Abstract
Pseudomonas aeruginosa is a critical priority pathogen due to its high resistance rates and limited treatment options. Rapid antimicrobial susceptibility testing (rAST) is essential to optimize therapy, particularly because resistance mechanisms in P. aeruginosa often arise from adaptive genetic expression rather than specific resistance genes. FASTinov, a flow cytometry-based rAST system, was developed to address this challenge by providing phenotypic susceptibility results within hours. In this study, we assessed FASTinov’s performance for rapid antimicrobial susceptibility testing (rAST) in 100 P. aeruginosa strains, using broth microdilution as the reference method. For clinical relevance, we also included the MicroScan WalkAway system as a comparator. FASTinov demonstrated a categorical agreement of 99% across the tested antibiotics, with particularly strong agreement for cephalosporins. Notably, FASTinov significantly reduced the time to results, delivering susceptibility data in an average of 3.15 h (CI 95% 3.13–3.17), compared to 28.88 h (CI 95% 20.71–37.04) for standard methods. Additionally, the system exhibited lower rates of major and very major errors compared to MicroScan, further reinforcing its reliability. These findings highlight FASTinov as a promising rAST system capable of providing rapid and accurate susceptibility profiles directly from bacterial colonies. Further research is warranted to evaluate its clinical impact on patient outcomes and cost-effectiveness.
Similar content being viewed by others
Introduction
Pseudomonas aeruginosa is a major opportunistic pathogen in both community and healthcare settings1. Infections caused by multidrug-resistant (MDR) P. aeruginosa strains are associated with increased mortality, prolonged hospital stays, and higher healthcare costs2,3. A key challenge in their management is the limited predictive value of molecular diagnostic methods, since resistance frequently arises from adaptive changes—such as AmpC overexpression, porin loss, or efflux pump activation—that are not detectable by genotypic assays4. This highlights the need for rapid phenotypic antimicrobial susceptibility testing (AST) methods capable of detecting functional resistance mechanisms in real time.
Among rapid AST commercially available systems, Accelerate Pheno provides phenotypic susceptibility results in approximately seven hours5, while the QMAC-dRAST System reports a total turnaround time of six to seven hours6. Additional technologies, including fluorescence-based assays, typically yield results within four to six hours7. Although these platforms represent a clear advancement over conventional methodologies, most remain dependent on bacterial growth and have been primarily validated using positive blood cultures. Importantly, their performance in P. aeruginosa remains insufficiently characterized, particularly when susceptibility testing is performed directly from bacterial colonies.
FASTinov (Porto, Portugal) has recently developed an innovative AST method based on flow cytometry. This system was designed to address the need for faster phenotypic resistance detection by enabling a growth-independent assessment of bacterial cell damage after exposure to antibiotics. The test is performed directly from isolated bacterial colonies and provides susceptibility results within 2–3 h, significantly reducing turnaround time compared to conventional methods. Previous studies have reported high sensitivity and specificity when using this system directly from positive blood cultures8,9. Its application for antimicrobial susceptibility testing in bacterial colonies, although described on instructions for use, remains insufficiently explored. This study aims to evaluate the performance of the FASTinov system for rapid antimicrobial susceptibility testing (rAST) from P. aeruginosa colonies, focusing on its accuracy in comparison to standard reference methods and the time required to obtain diagnostic results, with the goal of advancing its clinical applicability.
