTL will modify the root architecture of the plant allowing it to extract moisture from deeper layers of the soil. Furthermore, poor soil conditions, particularly phosphorus deficiency is a common problem in the rainfed rice ecology of India which requires adequate phosphate fertilizer for obtaining higher yield. In this current investigation, two QTL (DRO1 and DRO3) for deeper rooting and one QTL for phosphorus uptake (Pup1) were introgressed into a popular variety, ‘Maudamani’ through marker-assisted backcross breeding. The target QTL were transferred from a pre-breed genotype, CR3996-19-9-45-1 into the popular variety. Foreground selection was performed using the tightly linked microsatellite markers in the backcross generations progenies to select plants carrying the target QTL. Background selection in each backcross generations was performed using 123 polymorphic SSR markers spread across twelve chromosomes. In each backcross generations, plants carrying all three target QTL and with highest recurrent parent genome recovery percentage was used to backcross with the popular variety, Maudamani. In BC2F2 generation, five plants (CR6508-111-101-129, CR6508-111-101-267, CR6508-111-101-413, CR6508-111-101-537 and CR6508-111-101-713) were selected which were homozygous for all three target QTL. The pyramided lines were evaluated for deeper rooting, low phosphorus stress tolerance and various agro-morphologic including quality traits. Progenies of those five pyramided plants showed similarity with the recipient parent for the 14 agro-morphologic and quality traits in their BC2F3 and BC2F4 generations. The introgressed lines demonstrated significantly improved root traits, including increased number of deeper roots, longer root length and higher shoot and root dry weight, compared to Maudamani under both moisture-deficit and normal conditions. Furthermore, those lines exhibited enhanced phosphorus uptake and grain yield compared to the recipient parent, Maudamani under low phosphorus conditions.
Similar content being viewed by others
Introduction
Climate change and ever-increasing population are two major concerns that will be affecting the future food security of India and the globe as well. For the rainfed ecosystem, drought is a major limiting factor for increasing grain yield due to inadequate rainfall along with poor crop stand, low availability of P due to P-fixation, low water holding capacity and low nutrient use efficiency1. Root traits are key factors for drought tolerance in rice2,3. In general, rice is very susceptible to water-deficit condition due to its shallow rooting system which limits water uptake from deeper part of the soil4. A deep root system might enable drought avoidance in plants and improve the yield during drought conditions5,6,7,8. Additionally, poor soil conditions have led to several macro- and micro-nutrient deficiencies in rice. About 65% of rainfed rice in Asia is being cultivated on stress associated soils and one of them is phosphorus deficiency9.
Deeper rooting trait is helpful for drought avoidance and it is a complex trait which is influenced by combination of root length as well as root growth angle10. Thus, the combined study of root angle and length is crucial for studies of deeper root distribution in soil. Many studies have been carried out on root length, root thickness, root volume11,12,13,14 but very less on root growth angle10,15. DRO1 is a major QTL for root growth angle which is present on chromosome 9 and identified in the mapping population developed from shallow rooting cultivar ‘IR64’ and deep-rooting upland rice ‘Kinandang Patong’16. Ratio of deeper rooting (RDR) was measured using ‘basket method’16,17. DRO1 is mainly expressed in root apical meristem, thereby helps in changing root architecture. The DRO1-kp allele increases rice grain yield under drought stress, without negatively impacting yield in irrigated conditions, benefiting farmers in both scenarios18. QTL for wider root cone angle and RDR were identified in a panel population through LD-based association mapping19,20,21, marking the first report of genotypic variation in the nodal root growth angle (RGA) for upland rice. A functional DRO1 allele showed wide variations in RGA18, suggesting that along with DRO1 gene, some other genes also affect RGA in rice. Another major QTL DRO3 was detected in the long arm of chromosome 7 which was also involved in genetic pathway of DRO122. DRO3 was reported in the F2 mapping population generated from the cross between Kinandang Patong and a DRO1-NIL on chromosome 7. It was also concluded that DRO3 is involved in the genetic pathway of DRO122. Another QTL for deeper rooting (DRO2) was detected on chromosome 4 explaining 32.0–56.6% phenotypic variance for RDR23. In addition, two QTL were detected viz., DRO4 on chromosome 2 and DRO5 on chromosome 624. However, these three QTL (DRO2, DRO4, and DRO5) do not exhibit epistatic interactions with DRO1. These studies suggest that few QTL affect the deeper rooting in rice along with many minor QTL.
Phosphorus is an essential nutrient which plays an important part of several key plant structure compounds. Phosphorus is a nutrient important for root growth and development25,26,27,28,29, tillering30,31,32,33, early flowering and ripening. It also helps in stem strength, improved seed production, early maturity, and increased resistance to biotic and abiotic stresses34,35. The major reason for phosphorus deficiency is P-fixation in soil. In acidic soil, phosphorus precipitates as Fe or Al phosphates which are less soluble while in alkaline soil, it forms insoluble precipitates with calcium. Phosphorus gets absorbed in plants at neutral pH of 6.0 to 7.036,37. Phosphorus deficiency is more profound in rainfed ecology. Although this problem is overcome by the application of P-based fertilizers, however, it is expensive for marginal farmers. Majority of the direct seeded rice ecology in India faces phosphorus deficiency very frequently. The farmers usually ignore application of right quantity of phosphatic fertilizers due to its high cost38. Thus, development of phosphorus deficiency tolerance is considered an effective method and priority research area. To develop rice genotypes with better phosphorus uptake and to reduce the cost of fertilizers39, attempted to identify putative QTL for phosphorus deficiency using 98 backcross inbred lines (BILs) developed from a cross between ‘Nipponbare’ (Japonica variety) and ‘Kasalath’ (indica variety). They found that QTL on chromosome 2, 6 and 12 were good candidates for P-uptake. On further characterization, QTL mapping placed the Pup1 in a 3 cM interval flanked by markers S14025 and S1312640. Substitution mapping of Pup1 QTL on chromosome 12 explained 70% of the phenotypic variability41. The first attempt to develop and validate molecular markers for Pup1 QTL was done by Chin et al.42. The protein kinase cloned gene, OsPSTOL1 was identified which promotes extensive root growth, enabling enhanced uptake of P from the soil43. Successful introgression of OsPSTOL1 through marker-assisted backcross breeding has been attempted38,44,45,46,47.
The QTL DRO1 and DRO3 play a crucial role in altering root angle and length, enabling plants to develop deeper root system to access soil moisture from deeper layers of soil. By introducing these QTL into high-yielding varieties, it can enhance their resilience to sudden moisture-deficit condition, mitigating potential yield losses. Furthermore, the Pup1 QTL facilitates improved phosphorus uptake from the soil. This experiment focuses on developing pyramided lines by incorporating DRO1, DRO3, and Pup1 QTL into the popular rice variety ‘Maudamani’ to promote deeper rooting and efficient phosphorus uptake.
