Abstract
Campylobacteriosis is a significant zoonosis with major public health implications. This study aimed to investigate the prevalence, molecular characteristics, and antimicrobial resistance of Campylobacter spp. in animal (rectal swabs and milk), environmental (fecal, water, and wall swabs), and human samples (stool and hand swabs) in New Valley Governorate, Egypt. Among 573 samples analyzed, Campylobacter spp. prevalence was highest in rectal swabs (32.9%) and human stool samples (74.2%), with lower rates in fecal samples (25.9%), milk (25.7%), wall swabs (13.1%), and water (10%). All isolates showed 100% resistance to clindamycin, while being completely sensitive to imipenem and amikacin. Multidrug resistance was found in 90.1% of the isolates, and 16S rRNA was detected in 90% of randomly selected Campylobacter spp. The hipO, cadF, and ceuE genes were detected in 77.8%, 33.3%, and 22.2% of the isolates, respectively. Phylogenetic analysis of the 16S rRNA gene showed significant congruence either between the tested isolates and each other or with other isolates in the gene bank, confirming the zoonotic transmission of multi-drug-resistant Campylobacter spp. This highlights the urgent need for improved biosecurity on farms, better food handling practices, and heightened public health awareness to mitigate the risk of Campylobacteriosis.
Similar content being viewed by others
Introduction
Campylobacteriosis, a notable zoonosis, poses significant concerns for public health. The causative agent, Campylobacter, is a Gram-negative bacterium found in diverse environments, such as animal feces, water, and soil. This microorganism can inhabit a broad spectrum of animals, including farm animals, poultry, and wild species, which act as potential sources of human infection. Importantly, substandard hygiene practices in animal husbandry, poultry farming, and environmental management play crucial roles in the spread and proliferation of Campylobacter. On farms, insufficient biosecurity protocols, contaminated food and water supplies, and unhygienic animal living conditions can contribute to the dissemination of Campylobacter within and among animal groups. Furthermore, improper handling of waste and inadequate sanitation in livestock production facilities can result in environmental contamination, thereby increasing the risk of human exposure to the pathogen1,2,3,4. Campylobacter species can be shed in the feces of diseased, recovered, or asymptomatic animals, leading to contamination of the environment, which increases the transmission of infection between herds5,6.
Human campylobacteriosis primarily occurs through the consumption of contaminated food and beverages. Poultry and other meat products, particularly when undercooked or improperly managed, are major sources of infection. However, contaminated water, unpasteurized milk, and even fresh products can also transmit Campylobacter1,6,7. The consequences of Campylobacter infection can range from mild gastrointestinal illness to severe complications. While most cases resolve spontaneously, some individuals, such as young children, elderly individuals, and immunocompromised individuals, are at increased risk of developing severe infections, including Guillain–Barré syndrome and reactive arthritis8,9.
Polymerase chain reaction (PCR) is a powerful molecular technique for detecting and identifying Campylobacter species. 16S rRNA gene sequencing is a widely used method for species-level identification and differentiation10. Furthermore, PCR can be employed to detect the presence of virulence genes, which are crucial determinants of Campylobacter pathogenicity. These genes, such as the hipO, ceuE, and cadF genes, encode proteins involved in various virulence mechanisms, including iron acquisition, adhesion to host cells, and modulation of host immune responses. The presence or absence of these genes can provide valuable insights into the potential virulence and severity of Campylobacter infections10,11,12.
The emergence of antimicrobial resistance in Campylobacter poses a significant threat to public health. The indiscriminate use of antibiotics in both human and animal medicine has contributed to the development of drug-resistant strains, limiting treatment options and increasing the severity of infections13,14,15.
From a one health perspective, this study aimed to isolate Campylobacter from humans, animals and the environment. Then studying the sensitivity of isolates from different hosts to antibiotics. Finally, the genetic similarity of three representative isolates was studied to assess the potential of zoonotic transmission and support disease control strategies.
Methods
Ethical declaration
This study adhered to the ethical guidelines of the “Institutional Review Board” of New Valley University, Egypt (Approval Number: NVREC 0213-20,249). Informed consent was obtained from all farm owners prior to sample collection, ensuring their understanding of the study objectives and procedures.
Study area and design
The study was conducted in the New Valley Governorate, Egypt, from November 2023 to December 2024. This region encompasses a vast area with diverse agricultural practices, including livestock farming (Fig. 1).