Results
The performance of the FASTinov system was evaluated on 100 P. aeruginosa strains and compared to the reference broth microdilution method. In addition, the MicroScan WalkAway system was included as a comparator, as it is the routine method used in our hospital for susceptibility testing. Overall, FASTinov demonstrated high reliability, achieving an average categorical agreement (CA) of 99% across all tested antibiotics. The highest CA (100%) was observed for cefepime, piperacillin/tazobactam, ceftolozane/tazobactam, ceftazidime/avibactam, ciprofloxacin, and amikacin, with no major errors (ME) or very ME (VME) (Fig. 1A). However, meropenem and ceftazidime showed lower CA values of 95% and 97%, respectively. Meropenem displayed the highest discrepancy rate, primarily due to minor errors (3%), while ceftazidime exhibited 2.86% ME and 3.33% VME, indicating potential misclassification of resistant strains as susceptible, which could carry significant clinical implications. Similarly, MicroScan results were compared to the reference method, showing an overall CA of 97%. The highest CA (99%) was observed for ceftazidime, amikacin, and piperacillin/tazobactam, while meropenem had the lowest CA (93%), followed by cefepime (95%). Minor errors were most frequent with meropenem (4%), while major errors were observed for most antibiotics except ceftazidime and piperacillin/tazobactam. Very major errors were highest for meropenem (8%), followed by cefepime (3.7%) and ceftazidime (3.33%), raising concerns regarding the potential underestimation of resistance (Fig. 1A, Supplementary Table 1). To evaluate the diagnostic performance of FASTinov compared to MicroScan, we conducted a non-inferiority analysis using categorical agreement as the primary outcome measure (Fig. 1B). The difference in agreement was calculated for each antibiotic, along with 95% confidence intervals. A non-inferiority margin of − 5% was predefined. FASTinov demonstrated non-inferiority to MicroScan for all eight antibiotics tested. For most antibiotics, the difference in agreement favored FASTinov, with confidence intervals entirely above the − 5% margin. These findings highlight the strong performance of FASTinov for P. aeruginosa, with fewer discrepancies compared to MicroScan.
(A) Heatmaps display the performance of FASTinov and MicroScan for eight antibiotics, showing categorical agreement (CA), major errors (ME), and very major errors (VME) compared to the broth microdilution reference method. Color intensity corresponds to the percentage value, with harmonized scales across panels to facilitate visual comparison. Numeric values within each cell represent the percentage for each metric and technique. (B) Non-inferiority analysis comparing FASTinov and MicroScan. Difference in categorical agreement between FASTinov and MicroScan is shown for each antibiotic, with 95% confidence intervals. A non-inferiority margin of − 5% (red dashed line) was predefined. A positive value indicates higher agreement for FASTinov. (C) Comparative performance of FASTinov and MicroScan. Each panel includes two confusion matrices (left: FASTinov; right: MicroScan) and a central radar chart summarizing diagnostic performance metrics: sensitivity (Sen), specificity (Sep), positive predictive value (PPV), negative predictive value (NPV), and Cohen’s kappa coefficient (Kappa). The eight antibiotics evaluated are: (i) CAZ (ceftazidime), (ii) FEP (cefepime), (iii) C/A (ceftazidime–avibactam), (iv) C/T (ceftolozane–tazobactam), (v) MER (meropenem), (vi) P/T (piperacillin–tazobactam), (vii) CIP (ciprofloxacin), and (viii) AMK (amikacin).
In Fig. 1C, we can observe that FASTinov demonstrated consistently high sensitivity (≥ 97.3%) across all tested antibiotics in comparison with MicroScan, with perfect specificity (100%) for most agents except meropenem (88.5%), achieving overall high accuracy (≥ 98%) and strong agreement metrics, reflected in Youden index values reaching 100% for several antibiotics (Supplementary Table 2). The kappa coefficient further supported FASTinov’s reliability, exceeding 95 for most antibiotics, indicating almost perfect agreement with the reference method. In comparison, MicroScan exhibited similarly high sensitivity (≥ 98.5%), but specificity varied considerably, particularly for ceftolozane/tazobactam (66.7%) and ceftazidime/avibactam (62.5%), leading to lower Youden index values (62.5-66.7%) and reduced kappa coefficients (75.4–79.5) for these agents, suggesting moderate agreement (Supplementary Table 2). Despite both systems demonstrating strong overall performance, FASTinov achieved greater specificity, categorical agreement, and kappa values, highlighting its potential advantage in clinical applications where accurate resistance classification is crucial.