Results
Validation of donor and recipient parents for the target traits
The donor and recipient parents were validated for the presence or absence of the target quantitative trait loci before the start of the hybridization program. The amplification results showed presence of positive band size of 225 bp, 177 bp, 168 bp, 656 bp and 523 bp by the markers RM242, RM5508, RM28012, RM28073 and Pup1-K46, respectively indicating presence of DRO1, DRO3 and Pup1 in the CR3996-19-9-45-1 (Fig. S1). However, the recipient parent, Maudamani showed the absence of the target bands.
Parental polymorphism study
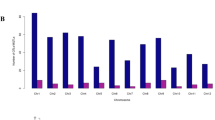
The parental polymorphism study was performed by genotyping 478 random SSR markers spread across all the twelve chromosomes of rice. Out of 478 markers, 123 SSR markers showed polymorphism between the parents (Maudamani and CR3996-19-9-45-1) were used for background selection in BC1F1 and BC2F1 generations (Fig. S2). The overall percentage of polymorphism found across all twelve chromosomes between the parents was 26.00% (Table S1).
Maker-assisted selection in F1 and BC1F1 progenies
The recipient parent, Maudamani was hybridized with CR3996-19-9-45-1 during wet season, 2021 and 410 F1 seeds were produced. After the hybridity test, 95 F1 plants were confirmed to be true hybrids using the trait specific markers during the dry season, 2022. One true F1 plant was backcrossed with the recipient parent, Maudamani in dry season, 2022 to generate BC1F1 seeds.
A total of one hundred eighty-nine BC1F1 seeds were generated from the backcross and were raised during the wet season, 2022. All the BC1F1 plants were genotyped using trait specific markers to detect the progenies carrying DRO1, DRO3 and Pup1 QTL and their combinations. Out of one hundred eighty-nine, 73 progenies showed heterozygous for RM242 (DRO1). The tracking for the QTL, DRO3 in those 73 positive progenies showed heterozygous for RM5508 (DRO3) in 29 progenies those carried both the DRO1 and DRO3 QTL. Also, the QTL tracked for Pup1 QTL using markers RM28102 (168 bp), RM28073 (656 bp) and Pup1-K46 (523 bp). Based on gene combination analysis, only 9 progenies were detected positive for all the three target QTL (Fig. S3). Background selection showed that the recurrent parent genome recovery varied from 70.10 to 81.03% with an average of 77.04% through SSR markers (Table 1). The highest genome recovery of 81.03% was detected in the progeny, CR6508-111 followed by CR6508-31 (80.11%) and CR6508-175 (79.35%) possessing the DRO1 + DRO3 + Pup1 QTL combinations. The progeny with highest genome recovery percentage with all the target traits and morphologically similar to recurrent parent was selected for backcrossing. The progeny CR6508-111 had all three target traits along with 81.03% genome recovery percentage and had phenotypic character similar to Maudamani. Therefore, CR6508-111 was backcrossed with Maudamani to generate BC2F1 progenies.
Marker-assisted selection in the BC2F1 and BC2F2 progenies
One hundred sixty-one BC2F1 seeds were generated and raised for selections in BC2F1 generation. The genotyping results of 161 BC2F1 progenies displayed heterozygosity for Pup1 QTL in 69 derived progenies. These individuals were further checked for the presence of DRO1 and DRO3 QTL using RM242 and RM5508 markers, respectively. Those sixty-nine individuals were checked for DRO1 QTL. A total of 25 progenies were found positive for DRO1. While checking for the presence DRO3 QTL in the positive progenies, 7 showed the presence of the 3 target QTL in heterozygous condition. Therefore, only 7 progenies were identified with all the three (DRO1 + DRO3 + Pup1) QTL combinations (Fig. S4). The background selections in those seven selected progenies showed recurrent parent genome recovery ranging from 82.14 to 93.26% with an average of 89.35% (Table 1). The highest genome recovery was 93.26% obtained from the progeny CR6508-111-101 followed by CR6508-111-94 (90.63%) and CR6508-111-141 (88.31%) possessing all the DRO1 + DRO3 + Pup1 QTL combination. Moreover, the morphological traits of CR6508-111-101 was more inclined towards Maudamani. Thereby, the seeds of BC2F1 line from CR6508-111-101 were self-bred for further evaluation in BC2F2 generation.
The identified BC2F1 plant was allowed for selfing to generate seeds for the BC2F2 generation. In BC2F2 generation, genomic DNA of 784 BC2F2 plants were subjected to PCR amplification using closely linked foreground markers. The presence of DRO1 QTL was tracked in the progenies using the marker RM242 and 181 progenies were detected to carry the QTL in homozygous condition. Those homozygous plants were checked for the presence of DRO3 QTL and 42 plants showed target bands for the presence of DRO1 + DRO3 in homozygous condition. While tracking for the third QTL, we could detect thirteen plants carrying DRO1 + Pup1 and 14 plants for DRO3 + Pup1. From banding pattern analysis, it was found that only five plants viz., CR6508-111-101-129, CR6508-111-101-267, CR6508-111-101-413, CR6508-111-101-537 and CR6508-111-101-713 were homozygous for the QTL DRO1 + DRO3 + Pup1 (Fig. 1). The final selected plants were subjected to phenotyping for deeper rooting, phosphorus uptake, grain quality and agro-morphological evaluation in dry season and wet season, 2024.
Cluster analysis with 14 agro-morphological and quality traits of two parental lines along with five pyramided lines revealed that all the five pyramided lines were identical to the recipient parent, Maudamani and were grouped in a single distinct cluster, while in another cluster the donor line CR3996-19-9-45-1 was placed (Fig. 2A). In the cluster-I, pyramided lines formed three different sub-clusters. The pyramided lines, CR6508-111-101-537 and CR6508-111-101-713 were placed in one sub-cluster with Maudamani. While the pyramided lines, CR6508-111-101-267 and CR6508-111-101-129 were placed in one group along with CR6508-111-101-413 revealing the greater similarity among them. However, all of the pyramided lines were present in one cluster with ‘Maudamani’ revealing their similarity with recipient parent ‘Maudamani’. Additionally, a dendrogram was generated using pyramided and parental lines based on the tracking of five foreground markers which grouped the recipient parent into one single cluster and the donor parent along with all pyramided lines in another cluster (Fig. 2B). Cluster I is further divided into three sub-clusters which includes CR3996-19-9-45-1 and CR6508-111-101-129 in one sub-cluster while other pyramided lines in another two sub-clusters. The pyramided line CR6508-111-101-129, CR6508-111-101-267, CR6508-111-101-413, CR6508-111-101-537 and CR6508-111-101-713 had highest genome recovery percentage and were almost similar to the popular parent, Maudamani (Fig. 2C).
