Abstract
The high-throughput Illumina sequencing technology was employed to decode the complete mitochondrial genome of isolate Jordan_1 black soldier fly (Hermetia illucens). The genome spans 15,699 base pairs (bp) with a GC content of 28.2%, and it includes 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. One InDel and 57 SNPs were revealed when compared to reference black soldier fly mitogenome. Phylogenetic tree was constructed using additional nine isolates from black soldier fly and Drosophila melanogaster as an out-group. The phylogenetic tree reveal that isolate Jordan_1 have common ancestor with an isolate from Kenya under the haplotype C. This study reveals the first report of black soldier fly mitogenome anywhere from the Middle East and North Africa region.
Similar content being viewed by others
Introduction
The black soldier fly (Hermetia illucens) has gained considerable attention in recent years due to its significant potential in waste recycling, animal feed production, and sustainable biomaterial development. As a species, H. illucens offers unique benefits for these industries owing to its rapid life cycle, high biomass conversion rate, and ability to thrive on a wide variety of organic substrates1 The development of genomic resources for this species has accelerated its commercial applications, particularly in insect farming, where it is increasingly used for converting organic waste into high-value products. Understanding the genome of the black soldier fly is critical for improving these processes and expanding the species’ utility across multiple sectors. A major application related to its ability to accumulate fat rapidly is of particular interest in the development of biodiesel, where the fly’s capacity to convert waste into lipids offers a promising solution for renewable energy production2. De novo transcriptome sequencing has provided molecular insights into the regulatory pathways that control fat accumulation in H. illucens, revealing key genes that could be targeted for genetic enhancement to improve lipid yields in industrial applications. Furthermore, studies on antimicrobial peptides produced by H. illucens larvae have identified novel compounds with potential therapeutic applications, underscoring the fly’s versatility as a resource for both environmental and medical advancements3.
The availability of a high-quality, chromosome-level genome assembly for H. illucens has been a milestone in the study of this species’ biology1. Such comprehensive genomic resources provided an essential foundation for examining the genetic underpinnings of its traits, including its capacity for rapid growth, waste conversion, and environmental adaptability. These traits are crucial for its roles in bioconversion systems and its potential use as a sustainable source of animal feed. The genome assembly has enabled more in-depth studies into the species’ evolutionary history, genetic diversity, and population structure.
In addition to these applications, H. illucens could serve as an intriguing model for genomic and evolutionary studies. Comparative genomic analyses revealed that this species exhibits a distinct genomic landscape shaped by its unique ecological niche as a decomposer of organic waste4. These analyses highlighted the importance of key genes associated with nutrient metabolism, fat accumulation, and stress responses, all of which contributed to its efficiency in waste recycling.
Furthermore, genetic manipulation tools, including CRISPR-based technologies, are being developed to further investigate and enhance these traits, paving the way for future genetic improvement for commercial applications5. Moreover, Kou et al.6 successfully established a highly efficient transgenic system for H. illucens, enabling precise genetic modifications that could enhance desirable traits such as growth rate, resistance to environmental stressors, and bioconversion efficiency. These developments open new avenues for optimizing black soldier fly farming operations and ensuring consistency in production, particularly under varying environmental conditions.
Further contributing to the genomic understanding of H. illucens are studies focused on its mitochondrial genome, which provide key insights into the evolutionary history of the species7. Mitochondrial genome analysis has allowed researchers to trace the phylogenetic relationships of the black soldier fly and explore its evolutionary adaptations to different environments. Therefore, the aim of this study was to characterize the mitogenome of and Jordanian isolate of black soldier fly as the first report covering the Middle East and North Africa region.
Materials and methods
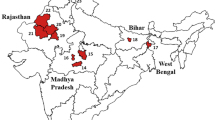
A comprehensive survey was conducted to assess the genetic and phenotypic diversity of black soldier fly populations across Jordan (Fig. 1), utilizing two primary collection methods: adult fly trapping and larvae collection from organic waste sites. The initial field survey, conducted from May 21–23, 2023, covered South Shuna, Aqaba, and Ghor Safi, using traps filled with spoiled apples, organic manure, and bird feed to attract adult insects. These traps were strategically placed at agricultural research stations, waste disposal sites, and wastewater treatment facilities. A subsequent survey, conducted from October 1–12, 2023, expanded the collection sites to Al-Dhuliel, Azraq, and Al-Muwaqqar, targeting diverse ecological contexts.