Time to results
The time required to obtain antimicrobial susceptibility testing results using FASTinov was estimated and compared with the standard routine workflow in our laboratory. The comparison was performed for 51 samples and revealed a significant reduction in the time required for FASTinov results compared to the routine workflow, which utilizes the MicroScan system. On average, FASTinov provided results in just 3.15 h (CI 95% 3.13–3.17), compared with the routine workflow that required an average of 28.88 h (CI 95% 20.71–37.04).
Discussion
Rapid and accurate antimicrobial susceptibility testing is essential for optimizing treatment strategies, particularly for P. aeruginosa. Genotypic methods are limited to detecting known resistance genes and cannot provide a comprehensive susceptibility profile, especially for P. aeruginosa. FASTinov, a flow cytometry-based system, was developed to address this gap by enabling rapid phenotypic resistance detection, not growth-dependent, directly from bacterial colonies. A multiparametric cell analysis including size, complexity and fluorescence of treated cells are compared with non-treated cells with a proprietary software. When performed from colonies bacteria need to grow in a broth for a short period in order to be in the exponential growth phase essential for most drug activity. In this study, FASTinov demonstrated superior categorical agreement (99%) compared to MicroScan (97%) and achieved superior specificity and kappa values, particularly for cephalosporins. Additionally, FASTinov met the FDA-recommended performance criteria, with very major errors (VME) below 1%, providing results in just 3 h, compared to the 18–24 h required for automated reading by the MicroScan system, plus the time needed to review the panels and generate the report in the patient’s medical record.
Recent literature emphasizes the critical role of rapid antimicrobial susceptibility testing in optimizing the treatment of infections caused by P. aeruginosa. Rates of inadequate therapy for these infections exhibit considerable variability, ranging from 24 to 80% depending on the clinical setting and patient population. For instance, Eklöf et al.10. reported that 80% of patients with pulmonary P. aeruginosa infections received inappropriate therapy, while Cillóniz et al.11. documented a rate of 64% for community-acquired pneumonia cases caused by this pathogen, rising to 77% in infections involving multidrug-resistant strains. Martinez-Nadal et al.12. observed that 24% of high-risk neutropenic patients with bacteremia caused by P. aeruginosa received inappropriate empirical therapy. Moreover, inadequate therapy has been also linked to extended hospital stays and higher healthcare costs13,14.
Given these findings, the implementation of rapid susceptibility testing emerges as a critical priority. However, most studies evaluating rAST methods include multiple bacterial species, often leading to limited P. aeruginosa sample sizes. Additionally, many studies focus solely on direct positive blood culture testing, whereas our study represents a unique approach by evaluating a rAST method directly from culture-grown colonies. Thus, the study performed by Pancholi et al.15. demonstrated that the Accelerate Pheno SYSTEM provides results within 5 h with a CA of ≥ 90% for 21 strains of P. aeruginosa, but with VME ≥ 5% for multiple antibiotics, including ceftazidime, piperacillin-tazobactam or amikacin. A more recent study by Sikorski et al.16. evaluated the performance of the Accelerate PhenoTest BC kit against a collection of 144 P. aeruginosa isolates, reporting a CA of ≥ 85%, with a VME of 7.9% for ceftazidime and minor errors of 10.4% for meropenem, indicating challenges in accurately detecting resistance for certain antibiotics. Other study evaluating the QMAC-dRAST system (QuantaMatrix) demonstrated a 92.9% concordance with the standard disk diffusion method for Gram-negative blood culture isolates, but among the 100 strains analyzed, only 9 were P. aeruginosa, limiting its applicability17. A recent study by Couchot et al.18. evaluated the Reveal rapid AST system using 200 P. aeruginosa strains, reporting a CA of 96.1% and very major errors (VME) of 1.6%, providing results within an average time of 6 h and 22 min. Building on these findings, our study demonstrated that the system achieves a higher CA than previously reported rAST methods, with superior performance across multiple antibiotics. In addition, given its ability to deliver results in just 3 h and its high agreement with microdilution, FASTinov presents itself as a promising clinical tool for rapid antimicrobial susceptibility testing, potentially improving early therapeutic decision-making and antimicrobial stewardship efforts. However, a potential limitation of the method is the need for strict adherence to incubation times and timely processing at each step, which requires the continuous involvement of a laboratory technician to ensure accuracy and reproducibility of the results.