(A) Dendrogram showing relatedness among five pyramided lines along with two parents (CR3996-19-9-45-1 and Maudamani) based on agro-morphological and quality traits; (B) Dendrogram showing the genetic relationship between lines based on five microsatellite markers and parents; (C) Percentage contribution of recurrent parent genome in the pyramided lines.
Analysis of genome introgression on the carrier chromosomes of the pyramided lines
Background analysis for recurrent parent genome recovery and genetic drag linked to donor segments was performed in the five pyramided lines using 123 background polymorphic markers and 5 foreground markers. The polymorphic markers for background analysis were carefully selected for all chromosomes to obtain maximum coverage in background screening. The foreground selection of the progenies in BC2F2 generation revealed that the five pyramided lines were homozygous for the target QTL. The DRO1 carrier chromosome 9 showed no linkage drag of donor fragment on either side of RM242 in all five pyramided lines (Fig. 3A). Similarly, there was no linkage drag observed in DRO3 and Pup1 carrying chromosome 7 and chromosome 12, respectively (Fig. 3B and C).
Graphical genotyping representation of QTL stacking and recipient parent’s genome recovery for the carrier chromosomes associated with deeper rooting and phosphorus uptake QTL in five pyramided lines (A) DRO1 QTL on carrier chromosome 9 (B) DRO3 QTL on carrier chromosome 7 (C) Pup1 QTL on carrier chromosome 12 present in BC2F2 progenies of Maudamani/ CR3996-19-9-45-1 1 (Legend A: CR3996-19-9-45-1; Legend B: Maudamani).
Evaluation of the pyramided lines for deeper rooting trait
Five pyramided lines along with parents were evaluated for deeper rooting under moisture-deficit conditions. During the stress period, both donor and pyramided lines exhibited slight leaf rolling (SES score 1), indicating a moderate response to drought. However, Maudamani showed a leaf rolling score of 7 indicating severe sensitive response to drought condition. Comparing the RDR percentage, all the pyramided lines had a greater number of roots in deeper angle as compared to Maudamani (12.50%). Shoot dry weight of CR3996-19-9-45-1 (12.26 g) was much higher than Maudamani (3.37 g) indicating the robust growth of CR3996-19-9-45-1 even under moisture deficit conditions which could be due to its greater RDR and longer root depth. Among the NIL lines, CR6508-111-101-129 had the highest RDR (71.20%) from all the pyramided lines followed by CR6508-111-101-267 (70.12%) and CR6508-111-101-713 (69.10%) suggesting that the presence of both deeper rooting QTL (DRO1 and DRO3) together leads to a higher number of roots with deep angle compared to absence of QTL in Maudamani (Table 2). The lines CR6508-111-101-413 and CR6508-111-101-713 showed higher root volume of 20.23 cm3 and 22.13cm3, respectively than the recipient parent, Maudamani (7.83 cm3). Comparing Maudamani and the pyramided lines, RDR along with all root traits were significantly higher for pyramided lines (Fig. 4; Table S2).
Evaluation of pyramided lines for phosphorus uptake trait
The screening results of the five pyramided lines (CR6508-111-101-129, CR6508-111-101-267, CR6508-111-101-413, CR6508-111-101-537, CR6508-111-101-713) along with the tolerant (CR3996-19-9-45-1) and susceptible (Maudamani) checks under P-deficient soil are presented in Table 2. It is clear that the pyramided lines had better concentration of phosphorus than Maudamani even at low P soil. Among all the pyramided lines, CR6508-111-101-129 (0.28%) had the highest phosphorus concentration in leaf (Table 2) followed by CR6508-111-101-267 (0.27%) and CR6508-111-101-413 (0.27%). Phosphorus concentration in Maudamani drastically decreased in low P soil (0.20%) as compared to normal P soil (0.35%) [Fig. 5; Table S3]. But the donor and pyramided lines performed better in P-deficient soil than that of Maudamani. The flowering of Maudamani was delayed by 20–24 days while the donor and pyramided lines flowered at similar interval when compared to normal conditions. Few pyramided lines flowered earlier than their normal flowering time which could be one of the avoidance mechanisms to tolerate low P in soil. The plant height and tiller number for pyramided lines were higher as compared to susceptible parent under low P conditions (Table 3). Spikelet fertility for pyramided lines was much better than Maudamani. Total filled grain number for all pyramided lines was much higher than Maudamani. Whereas, there was drastic reduction (> 88%) in total filled grains in Maudamani as compared to normal P soil. The adverse effect of low P on Maudamani could be deduced from its high chaff number (61.67). When comparing the panicle weight, pyramided lines showed better weight than that of Maudamani (Fig. S6). Higher panicle weight was observed in CR6508-111-101-129 (2.68 g) followed by CR6508-111-101-413 (2.55 g) and CR6508-111-101-267 (2.26 g). There was better root growth in Pup1 pyramided lines under low P conditions. The root volume and root dry weight was higher in pyramided lines as compared to Maudamani (Fig. S8). CR6508-111-101-413 had highest RDW (8.55 g) among all pyramided lines (Table 3). The evaluation of the pyramided and parental lines for grain yield revealed that the five improved lines had significantly higher yield than the recurrent parent, ‘Maudamani’ (Table 2). There was better grain yield in pyramided lines as compared to recurrent parent. Among the pyramided lines, all lines showed higher grain yield than the donor parent, CR3996-19-9-45-1. The highest grain yield per plant was recorded from CR6508-111-101-129 (18.51 g) followed by CR6508-111-101-413 (17.60 g) and CR6508-111-101-267 (17.33 g). Maudamani showed approximately 76.62% reduction in yield under P-deficient condition compared with the yield under normal-P condition. There was slight decrease in grain yield per plant among the pyramided lines in P-deficient condition as compared to normal-P condition (Fig. S7). The grain yield reduction in pyramided lines ranged from 22.84% (CR6508-111-101-129) to 39.97% (CR6508-111-101-713).