Total genomic DNA from the Jordan_1 black soldier fly was extracted using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) following the manufacturer’s instructions. Voucher specimens including DNA are available at the black soldier fly lab, National Agricultural Research Center (NARC), Jordan. Genomic DNA was sequenced using TruSeq Nano DNA with 350 bp paired-end reads (Illumina, USA). Reads were paired and mapped to the black soldier fly reference mitogenome available in the GenBank, which was an isolate from China, accession NC_0352328. CLC Genomics Workbench (Redwood City, CA) was used for read pairing, trimming, assembly and annotation of Jordan_1 black soldier fly mitogenome. One complete contig was successfully assembled based on mapped reads, which was followed by sequence annotation. The Jordan_1 black soldier fly mitogenome was visualized using OGDRAW9. Both Single Nucleotide Polymorphisms (SNPs) and Insertions and Deletions (InDels) they were identified using CLC Genomics Workbench software by pairwise alignment between the reference genome (NC_035232) and Jordan_1 black soldier fly mitogenome.
For phylogentic analysis, mitogenomes of Jordan_1 black soldier fly (Hermetia illucens) (PQ498594) and nine isolates from different countries7 including the reference mitogenome form China (NC_035232) were aligned using the multiple alignment (CLC Genomics Workbench, Redwood City, CA). As an out-group, the Drosophila melanogaster mitogenome (NC_024511) was used. Aligned sequences were used to generate 1000 replicates using the SEQBOOT function available in PHYLIP10. Bootstrapped data were subjected to the maximum likelihood and a consensus tree was generated.
Results
After conducting the first field survey (May 21–23, 2023), a limited number of H. illucens adults (fewer than 6) were collected, however, attempts to rear them were unsuccessful. On the other hand, during the second survey (October 1–12, 2023), H. illucens adults and larvae were successfully collected and reared, with notable findings from South Shuna, Al-Dhuliel, and Azraq, where the isolate Jordan_1 black soldier fly (Hermetia illucens) was collected. Unique details of insect body (both female and male insects) were documented from both abdominal and dorsal views (Fig. 2).
Reference images for isolate Jordan_1 black soldier fly (Hermetia illucens). A: Female insect with abdominal view; B. Female insect with dorsal view; C: Male insect with abdominal view; D. Male insect with dorsal view. Ruler scale in cm. Photo courtesy of The National Agricultural Research Center (NARC), Jordan.
The performed genome skimming resulted in high coverage (2300 x) for the entire mitogenome as revealed by sequencing depth plot (Fig. 3). The assembled mitogenome of black soldier fly Jordan_1 isolate was able to resolve all complex gene structures including gene overlapping such as those for tRNA-Trp and tRNA-Cys (Fig. 4). The length of the obtained circular genome of Jordan_1 black soldier fly was 15,699 base pairs (bp) (Fig. 5) with 28.2% GC content. The genome spans 37 genes including: 13 protein-coding genes, 22 tRNA, and two rRNA. All genes were H-strand encoded except ND1, ND4, ND5, two rRNA and eight tRNA genes, which were L-strand encoded.
When compared to the reference black soldier fly mitogenome (Accession number NC_035232), the Jordan_1 black soldier fly mitogenome showed fifty seven SNPs and five InDels (Table 1). There were 4 SNPs and 3 InDels in intergenic regions, while 49 SNPs and 2 InDels were recorded for 13 mitochondrial genes.
However, most detected SNPs were silent; they did not change coded amino acids in protein coding genes in the mitogenome of Jordan_1 black soldier fly (PQ498594) as compared to the reference genome (NC_035232). Nonetheless, amino acid changes were detected in five genes (Fig. 6). ND2 showed three SNPs with amino acid changes (T > I, L > I, V > L). Likewise, three SNPS with changed amino acids were detected in ND5 (V > I, R > G, V > I). Moreover, each of the following amino acid changes; T > I, I > M and S > G could be found in ND4, CYTB and ND4 genes, respectively.
Pairwise alignment of major SNPs causing amino acid changes for some coding proteins genes in Jordan_1 black soldier fly mitogenome (PQ498594) as compared to the reference geneome (NC_035232). Genes are ND2 (NADH dehydrogenase subunit 2), ND5 (NADH dehydrogenase subunit 5), ND4 (NADH dehydrogenase subunit 4), CYTB (Cytochrome c oxidase subunit III) and ND1 (NADH dehydrogenase subunit 1).
The phylogenetic analysis revealed four major clades covering the known four major haplotypes of black soldier fly (Fig. 7). The out group mitogenome of Drosophila melanogaster (NC_024511) was nicely separated from the entire black solider isolates used to build this phylogenetic tree. The C haplotype was the biggest clade with isolates from Jordan, Kenya, UK and USA. Haplotype C calde was divided into two major sub-clades, where the Jordanian isolate was clustered with the Kenyan one. Followed by haplotype A clade, with insect isolates from France and Taiwan. Most resolved branches were very robust showing 100% bootstrap values.