In conclusion, our study showed the FASTinov system as a promising tool for guiding early and targeted antimicrobial therapy for P. aeruginosa infections. Compared to MicroScan, FASTinov demonstrated higher accuracy, faster turnaround times, and fewer errors. Its ability to perform susceptibility testing directly from bacterial colonies represents a significant advancement, expanding its applicability to a wider range of clinical samples and enhancing its utility, particularly in severe infections. However, further studies are needed to assess the real-world clinical impact of FASTinov, specifically its role in optimizing targeted therapy and improving patient outcomes. Additionally, as rapid AST systems typically incur higher costs than conventional susceptibility testing methods, a thorough cost-effectiveness analysis will be crucial to determining its feasibility for routine implementation in clinical microbiology laboratories.
Materials and methods
Bacterial strains
A total of 100 P. aeruginosa strains were included in the study, consisting of 99 clinical isolates collected from samples processed at the Microbiology Laboratory of Virgen del Rocío University Hospital (Seville, Spain) between 2021 and 2024, and one reference strain (P. aeruginosa ATCC 27853). All clinical strains were identified using MALDI-TOF mass spectrometry (Bruker, Germany).
Antimicrobial susceptibility testing
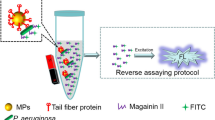
Antimicrobial susceptibility testing was determined by three different methods: (i) Flow Cytometry Assay (FASTinov): A novel flow cytometry-based method was also evaluated with the aim of reducing the time needed to obtain an antibiotic sensitivity report (Fig. 2A). FASTinov is a growth-independent AST method in which each well of the panel contains a specific antimicrobial combined with selected fluorescent probes that target early cellular damage. After a short incubation period, susceptible bacteria exhibit structural lesions, leading to altered fluorescence signals, whereas resistant bacteria maintain their integrity, showing no significant change. These differential patterns allow for rapid interpretation of susceptibility profiles without relying on bacterial replication or growth (Supplementary Fig. 1). To apply this principle in our experimental workflow, 2–3 colonies of P. aeruginosa were selected and incubated in 7 mL of MHB for 1.5 h at 37 °C with shaking at 250 rpm. The samples were then centrifuged (5.000 rpm 5 min), the supernatant was discarded, and the pellet was resuspended in 1 mL of saline to achieve a suspension adjusted to 0.65 McFarland. From this suspension, 100 µL were transferred to 7 mL of MHB and used to inoculate GRAMNEGcryo1 panels by adding 100 µL to each well. The entire preparation process, from centrifugation to panel inoculation, takes approximately 10–15 min. The panels were incubated at 37 °C for 1 h with shaking at 250 rpm. Following incubation, flow cytometric analysis was performed, (Fig. 2B), a process that requires approximately 25 min. (ii) MicroScan WalkAway system: Susceptibility profiles for all strains were initially determined using the NMR1 panel on the MicroScan WalkAway system (Beckman Coulter, USA), the routine method employed in our hospitals for antimicrobial susceptibility testing. (iii) Broth Microdilution: Minimum inhibitory concentrations (MICs) were assessed by the broth microdilution method using Mueller-Hinton broth (MHB), following the guidelines established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2024, Breakpoint tables for interpretation of MICs and zone diameters, http://www.eucast.org). All MIC values for the included isolates are provided in Supplementary Table 3.
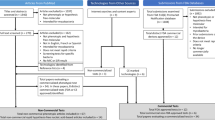
(A) Schematic comparison of the conventional antimicrobial susceptibility testing (AST) workflow and the FASTinov-based rapid AST (rAST) workflow for Pseudomonas aeruginosa infections. The conventional workflow requires overnight incubation for AST results, whereas the FASTinov rAST workflow performs susceptibility testing directly from isolated colonies, significantly reducing the turnaround time. (B) Overview of the FASTinov rAST workflow, showing the main steps from colony preparation to fluorescence-based detection of antimicrobial susceptibility. Original figure created with BioRender.com.