Evaluation of pyramided lines for agro-morphological and quality traits
The pyramided lines along with parental lines were evaluated for various agro-morphological and quality traits in BC2F3 and BC2F4 generations. The pooled mean data of two seasons for most of the traits of all pyramided lines were nearer to the estimated phenotypic value of the recurrent parent, ‘Maudamani’ (Table 4). Days to 50% flowering was 4–7 days more than Maudamani (110 days) in normal condition. Tiller number of all the pyramided lines were more than Maudamani (7.17) (Fig. S9). Comparing the panicle weight and grain number per panicle, it ranged from 6.53 to 6.95 g and 220.56-265.54, respectively (Fig. 6). However, the line CR6508-111-101-537 (24.1 g) had higher 1000-grain weight compared to Maudamani (23.08 g). The trend for L/B ratio showed that all pyramided lines were almost similar to Maudamani. Many of the grain quality characters of Maudamani such as milling, head rice recovery, amylose content and gel consistency were nearer in all the pyramided lines. Among all the pyramided lines, CR6508-111-101-129 showed highest plot yield (7.43 t/ha) followed by CR6508-111-101-267 (7.33 t/ha) and CR6508-111-101-713 (7.26 t/ha) which was more than yield of Maudamani (7.12 t/ha). The line CR6508-111-101-129 showed > 4% increase in yield compared to recipient parent. There was no yield penalty in any pyramided line as compared to Maudamani except for CR6508-111-101-537 (6.95 t/ha).
From the placement pattern in the quadrants observed in genotype-by-trait biplot diagram (Fig. 7) which was generated using 14 agro-morphological and quality traits using the five pyramided lines along with their parents indicated the similarity among pyramided lines and Maudamani for all traits under the study. The biplot places the donor parent away from the origin and in a separate quadrant (Quadrant III) from pyramided line indicating less or no drag from donor parent except for the target traits. All the pyramided lines were placed in quadrant I and II along with Maudamani indicating minor variations among them. However, Maudamani was placed in quadrant II with pyramided line CR6508-111-101-537 representing the similarity between them. The pyramided lines in quadrant I and quadrant II showed similarity for the studied traits along with the recipient parent, Maudamani and are good candidates for further evaluation and release as variety in various parts of the country. The variation observed for the first principal component was 58.16%, while 19.27% was explained for the second component.
Panicle photographs of parents (CR3996-19-9-45-1 and Maudamani) and their pyramided lines evaluated in BC2F4 generation during wet season, 2024. Numbers in the figure indicate the pyramided lines, 1: CR6508-111-101-129, 2: CR6508-111-101-267, 3: CR6508-111-101-413, 4: CR6508-111-101-537, 5: CR6508-111-101-713.
Genotype-trait biplot diagram of five pyramided lines carrying DRO1, DRO3 and Pup1 QTL along with the parents for the first two principal components. DFF: Days to 50% flowering (Days); PH: Plant height (cm); TN: Tiller Number; PL: Panicle length (cm), PW: Panicle weight (g); GN: Grain Number/Panicle; CN: Chaff Number/Panicle; TGW: 1000 Grain Weight (g); L/B: L/B Ratio; M: Milling (%); HRR: Head Rice Recovery (%); AC: Amylose Content (%); GC: Gel Consistency (mm) and PY: Plot Yield (t/ha).
Discussion
Trait improvement through classical breeding approach is very effective for those traits showing a clear phenotype with high inheritance. But, for few traits, phenotypes are not clear for distinguishing the progenies. Under such situation, marker-assisted backcross breeding (MABB) is the precise selection approach for improvement of such virtual traits. The rice cultivar Maudamani is a popular variety for the mid duration irrigated ecology, but not suitable for rainfed direct seeded situation due to its shallow rooting character and susceptibility to P-deficiency under such ecology. We were successful in developing the pyramided lines with deeper rooting and low phosphorus tolerance traits through MABB without compromising the yield and other main features of the popular variety.
Marker assisted backcross breeding is widely used for incorporating various genes/QTL for variety of traits. The efficacy of backcross breeding is greatly increased by the use of molecular markers48,49. In the current investigation, crosses were made between Maudamani and CR3996-19-9-45-1 (donor line carrying DRO1, DRO3 and Pup1 QTL) and finally five plants were selected which carried all the three QTLs in homozygous condition. Earlier few examples of Pup1 transfer alone or in combination with other QTL were reported by researchers38,47. However, transfer of other traits through marker assisted breeding have been reported by many workers38,50,51,52,53,54. The three target QTL were transferred into the recipient variety simultaneously. This is the first successful case of combining deeper rooting with better phosphorus uptake traits in case of rice. Additionally, the time taken for developing the variety was quite low compared to the conventional breeding approach.
Background selection is an important step in marker-assisted backcross breeding which serves a dual purpose: maximizing genome recovery of the desired parent and effectively mitigating the undesirable phenomenon of linkage drag originating from the donor parent. This helps to accelerate the breeding process by selecting progenies with maximum similarity to recipient parent and reducing the number of backcross generations. Here, a total of 123 polymorphic microsatellite markers distributed across twelve chromosomes were used for background selection to select plant with highest recurrent parent genome recovery percentage. It is expected that the undesirable genetic segments from the donor parent may come as linkage drag to the derived lines as the unlinked loci in during backcross progenies selection. In this molecular breeding effort, we used the donor parent of a popular variety background derived line, Naveen and therefore such effects were not observed. The graphical representation of the genotyping data on the chromosomes carrying the target QTL depicting the linkage drags are shown in the diagram (Fig. 3). The figure shows no linkage drags on the chromosomes carrying the 3 target QTL. Low linkage drag was also reported by many previous researchers in their research findings with recommendation of using elite donor parents in marker-assisted breeding54,55,56,57,58,59.
The introgressed lines were phenotyped for deeper rooting under moisture-deficit and normal conditions. The comparison between parental lines and introgressed lines showed an increase in RDR in the introgressed lines as compared to Maudamani. Among all the pyramided lines, CR6508-111-101-129 had the highest RDR of 71.20% suggesting the presence of both the DRO QTL increased the root angle in rice. The depth of roots and ratio of deeper rooting in case of pyramided lines carrying DRO1 + DRO3 were deeper than the recipient variety, Maudamani (Table 2).The pyramided lines showed higher root and shoot biomass compared to the recipient parent under water stress condition. These results are in agreement with the results obtained by16,38,60,61,62,63 which resulted due to the root system architecture and increased rice yield under drought conditions.
In India, large rice areas are either deficient or unavailable for phosphorus. Therefore, either proper phosphorus application or tolerant varieties under low soil phosphorus are required for the country. In this investigation, the pyramided lines carrying the Pup1 locus showed almost similar P-uptake under normal and deficient phosphorus condition (Table 2). The recipient parent, Maudamani showed higher P-uptake under normal P-level but low uptake under deficient conditions (Table 2). The results confirmed our expected results for P-tolerance trait under low phosphorus condition in the soil in presence of Pup1 QTL. Similar results on low P-tolerance were reported earlier by earlier researchers in rice43,44,61,64,65,66,67,68. The root dry weight and shoot dry weight were higher in Pup1 carrying pyramided lines compared to the recipient parent, Maudamani under low P condition (Table 3). The higher root dry weight in the pyramided lines suggested better root development under low P condition which might have helped in acquisition of phosphorus from phosphorus limiting soil. Earlier publications are in agreement with the current findings of better root growth of Pup1 pyramided lines under P-limiting conditions8,43,45,67,69. Among the pyramided lines, all lines showed higher grain yield under P-deficient conditions than the recurrent parent, Maudamani. Even under normal P condition, there was no negative effect of Pup1 and the pyramided lines showed at par yield with Maudamani (Table 2). The findings that Pup1 introgressed line performed both in P-normal and P-deficient condition was similar to the previous findings45,47,65. Rice lines exhibiting strong performances under both normal and low soil phosphorus conditions, represent promising candidates for deployment as enhanced versions of Maudamani.