Sampling
The total number of samples examined is 573 from animals, humans and the environment. Animal samples were firstly, rectal swabs from cows (n = 62), sheep (n = 75) and goats (n = 70). Secondly, the milk samples were from cows (n = 41), sheep (n = 36) and goats (n = 36). As for the human samples, they included: Firstly, 66 stool samples were collected from patients with intestinal disorders who frequent hospitals and laboratories in the study area. Secondly, 46 hand swabs from farm workers and people in contact with animals or their products. As for the environmental samples, they were as follows: 116 fecal samples, 10 water samples from animal courses and 15 wall swabs from animal residences. The samples were withdrawn according to the Campylobacter isolation protocol. All samples were collected under aseptic conditions, labelled and transferred in an ice box to the microbiology lab, Faculty of veterinary medicine New valley university with minimum delay as possible.
Bacteriological and biochemical analysis of Campylobacter spp.
Campylobacter spp. were isolated according to the ISO 10,272–1 method16. Briefly, samples were enriched in Bolton broth (M1592 Himedia, India) supplemented with selective antibiotics and lysed horse blood, then incubated under microaerophilic conditions at 41.5 ± 1 °C for 48 h. Enriched cultures were streaked onto modified charcoal cefoperazone deoxycholate agar (M887I Himedia, India), and incubated under microaerophilic conditions (e.g., gas pack system) at 41.5 °C for 48 h to isolate presumptive Campylobacter colonies. Biochemical identification was carried out to the suspected colonies.
Molecular identification and characterization of Campylobacter spp.
PCR was performed to the randomly selected samples to perform molecular confirmation through the detection of the 16S rRNA gene for species identification and virulence-associated genes (hipO, ceuE, and cadF) via specific primers (Table 1)17,18,19,20. Genomic DNA was extracted via a QIAamp DNA Mini (catalog no. 51304). Emerald Amp GT PCR master mix (Takara, Code No. RR310A Kit) was used. The PCR products were analyzed via agarose gel electrophoresis and visualized via UV transillumination. The marker for the PCR products was a 100 bp DNA ladder.
Phylogenetic analysis
Phylogenetic analysis was performed on a representative subset of isolates (one animal, one environmental, and one human isolate) on the basis of 16S rRNA gene sequences (Table 1)21. Sequence alignment was performed via CLUSTAL22, and phylogenetic trees were constructed via MEGA623 via the maximum likelihood, neighbor-joining, and maximum parsimony methods.
Antibiotic resistance profile of isolated Campylobacter spp.
Antimicrobial susceptibility testing was performed via the disc diffusion method. The CLSI (2020) and EUCAST (2021) recommended guidelines breakpoints were used for macrolides, while CLSI breakpoints for Enterobacteriaceae were used for other antibiotics24,25. The antimicrobial agents evaluated are listed in Table 2. The multidrug resistance (MDR) index was calculated26.
Statistical analysis
Statistical analysis was performed via SPSS, version 25 (IBM Corp., 2013). The data were analyzed via chi-square tests, and the significance level was set at p < 0.05. The data were treated as a complete randomization design according to Steel et al27.
Results
Prevalence
A total of 181 Campylobacter spp. isolates were recovered from 573 examined samples. The prevalence of Campylobacter spp. was at Animal samples level 32.9% in rectal swabs and 25.7% in milk samples. Additionally, it was at Human samples level 74.2% in stool samples and 4.4% in hand swabs. Moreover, at Environmental samples level the prevalence was 25.9% in fecal samples from animal litter, 10% in water samples, and 13.3% in wall swabs, as shown in (Table 3).
Molecular characterization
Ten randomly selected isolates were evaluated by PCR for the presence of 16S rRNA and virulence genes (hipO, ceuE, and cadF). The prevalence of 16S rRNA was 90% (9/10), whereas the prevalence of virulence genes was 77.8% (7/9) for hipO, 22.2% (2/9) for ceuE, and 33.3% (3/9) for cadF, as shown in Table 4 and Fig. 2.
Antimicrobial resistance
A total of 181 Campylobacter spp. isolates were evaluated for antibiotic susceptibility to 12 commonly used antibiotics in the study area. All the isolates were resistant to clindamycin. In contrast, the highest sensitivity was observed for imipenem and amikacin (100% sensitivity each). The isolates exhibited 96.1% sensitivity to gentamicin, 94.5% sensitivity to doxycycline, and 71.8% sensitivity to ciprofloxacin (Table 5). The mean MAR indices of Campylobacter spp. isolated from animal, environment and human samples were 0.393, 0.481, and 0.416, respectively (Table 6). Notably, the highest MAR index was observed in environmental isolates.
Phylogenetic analysis
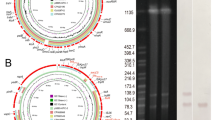
The 16S rRNA genes of three randomly selected isolates have been deposited in GenBank under accession numbers: PQ780749 for fecal isolates, PQ780750 for milk isolates, and PQ780748 for human stool isolates (Figs. 3 and 4). Phylogenetic analysis of sequences revealed high genetic similarity between the three isolates ranging from 99.8 to 100%. Additionally, 99.7–100% similarity was found with many other isolates in GenBank.