The FASTinov panels are proprietary dried 96-well microplate format with the antimicrobials spanning breakpoint concentrations according to EUCAST criteria and a fluorescent dye: 2 controls (C1-non-treated cells and C2- dead cells), ceftazidime 8 µg/ml, cefepime 8 µg/ml, piperacillin/tazobactam 16/4 µg/ml, meropenem 2 and 8 µg/ml, ceftolozane/tazobactam 4/4 µg/ml, ceftazidime/avibactam 8/4 µg/ml, ciprofloxacin 0.5 µg/ml and amikacin 16 µg/ml.
Data analysis
Interpretation of flow cytometry data was performed using BioFASTast software, developed by FASTinov. A multiparametric analysis is performed comparing size (SSC), complexity (FSC) and intensity of fluorescence of the treated cells with non-treated cells (C1) (Supplementary Fig. 2). Cut-off values were defined to classify the susceptibility of the strains based on the analysis of a lot of well characterized strains. The program reports whether the strain is “susceptible” (S), “susceptible to increased doses” (I; EUCAST), “intermediate” (I; CLSI) or “resistant” (R). Another control (C2) with a killing agent is present in the panel to ensure that the fluorescent dyes perform as expected. To evaluate diagnostic testing accuracy, we assessed multiple parameters, including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and categorical agreement (CA) between the results of FASTinov, MicroScan and broth microdilution methods19. In addition, data from the laboratory information system (LIS) were used to determine the time elapsed from the identification of P. aeruginosa by MALDI-TOF to the reporting and availability of antimicrobial susceptibility results in the patient’s medical record.
To assess the comparative performance of FASTinov and MicroScan against the reference standard (broth microdilution), we conducted a non-inferiority analysis for each antibiotic tested. Susceptibility results were categorized as “Susceptible” or “Resistant”, considering both “S” and “I” as susceptible. The categorical agreement between each method and microdilution was calculated. The difference in agreement (FASTinov minus MicroScan) was computed along with the corresponding 95% confidence interval (CI) using the Wald method for two independent proportions. A non-inferiority margin (Δ) of − 5% was pre-specified.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request. Due to ethical and privacy considerations, some data may be subject to access restrictions. All relevant processed data supporting the findings of this study are included in the manuscript.
References
Lister, P. D., Wolter, D. J. & Hanson, N. D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev.22, 582–610 (2009).
Tabak, Y. P. et al. Incremental clinical and economic burden of suspected respiratory infections due to multi-drug-resistant Pseudomonas aeruginosa in the United States. J. Hosp. Infect.103, 134–141 (2019).
Recio, R. et al. Predictors of mortality in bloodstream infections caused by Pseudomonas aeruginosa and impact of antimicrobial resistance and bacterial virulence. Antimicrob. Agents Chemother.64, e01759–e01719 (2020).
Pang, Z., Raudonis, R., Glick, B. R., Lin, T.-J. & Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv.37, 177–192 (2019).
Marschal, M. et al. Evaluation of the accelerate pheno system for fast identification and antimicrobial susceptibility testing from positive blood cultures in bloodstream infections caused by Gram-negative pathogens. J. Clin. Microbiol.55, 2116–2126 (2017).
Rosselin, M., Prod’hom, G., Greub, G. & Croxatto, A. A performance evaluation of the Quantamatrix QMAC-dRAST system for rapid antibiotic susceptibility testing directly from blood cultures. Microorganisms10, 1212 (2022).
Radhakrishnan, R., Rajesh, J., Dinesh, N. S., Thangavelu, C. P. & Sankaran, K. Antibiotic susceptibility testing based on fluorescein quenching by bacteria: Application to urinary tract infection. Sci. Rep.10(1), 4058 (2024).
Pina-Vaz, C. et al. A multisite validation of a two hours antibiotic susceptibility flow cytometry assay directly from positive blood cultures. BMC Microbiol. 24, 187 (2024).