The developed pyramided lines carrying the 3 target QTL were similar to the recipient parent for many traits. The constructed dendrogram based on the phenotypic traits grouped the pyramided and parental lines into two main clusters with similarity among the members of the clusters (Fig. 2A). The biplot diagram prepared on the basis of 14 studied morpho-quality traits placed the pyramided and recipient parent in quadrant I and II indicating very less variations among the developed lines (Fig. 7). The pooled evaluation results of the pyramided lines for grain yield and grain quality traits showed higher yield in pyramided lines viz., CR6508-111-101-129, CR6508-111-101-267, CR6508-111-101-413, CR6508-111-101-537 and CR6508-111-101-713 than the recipient parent, Maudamani (Table 4). Pyramided lines which were similar morpho-quality traits and on par or better in yield than the recipient parent was also reported many gene-pyramiding works51,70,71. Overall, the pyramided lines showed comparable or superior performance in terms of yield and agro-morphologic traits. This suggested that the introgression of QTL from the donor line into the Maudamani background did not negatively impact its inherent yielding ability. Instead, it resulted in the development of near-isogenic lines that retained yield potential of Maudamani while incorporating the stress resilience conferred by the donor QTL. Similar findings where the developed lines showed either at par or superior performance to their recipient parent were reported by38,45,47,65,66,68,69.
Materials and methods
Plant materials
Maudamani (CR Dhan 307), a high-yielding rice variety developed by ICAR-Central Rice Research Institute (CRRI), Cuttack for irrigated conditions, was used as the recipient parent in this breeding program. Despite its high yield potential, Maudamani is highly susceptible to water-deficit condition and P deficiency. The donor line carrying DRO1, DRO3 and Pup1 was a pre-bred line developed previously by the team at ICAR-CRRI, Cuttack in the background of Naveen using the landrace donor line, Surjyamukhi which was designated as ‘CR3996-19-9-45-1. Both the parents were validated for presence of DRO1, DRO3 and Pup1 QTL using all closely linked markers of deeper rooting and phosphorus uptake traits (Table 5).
Breeding program
To improve its drought avoidance and phosphorus uptake ability, Maudamani was crossed with the donor parent pre-bred line ‘CR3996-19-9-45-1’, possessing DRO1, DRO3, and Pup1 QTL (Fig. 8). True F1 individuals were identified using SSR markers and then backcrossed with Maudamani. Starting from BC1F1, molecular markers closely linked to the target QTL were employed for PCR amplification to select plants carrying the tolerant alleles for all three QTL. Additionally, polymorphic markers were used to ensure that the selected plants maintained high genome content from the recurrent parent, Maudamani. The progeny carrying the target QTL and with highest genome content of recipient genome was crossed in each backcross generations with the recipient parent, Maudamani. This process was repeated till the BC2F1 generation. Further, BC2F1 plants were selfed to generate BC2F2 seeds. Homozygous pyramided lines were further selected using the target QTL foreground markers and selfed to produce the BC2F3 progenies. Evaluations for deeper rooting, phosphorus uptake and agro-morphological-quality traits were carried out in both BC2F3 and BC2F4 generations. The promising pyramided lines were subjected to seed multiplication and advanced yield trials.
Phenotyping for deeper rooting genotypes in microplot under moisture deficit condition
A controlled experiment was carried out in a manually raised brick-structured tank of 15 feet inside length, 4.5 feet inside width, 3 feet height above ground and 1.5 feet below ground with low proportion of sand to cement of 20:1 so that it was easy to dismantle the structure (Fig. 9). The soil was filled in the tank up to 3feet. The soil was treated with insecticide and fungicide to disinfect the soil from any pest and diseases. The tanks were watered for two days to allow proper compaction of soil and for proper levelling. Phenotyping for deeper rooting was performed to determine RDR (an index for root growth angle) using basket method16 with minor modifications. The maximum and minimum temperature data at ICAR-CRRI were taken from the Institute’s weather station. Open steel baskets of 7.5 cm upper diameter and 5 cm in height having 2 mm mesh size were placed inside the soil with a gap of 15 cm between two baskets. Three tensiometers (two 15 cm depth and one 30 cm depth) were installed in the tank to check the water status of the soil. In the middle of each basket, four to five seeds were sown. After germination, the plants were thinned to one plant per basket. The tank was fertilized with regular doses of NPK fertilizers and insecticide was sprayed to keep the plant healthy free from pests and diseases. Tensiometer probe readings were taken regularly during the stress period. After 30 DAS, stress was imposed on the tank by withdrawing water till the tensiometer reading reached − 55 kPa and − 33 kPa (15 cm and 30 cm, respectively) (Fig. S5). During the stress period, the tank was covered with transparent polythene sheet (200 GSM) to prevent from rain and even allow light to pass through it.
After the stress period, the tank was dismantled carefully brick by brick, allowing the soil to remain compact without falling off. A water spray pipe with moderate water pressure was used to remove the soil from the root zone. Plant height and maximum root depth were recorded. The baskets along with intact roots were extracted from the soil carefully by continuously spraying water. Then the roots with basket were washed in tap water to remove the extra soil. Then the root length, root angle and ratio of deeper rooting (RDR) were estimated. The ratio of deep rooting was calculated by taking the total number of roots that penetrated the lower part of the mesh (≥ 50˚ from the horizontal, centred on the stem of the rice plant) divided by the total number of roots entered the whole mesh16. Roots were scanned in the root scanner (EPSON 11000X) and analysed using WinRHIZO Pro 2013e (LA 2400, Regent Instruments INC.). Root volume, total root length, total surface area and average diameter of root were recorded by WinRHIZO software.
Phenotyping for deeper rooting under moisture-deficit stress. (A) Direct sowing through open mesh wire basket in controlled tank; (B) Plants grown at seedling stage; (C) Plant response in moisture-deficit condition; (D) Dismantling of the tank and use of high-pressure water spray to remove the soil; (E) Root distribution in soil; (F) Extraction of roots along with basket; (G) Observation of root length and root angle distribution affected by the stress.