Maximum likelihood phylogenetic tree of mitogenomes (deprived of D-Loop regions) of isolate Jordan_1 black soldier fly (Hermetia illucens) (PQ498594) and nine isolates from different countries7 including the reference mitogenome form China (NC_035232). As an out-group, the Drosophila melanogaster (NC_024511) mitogenome was used. Percentage bootstrap values are given on each branch (1000 runs). Haplotypes as described in Guilliet et al.7.
Discussion
This study delivers the complete mitochondrial genome of the black soldier fly (Hermetia illucens) isolate_1 from Jordan, marking an important step in understanding the genetic diversity and potential applications of this species. This is the first report of mitogenome of the black soldier fly from the entire region of Middle East and North Africa, making it novel and crucial for future studies in the region and the world. Black soldier flies have gained significant attention for their roles in waste recycling, feed production, and biofuel potential. Prior studies, including Zhan et al.4, have highlighted the adaptability of H. illucens to various environments and substrates, and the availability of its genomic data supports further advancements in industrial applications. The 15,699 bp genome with a 28.2% GC content and 57 SNPs compared to the reference genome underscores the genetic variation present within the species, which potentially be could have been contributed by natural and artificial selection forces on this insect. In fact, major amino acid changes were evident in the present mitogenome of the black soldier fly (Hermetia illucens) isolate_1 from Jordan (Fig. 6). These evolutionary changes are evident for a potential vast array of SNPs and InDels along the entire genome, which would have huge implications for environmental adaptation and climate change mitigation. This is an important direction for whole genome analysis in a pan genome study.
Building on the genetic insights provided by this study, the importance of high-quality genome assembly is further emphasized by recent work, such as that by Generalovic et al.1, which provided a chromosome-level genome analysis for H. illucens. Such comprehensive genomic resources are critical in understanding the organism’s gene functions related to major applications such as fat accumulation2, nutrient recycling, and environmental resilience. The phylogenetic analysis conducted in this study supports the hypothesis of global genetic diversity among H. illucens populations, with the Jordan isolate showing a close relationship with the Kenyan isolate. This finding aligns with Guilliet et al.7, who proposed the existence of several haplotypes within H. illucens, suggesting that populations have adapted to diverse ecological niches over time. The presented phylogenetic tree was resolved into two main clades (Fig. 4), the first includes six isolates from Kenya, UK, USA, China (reference sequence), Ghana and Jordan. All of which belong to the haplotype C7. Nonetheless, it is proposed to have two sub-haplotypes within haplotype C (Fig. 6). The first would be C.1 including black soldier fly isolates from Jordan and Kenya, as they are clustered in an unique separated sub-clade, while the remaining isolates would be designated as C.2 (from UK, USA, China and Ghana). The second major clade clustered separated isolates; isolates from France and Taiwan in a sub-clade under A haplotype, a branch for an isolate from Guyana under H haplotype and finally a branch with a black soldier fly from Mexico. The clustering and separation of all isolates completely agrees with earlier findings (Guilliet et al.7. While the isolate Jordan_1 black soldier fly seems to have common ancestor with the isolate from Kenya, clustering based on geographical distribution was not observed, this finding is consistence with the fact that black soldier fly would move readily with old and new transportation means across the world.
The mitochondrial genome analysis also provides insights into the evolutionary history of H. illucens, a crucial aspect for both ecological and industrial applications. Guilliet et al.7 emphasized the role of mitochondrial genomes in tracing evolutionary adaptations, and this study’s phylogenetic tree supports the proposed sub-haplotypes within the species. Understanding these evolutionary pathways may improve the species’ application in industrial processes where genetic diversity could enhance resilience to environmental stressors and fluctuations in waste composition.
This research also contributes to the growing body of knowledge surrounding gene functions related to energy and nutrient conversion. Sukmak et al. (2024) explored nutrient-related gene functions in H. illucens, revealing pathways that could be optimized for enhanced bioconversion efficiency. Moreover, Cai et al.5 demonstrated the commercial implications of genetic variation in captive H. illucens populations. Therefore, the identification of major SNPs and InDels in protein-coding genes and tRNA genes in the Jordanian isolate’s mitogenome would be a sign for possible SNPs and InDels along the entire genome adds to this knowledge base, allowing researchers to pinpoint genes that may contribute to its effectiveness as a waste recycler. The distinct gene arrangement and SNPs identified in this study could serve as genetic markers for selective breeding, furthering industrial efficiency.
Furthermore, the presence of specific genes associated with fat accumulation in the mitogenome makes H. illucens a promising candidate for biodiesel production. Zhu et al.2 discussed the molecular basis of fat accumulation in H. illucens, and the genome data from this study could aid in selecting strains with high lipid production potential. The unique genomic characteristics of the Jordan isolate could thus inform the development of biofuel-oriented breeding programs that optimize lipid yield in industrial settings.