Silva-Dias, A. et al. Evaluation of FASTinov ultrarapid flow cytometry antimicrobial susceptibility testing directly from positive blood cultures. J. Clin. Microbiol. 59, e0054421 (2021).
Eklöf, J., Gliese, K. M., Ingebrigtsen, T. S., Bodtger, U. & Jensen, J.-U.-S. Antibiotic treatment adequacy and death among patients with Pseudomonas aeruginosa airway infection. PLoS One14, e0226935 (2019).
Cillóniz, C. et al. Community-acquired pneumonia due to multidrug- and non-multidrug-resistant Pseudomonas aeruginosa. Chest150, 415–425 (2016).
Martinez-Nadal, G. et al. Inappropriate empirical antibiotic treatment in high-risk neutropenic patients with bacteremia in the era of multidrug resistance. Clin. Infect. Dis.70, 1068–1074 (2020).
Riccobene, T. et al. Outcomes of inadequate empiric therapy and timing of newer antibacterial therapy in hospitalized adults with culture-positive enterobacterales and Pseudomonas aeruginosa: a multicenter analysis. BMC Infect. Dis. 24, 810 (2024).
Xiao, S., Liang, X., Han, L. & Zhao, S. Incidence, antimicrobial resistance and mortality of Pseudomonas aeruginosa bloodstream infections among hospitalized patients in china: a retrospective observational multicenter cohort study from 2017 to 2021. Front. Public. Health. 11, 1294141 (2023).
Pancholi, P. et al. Multicenter evaluation of the accelerate phenotest BC kit for rapid identification and phenotypic antimicrobial susceptibility testing using morphokinetic cellular analysis. J. Clin. Microbiol. 56, e01329–e01317 (2018).
Sikorski, A. et al. Performance of Antipseudomonal β-Lactams on the Accelerate PhenoTest BC Kit against a Collection of Pseudomonas aeruginosa Isolates. J. Clin. Microbiol. https://doi.org/10.1128/jcm.01781-20 (2021).
Grohs, P. et al. Comparative evaluation of the QMAC-dRAST V2.0 system for rapid antibiotic susceptibility testing of Gram-negative blood culture isolates. J. Microbiol. Methods172, 105902 (2020).
Couchot, J. et al. Evaluation of the reveal rapid AST system to assess the susceptibility of Pseudomonas aeruginosa from blood cultures. Eur. J. Clin. Microbiol. Infect. Dis.42, 359–363 (2023).
Yusuf, E., Schijffelen, M. J. & Leeflang, M. How to verify and validate a clinical microbiology test before it can be used in routine diagnostics: A practical guide. Clin. Microbiol. Infect.30, 1261–1269 (2024).
Acknowledgements
JMO-R. is supported by the Subprograme Sara Borrell, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spain (CD21/00098). GM-G has received funding from the Andalusia Government in the grants for human resources reinforcement in the research activity (Acción B de refuerzos de larga duración). This study has been funded by Instituto de Salud Carlos III (ISCIII) through the project “PI23/01760” and by national funds through FCT - Fundação para a Ciência e a Tecnologia, I.P., within CINTESIS, R&D Unit (reference UIDP/4255/2020).
Author information
Authors and Affiliations
Contributions
Conception and design: CC-M, GM-G. Protocol development: CC-M, GM-G, CP-V. Analysis and interpretation of the results: CC-M, JO-R, CP-V, GMG. First draft of the manuscript: CC-M, GM-G, JAL. Reading and final approval of the manuscript: All authors.
Corresponding authors
Ethics declarations
Competing interests
CP-V is a co-founder of FASTinov, S.A. The company provided full funding for the materials required to conduct this study and participated in the design of the study protocol. Other authors declare no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cintora-Mairal, C., de la Rosa, J.M.O., Pina-Vaz, C. et al. Evaluation of FASTinov for rapid antimicrobial susceptibility testing in Pseudomonas aeruginosa. Sci Rep 15, 28649 (2025). https://doi.org/10.1038/s41598-025-12137-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12137-w