Phenotyping for phosphorus uptake under phosphorus-deficient microplot
All the selected pyramided lines and their parental checks were screened in a tank (30feet length, 5 feet width and 4 feet height) containing low phosphorus soil at ICAR-CRRI, Cuttack. Soil sampling from five random places in the phosphorus deficient soil tank was done to evaluate the soil pH and NPK content in the tank before sowing73.
After proper soil treatment, seeds of Maudamani, CR3996-19-9-45-1 along with their NILs were direct-seeded in the tank using five replications with a spacing of 20 × 15 cm (Fig. 10). Fifteen days after sowing, the seedlings were thinned to one seedling per hill. Low phosphorus in the tank was maintained without the application of any external P fertilizers. Recommended cultural practices were followed to raise a healthy crop. Additionally, need based plant protection practices were taken during the crop seasons for the control of insect pests and diseases.
At dough stage, leaves from three replications of parental and pyramided lines were collected and oven dried at 80˚C for three days. The dried samples were powdered and quantification for P-concentration was done. Wet digestion was done in Advanced Microwave Digestion System (Milestone ETHOS™ EASY) using 5 ml of HNO3 and 2 ml of H2O2 for 0.5 g of powdered sample74. The digest was then used for quantification of phosphorus using Vanadomolybdate method75. The intensity of yellow colour is directly proportional to P-concentration measured using UV Spectrophotometer. Morphological data including grain yield per plant was taken for all five pyramided lines along with their parents at maturity in P-deficient as well as P-normal soil conditions.
Evaluation of the pyramided lines for agro-morphological, grain quality and yield traits
The selected genotypes along with parents were grown in the experimental field at ICAR-CRRI, Cuttack, India during the dry and wet seasons, 2024. Seeds were sown on seed bed and thirty days old seedlings were transplanted in the main field with a spacing of 20 cm×15 cm with randomized block design in three replications. Normal dosage of fertilizer at 80:40:40 kg/ha of N: P:K was applied in field. Normal agronomic practices and need based plant protection measures were taken up. Days to 50% flowering was recorded along with agro-morphological and quality traits like plant height (cm), tiller number, panicle length (cm), panicle weight (g), grain number per panicle, chaff number per panicle, 1000-grain weight (g), L/B ratio, milling (%), head rice recovery (%), amylose content (%), gel consistency (mm) and plot yield (t/ha) were recorded from replicated trials.
Statistical analysis
The general statistical analysis like mean, standard deviation of the phenotypic data was calculated by using Excel 2013. The estimated morpho-quality traits recorded from the pyramided and parental lines were analysed using SAS 2008, version 9.276. The Principal Component Analysis (PCA) was performed for the pyramided and parental lines using multivariate analysis (Past Software version 4.03) data for the studied 14 morphological and quality traits. A scatter plot was drawn by using the Principal Component 1 (PC1) and Principal Component 2 (PC2). The Eigen value and percentage of variance were generated by the interaction of a variance covariance matrix. The interactions among the studied traits were depicted through a biplot graph in the matrix. All the plots and results of PCA were generated as per the standard procedure followed in earlier publications38,77,78. Genotypic data were sorted by using Excel 2013, and graphs of recurrent parent genome recovery were drawn by using GGT 2.079.
Conclusions
The backcross derived lines from Maudamani namely CR6508-111-101-129, CR6508-111-101-267, CR6508-111-101-413, CR6508-111-101-537 and CR6508-111-101-713 carrying DRO1, DRO3 and Pup1 had deeper rooting and tolerance to low phosphorus stress along with better in grain yield than the recipient parent, Maudamani. The higher yield harvested from the pyramided lines under water stressed and low-P condition may be due to deeper rooting and better P-uptake in the pyramided lines than the recipient variety. The popular variety, Maudamani showed higher P-uptake under normal soil P-level but low uptake under deficient conditions. The results confirmed our expected results for P-tolerance trait under low phosphorus condition in the soil and better yield under water stressed situation. In addition, no yield penalty was observed in the pyramided lines due to the introgression of three QTL in the background. The main agro-morphological and quality traits of the variety such as like tiller number, panicle length, panicle weight, grain number per panicle, 1000-grain weight, L/B ratio, milling, head rice recovery, amylose content, gel consistency along with yielding ability were retained in the pyramided lines. The best pyramided, CR6508-111-101-129 which had higher yield and better performance in stress conditions will be nominated for the national trial for variety release. In addition, the pyramided lines carrying DRO1, DRO3 and Pup1 QTL may serve as potential donors for deeper rooting and low-P tolerance traits in the future breeding programs. This investigation validated the application of marker-assisted selection for transfer of complex traits such as deeper rooting and low phosphorus stress tolerance in rice.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Dianga, A. I., Musila, R. N. & Joseph, K. W. Rainfed rice farming production constrains and prospects, the Kenyan situation. Integr. Adv. Rice Res. 12, 523 (2021).
Ilyas, M. et al. Drought tolerance strategies in plants: a mechanistic approach. J. Plant Growth Regul. 40, 926–944 (2021).
Aslam, M. M. et al. Recent insights into signaling responses to Cope drought stress in rice. Rice Sci. 29 (2), 105–117 (2022).
Kondo, M. et al. Genotypic and environmental variations in root morphology in rice genotypes under upland field conditions. In Roots: The Dynamic Interface between Plants and the Earth: The 6th Symposium of the International Society of Root Research, 11–15 November 2001, Nagoya, Japan 189–200 (Springer Netherlands, 2003).
Yoshida, S. & Hasegawa, S. The rice root system: its development and function. Drought Resist. Crops Emphas. Rice. 10, 97–134 (1982).
de Dorlodot, S. et al. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12 (10), 474–481 (2007).
Miyazaki, A. & Arita, N. Deep rooting development and growth in upland rice NERICA induced by subsurface irrigation. Plant. Prod. Sci. 23 (2), 211–219 (2020).
Pandit, E., Panda, R. K., Sahoo, A., Pani, D. R. & Pradhan, S. K. Genetic relationship and structure analysis of root growth angle for improvement of drought avoidance in early and mid-early maturing rice genotypes. Rice Sci. 27 (2), 124–132 (2020).
Haefele, S. M. & Hijmans, R. J. Soil quality in rice-based rainfed lowlands of Asia: characterization and distribution. In Proceedings of the 26th International Rice Research Conference,October 9–12, New Delhi, India (eds. Aggarwal, P. K. et al.) 297–308 (2007).
Araki, H., Morita, S., Tatsumi, J. & Iijima, M. Physiol-morphological analysis on axile root growth in upland rice. Plant. Prod. Sci. 5 (4), 286–293 (2002).