Moreover, advancements in genetic manipulation techniques, as described by Zhan et al.4 and Kou et al.6, could enable targeted enhancements in the Jordan isolate of H. illucens. This study’s detailed genomic data provide a foundation for utilizing CRISPR or other gene-editing technologies to improve traits such as bioconversion efficiency, stress tolerance, and reproductive rates. By integrating these advancements with the specific genetic information from the Jordan isolate, researchers can tailor black soldier fly strains for specific industrial needs.
The study’s findings also have potential implications for enhancing feed safety and quality. As indicated by Khamis et al.11, understanding the genetic makeup and microbiome of H. illucens can improve its safety as animal feed. The genetic variation and SNPs identified in the Jordanian isolate offer a foundation for monitoring and controlling genetic diversity in captive populations, ensuring consistency in feed quality and reducing the risk of introducing undesirable traits in commercial settings.
Conclusion
In conclusion, the complete mitogenome of the Jordan isolate of H. illucens offers a valuable resource for genetic, ecological, and industrial applications. This study enhances our understanding of the species’ genetic diversity, provides insights into its evolutionary relationships, and opens up possibilities for genetic improvement programs targeting bioconversion efficiency, feed quality, and biodiesel production. The findings are consistent with existing genomic studies on H. illucens and underscore the potential of integrating mitochondrial data in the selective breeding and genetic engineering of this versatile insect.
Data availability
The genome sequence data of Jordan_1 black soldier fly (Hermetia illucens) that support the findings of this study are openly available in GenBank of NCBI under the accession no. PQ498594.
References
Generalovic, T. N. et al. D. A high-quality, chromosome-level genome assembly of the black soldier fly (Hermetia illucens L.). G3 11(5), jkab085 (2021).
Zhu, Z. et al. De Novo transcriptome sequencing and analysis revealed the molecular basis of rapid fat accumulation by black soldier fly (Hermetia illucens, L.) for development of insectival biodiesel. Biotechnol. Biofuels. 12, 1–16 (2019).
Park, S. I., Kim, J. W. & Yoe, S. M. Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae. Dev. Comp. Immunol. 52 (1), 98–106 (2015).
Zhan, S. et al. Genomic landscape and genetic manipulation of the black soldier fly Hermetia illucens, a natural waste recycler. Cell Res. 30 (1), 50–60 (2020).
Cai, Z. et al. Whole-genome sequencing of two captive black soldier fly populations: implications for commercial production. Genomics 110891 (2024).
Kou, Z. et al. Establishment of highly efficient Transgenic system for black soldier fly (Hermetia illucens). Insect Sci. 30(4), 888–900 (2023).
Guilliet, J., Baudouin, G., Pollet, N. & Filée, J. What complete mitochondrial genomes tell Us about the evolutionary history of the black soldier fly, Hermetia illucens. BMC Ecol. Evol. 22(1), 72 (2022).
NCBI. https://www.ncbi.nlm.nih.gov (2024).
Greiner, S., Lehwark, P. & Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1), W59–W64 (2019).
Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the Author (Department of Genome Sciences, University of Washington, 2004).
Khamis, F. M. et al. Insights in the global genetics and gut Microbiome of black soldier fly, Hermetia illucens: implications for animal feed safety control. Front. Microbiol. 11, 1538 (2020).
Funding
This research was financially supported by the USAID through the project “Conversion of Wastewater Treatment Sludge to Biofuels and High-Value Added Products”. The Jordanian national owner is the Center on Agrarian Reform and Rural Development for the Near East (CARDNE), and implemented in collaboration with the Jordanian National Agricultural Research Center (NARC).
Author information
Authors and Affiliations
Contributions
The authorship contributions for this study are as follows: A.A. (Abeer Albalawneh) was involved in all aspects, including conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing (original draft and review & editing), visualization, supervision, project administration, and funding acquisition. B.A.-S. (Banan Al-Shagour), S.A. (Sami Alarsan), and M.S. (Monther Sadder) contributed to conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing (original draft and review & editing), visualization, and supervision. H.H. (Heba Hasan), and M.D. (Mai Deab) contributed to conceptualization and methodology. A.S. (Amer Sweity) contributed to methodology by field survey. B.A. (Belal Alshorman), M.A. (Majdi Aledwan), and M.A.D. (Mousa Abu Deah) participated in the methodological component, specifically in collecting insect samples, while S.A.Z. (Saja Abu Znaimah), Y.A. (Yazan AlBalawnah), E.N. (Ehab Neimat), and A.E. (Ahmed Elwan) contributed to methodology by supporting the growth and cultivation of insect samples. Where N.H. (Nizar Haddad) was contributed in the funding acquisition. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Albalawneh, A., Al-Shagour, B., Alarsan, S. et al. Complete mitochondrial genome of a novel black soldier fly isolate from Jordans. Sci Rep 15, 27817 (2025). https://doi.org/10.1038/s41598-025-12915-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12915-6