Zhang, W. P., Shen, X. Y., Wu, P., Hu, B. & Liao, C. Y. QTLs and epistasis for seminal root length under a different water supply in rice (Oryza sativa L). Theor. Appl. Genet. 103, 118–123 (2001).
Obara, M., Takeda, T., Hayakawa, T. & Yamaya, T. Mapping quantitative trait loci controlling root length in rice seedlings grown with low or sufficient supply using backcross Recombinant lines derived from a cross between Oryza sativa L. and Oryza glaberrima Steud. Soil. Sci. Plant. Nutr. 57 (1), 80–92 (2011).
Wu, W. & Cheng, S. Root genetic research, an opportunity and challenge to rice improvement. Field Crops Res. 165, 111–124 (2014).
Khahani, B., Tavakol, E., Shariati, V. & Rossini, L. Meta-QTL and ortho-MQTL analyses identified genomic regions controlling rice yield, yield-related traits and root architecture under water deficit conditions. Sci. Rep. 11 (1), 6942 (2021).
Abe, J. & Morita, S. Growth direction of nodal roots in rice: its variation and contribution to root system formation. Plant. Soil. 165, 333–337 (1994).
Uga, Y., Okuno, K. & Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62 (8), 2485–2494 (2011).
Kato, Y., Abe, J., Kamoshita, A. & Yamagishi, J. Genotypic variation in root growth angle in rice (Oryza sativa L.) and its association with deep root development in upland fields with different water regimes. Plant. Soil. 287, 117–129 (2006).
Uga, Y. et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45 (9), 1097–1102 (2013).
Bettembourg, M. et al. Genome-wide association mapping for root cone angle in rice. Rice 10, 1–17 (2017).
Lou, Q. et al. Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 66 (15), 4749–4757 (2015).
Vinarao, R. et al. Stable and novel quantitative trait loci (QTL) confer narrow root cone angle in an aerobic rice (Oryza sativa L.) production system. Rice 14, 1–12 (2021).
Uga, Y. et al. A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice 8, 1–8 (2015).
Uga, Y. et al. A major QTL controlling deep rooting on rice chromosome 4. Sci. Rep. 3 (1), 3040 (2013b).
Kitomi, Y. et al. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice 8, 1–12 (2015).
Mori, A. et al. The role of root size versus root efficiency in phosphorus acquisition in rice. J. Exp. Bot. 67 (4), 1179–1189 (2016).
Vejchasarn, P., Lynch, J. P. & Brown, K. M. Genetic variability in phosphorus responses of rice root phenotypes. Rice 9, 1–16 (2016).
Deng, Y. et al. Tolerance to low phosphorus in rice varieties is conferred by regulation of root growth. Crop J. 8 (4), 534–547 (2020).
Ding, Y. et al. Mechanism of low phosphorus inducing the main root lengthening of rice. J. Plant Growth Regul. 40, 1032–1043 (2021).
Yang, G. et al. Response of root development and nutrient uptake of two Chinese cultivars of hybrid rice to nitrogen and phosphorus fertilization in Sichuan province, China. Mol. Biol. Rep. 48, 8009–8021 (2021).
Massawe, P. & Mrema, J. Effects of different phosphorus fertilizers on rice (Oryza sativa L.) yield components and grain yields. Asian J. Adv. Agricultural Res. 3 (2), 1–13 (2017).
Hasanuzzaman, M., Ali, M. H., Karim, M. F., Masum, S. M. & Mahmud, J. A. Response of hybrid rice to different levels of nitrogen and phosphorus. Int. Res. J. Appl. Basic. Sci. 3 (12), 2522–2528 (2012).
Kumar, S. et al. Transcriptome analysis of a near-isogenic line and its recurrent parent reveals the role of Pup1 QTL in phosphorus deficiency tolerance of rice at tillering stage. Plant Mol. Biol. 109 (1), 29–50 (2022).
Kekulandara, D. S. et al. Development of high yielding rice varieties tolerant to phosphorus deficiency. Trop. Agric. Res. 35, 2 (2024).
Shrestha, J., Kandel, M., Subedi, S. & Shah, K. K. Role of nutrients in rice (Oryza sativa L.): a review. Agrica 9 (1), 53–62 (2020).
Bhatta, B. B. et al. Improvement of phosphorus use efficiency in rice by adopting image-based phenotyping and tolerant indices. Front. Plant Sci. 12, 717107 (2021).
Muindi, E. D. M. Understanding soil phosphorus. Int. J. Plant. Soil. Sci. 31 (2), 1–18 (2019).
Shen, J. et al. Phosphorus dynamics: from soil to plant. Plant Physiol. 156 (3), 997–1005 (2011).
Barik, S. R. et al. Transfer of stress resilient QTLs and panicle traits into the rice variety, Reeta through classical and Marker-Assisted breeding approaches. Int. J. Mol. Sci. 24 (13), 10708 (2023).
Wissuwa, M., Yano, M. & Ae, N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L). Theor. Appl. Genet. 97, 777–783 (1998).
Wissuwa, M., Wegner, J., Ae, N. & Yano, M. Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet. 105, 890–897 (2002).
Wissuwa, M., Gatdula, K. & Ismail, A. Candidate gene characterization at the Pup1 locus: a major QTL increasing tolerance of phosphorus deficiency. nt. Rice Res. Inst. 2005, 83 (2005).
Chin, J. H. et al. Development and application of gene-based markers for the major rice QTL phosphorus uptake 1. Theor. Appl. Genet. 120, 1073–1086 (2010).
Gamuyao, R. et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488 (7412), 535–539 (2012).
Chin, J. H. et al. Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application. Plant Physiol. 156 (3), 1202–1216 (2011).
Anila, M. et al. Breeding lines of the Indian mega-rice variety, MTU 1010, possessing protein kinase OsPSTOL (Pup1), show better root system architecture and higher yield in soils with low phosphorus. Mol. Breeding. 38, 1–9 (2018).
Chithrameenal, K. et al. Genetic enhancement of phosphorus starvation tolerance through marker assisted introgression of OsPSTOL1 gene in rice genotypes harbouring bacterial blight and blast resistance. PLoS One 13 (9), e0204144 (2018).
Swamy, H. M. et al. Marker assisted improvement of low soil phosphorus tolerance in the bacterial blight resistant, fine-grain type rice variety, improved Samba Mahsuri. Sci. Rep. 10 (1), 21143 (2020).
Collard, B. C. Y. & Mackill, D. J. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R Soc. 363, 557–572 (2008).
Hasan, M. M. et al. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 29 (2), 237–254 (2015).
Pradhan, S. K. et al. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘swarna’through marker-assisted backcross breeding. Sci. Rep. 9 (1), 12810 (2019).
Mohapatra, S. et al. Development of submergence-tolerant, bacterial blight-resistant, and high-yielding near isogenic lines of popular variety, ‘swarna’ through marker-assisted breeding approach. Front. Plant Sci. 12, 672618 (2021).
Mishra, A. et al. Genetics, mechanisms and deployment of brown planthopper resistance genes in rice. CRC. Crit. Rev. Plant Sci. 41 (2), 91–127 (2022).
Pradhan, K. C. et al. Incorporation of two bacterial blight resistance genes into the popular rice variety, Ranidhan through marker-assisted breeding. Agriculture 12 (9), 1287 (2022a).
Mohapatra, S. et al. Molecular breeding for incorporation of submergence tolerance and durable bacterial blight resistance into the popular rice variety ‘ranidhan’. Biomolecules 13 (2), 198 (2023).
Ahmed, F. et al. Recurrent parent genome recovery in different populations with the introgression of Sub1 gene from a cross between MR219 and Swarna-Sub1. Euphytica 207, 605–618 (2016).
Arya, K. V. & Shylaraj, K. S. Introgression of Sub1 QTL (Submergence tolerant QTL) into the elite rice variety Jaya by marker assisted backcross breeding. J. Trop. Agric. 56, 2 (2018).
Chukwu, S. C. et al. Marker-assisted introgression of multiple resistance genes confers broad spectrum resistance against bacterial leaf blight and blast diseases in Putra-1 rice variety. Agronomy 10 (1), 42 (2019).
Pradhan, K. C. et al. Development of broad spectrum and durable bacterial blight resistant variety through pyramiding of four resistance genes in rice. Agronomy 12 (8), 1903 (2022b).
Merugumala, G. R. et al. Marker assisted breeding of Sub1 introgressed rice (Oryza sativa L.) lines and identification of stable variety MTU 1232 suitable for flood prone ecosystem. Plant Sci. Today 11(2) (2024).
Arai-Sanoh, Y. et al. Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Sci. Rep. 4 (1), 5563 (2014).
Panda, R. K. et al. Response of physiological and biochemical parameters in deeper rooting rice genotypes under irrigated and water stress conditions. ORYZA-An Int. J. Rice. 53 (4), 422–427 (2016).
Ramalingam, P., Kamoshita, A., Deshmukh, V., Yaginuma, S. & Uga, Y. Association between root growth angle and root length density of a near-isogenic line of IR64 rice with DEEPER ROOTING 1 under different levels of soil compaction. Plant. Prod. Sci. 20 (2), 162–175 (2017).
Zhao, Y. et al. INDITTO2 transposon conveys auxin-mediated DRO1 transcription for rice drought avoidance. Plant. Cell. Environ. 44 (6), 1846–1857 (2021).
Anila, M. et al. Marker-assisted introgression of Pup1, a major QTL associated with tolerance to low soil phosphorus into the elite rice variety MTU1010. Prog Res. 9, 735–738 (2014).
Anila, M. et al. Marker assisted introgression of Pup1, A major QTL associated with tolerance to low soil phosphorus into elite rice varieties. ICAR-IIRR Newsl. 15 (1), 26–29 (2017).
Pandit, E. et al. Molecular marker and phenotypic analyses for low phosphorus stress tolerance in cultivars and landraces of upland rice under irrigated and drought situations. Indian J. Genet. Plant. Breed. 78 (01), 59–68 (2018).
Madhusudan, N. et al. Stacking of Pup1 QTL for low soil phosphorus tolerance and bacterial blight resistance genes in the background of APMS6B, the maintainer line of rice hybrid DRRH-3. Euphytica 218 (4), 37 (2022).
Navea, I. P., Han, J. H., Shin, N. H., Lee, Y. J. & Chin, J. H. Development of rainfed-adapted, fertilizer-efficient temperate rice varieties by Pup1 Introgression. In Proceedings of the Korean Society of Crop Science Conference, The Korean Society of Crop Science 272 (2022).
Mahender, A., Anandan, A., Pradhan, S. K. & Singh, O. N. Traits-related QTLs and genes and their potential applications in rice improvement under low phosphorus condition. Arch. Agron. Soil. Sci. 64 (4), 449–464 (2018).
Pradhan, S. K. et al. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety. Jalmagna Rice. 8, 1–14 (2015).
Pandit, E. et al. Marker-assisted backcross breeding for improvement of submergence tolerance and grain yield in the popular rice variety ‘maudamani’. Agronomy 11 (7), 1263 (2021).
Pradhan, S. K. et al. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 106 (7), 710–718 (2016).
Bray, R. H. & Kurtz, L. T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 59 (1), 39–46 (1945).
Kumar, A. et al. The diversity of phytic acid content and grain processing play decisive role on minerals bioavailability in rice. J. Food Compos. Anal. 115, 105032 (2023).
Nayak, A. K. et al. Modern techniques in soil and plant analysis. Kalyani, New Delhi, India 272 (2016).
SAS Institute Inc. SAS®9.2 Language Reference: Concepts 626–978 (SAS Institute Inc., 2010).
Pandit, E. et al. Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in indica rice. Front. Plant Sci. 8, 552 (2017).
Mohanty, S. P. et al. Mapping the genomic regions controlling germination rate and early seedling growth parameters in rice. Genes 14 (4), 902 (2023).
Van Berloo, R. Computer note. GGT: software for the display of graphical genotypes. J. Hered. 90 (2), 328–329 (1999).
Deshmukh, V., Kamoshita, A., Norisada, M. & Uga, Y. Near-isogenic lines of IR64 (Oryza sativa subsp. Indica cv.) introgressed with DEEPER ROOTING 1 and STELE TRANSVERSAL AREA 1 improve rice yield formation over the background parent across three water management regimes. Plant. Prod. Sci. 20 (3), 249–261 (2017).
Acknowledgements
The authors acknowledge the support of the Director, ICAR-Central Rice Research Institute, Cuttack for providing all the necessary facilities.
Author information
Authors and Affiliations
Contributions
A.M. Genotyped, phenotyped the progenies for the target traits and prepared Tables; S.R.B. Data analysis and genotyping work; D.L. Phenotyping the progenies for the target traits; E.P. Prepared Figs. 1, 2, 3, 4 and 5, graphs and edited the draft manuscript; L.B. Data analysis and monitoring; S.K.D. Phenotyping and monitoring work; S.S. Genotyping work; S.D. Data analysis and article correction; S.K.P. Conceived the idea, supervision, fund arrangement, Figures, manuscript write up; All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mishra, A., Barik, S.R., Lenka, D. et al. Transfer of deeper rooting and phosphorus uptake QTL into the popular rice variety ‘maudamani’ via marker-assisted backcross breeding. Sci Rep 15, 25418 (2025). https://doi.org/10.1038/s41598-025-10951-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-10951-w