Discussion
Campylobacteriosis is a major global public health concern and the leading cause of bacterial gastroenteritis, affecting millions of people annually and contributing to severe complications such as Guillain–Barré syndrome and reactive arthritis. Its burden extends beyond human health, causing infections in livestock, contaminating food and water sources, and imposing significant economic losses on healthcare systems, agriculture, and the food industry5,28,29.
The present results revealed that the prevalence of Campylobacter spp. in animal rectal swabs was 32.9%. These results were similar to those obtained in Kenya (30.9%)30. However, it was higher than the results of previous studies in Assiut, Egypt and Kenya, where the prevalence rates were 20.1% and 15.7%, respectively31,32. In contrast, it was lower than the prevalence reported in the United States, Bahama, and Mexico (80.7%)33, and 59.2% in northern Spain34.
The prevalence of Campylobacter spp. in milk was 25.7%. These results were similar to those reported in Italy (24.9%)35. However, lower prevalences of 12.6% and 2% were reported in Iraq and Egypt, respectively36,37. On the other hand, a higher prevalence rate was reported in Iran (65.8%)38.
The prevalence of Campylobacter in stool samples was 74.2%, which is similar to findings previously -reported in Egypt (76.9%)39. However, it was higher than other results in Egypt and Iran, where the prevalence rates were 66.6% and 8.4%, respectively6,8. In contrast, it is lower than the prevalence reported in other Egyptian studies (90.9%)40.
The results revealed that the prevalence of Campylobacter in hand swabs was 4.4%, which was similar to findings reported among handlers of raw poultry or meat9. However, this value was higher than that reported in a previous study in Ethiopia (1.8%)41. In contrast, it was lower than the prevalence (11.9%) reported in China42, whereas another study could not isolate Campylobacter from examined hand swab samples in Egypt43. There was a highly significant difference between the human stool and hand swabs (p < 0.001).
The prevalence of Campylobacter spp. in environmental fecal samples was 25.9%. These results were very close to those of two studies in Canada and Bangladesh (27% and 26.7%, respectively)29,44. However, it is lower than the prevalence reported in other studies in Sweden (78%) 7.
The prevalence of Campylobacter spp. in the water samples was 10%. These findings are similar to those reported in Malaysia (11.1%)45; however, these findings are lower than the prevalence reported in another study in South Africa (21.7%)3. Notably, no Campylobacter was detected in water samples in Bangladesh5. Moreover, the prevalence of Campylobacter spp. in wall swabs was 13.3%. This finding was similar to the prevalence of 15% reported in Canada46, and a previous study reported a prevalence of 15% in poultry farm environments47. However, it was lower than the prevalence reported in Malaysia (33.3%)45.
PCR is a valuable tool for the molecular confirmation of Campylobacter spp., including species identification and the detection of virulence genes10,11. In the present study, 90% of the randomly selected isolates were positive for the 16S rRNA gene. Among the positive isolates, 77.8% (7/9) harbored the hipO gene, 22.2% (2/9) harbored the ceuE gene, and 33.3% (3/9) harbored the cadF gene. These findings are consistent with previous studies reporting the presence of the hipO gene in 75% of isolates48. However, other studies have reported higher detection rates for the ceuE, hipO, and cadF genes49,50. The distribution of genes reflects both species identity and isolation source and the differences between them are always statistically significant. In general, it can be said that The variation in the distribution of these genes may be attributed to factors such as sample sources (e.g., animal. Environment, food and human) Campylobacter serotype, geographical location and genetic mutations12,48,49.
The high genetic similarity between the three isolates implies that cattle are an important reservoir for human campylobacteriosis. Additionally, these results showed similarities ranging from 99.7 to 100% with many isolates of human and animal origin, such as human stool isolates from the United Kingdom, human stool isolates from a child in Brussels, Belgium, and broiler cloacal swabs from Sweden (Accession Nos. AL111168, HE978252 and AF550627). The phylogenetic relatedness of C. jejuni isolates from animals and humans further supports cross-species transmission of this pathogen. These findings are in agreement with those of previous studies that documented zoonotic transmission of Campylobacter51,52.
The present data revealed that Campylobacter isolates presented 100% resistance to clindamycin, followed by high resistance rates to erythromycin, amoxicillin-clavulanic acid (85.64%), and azithromycin (70.2%). These findings are consistent with those of several previous studies15. However, they contradict other studies that reported higher susceptibility to these antibiotics53. Conversely, the highest sensitivity levels were observed for imipenem, amikacin (100%), gentamicin (96.1%), doxycycline (94.5%), and ciprofloxacin (71.8%). These results align with other studies that reported high sensitivity to imipenem, amikacin, and gentamicin53. Our results revealed that 90.1% of the Campylobacter spp. isolates exhibited multidrug resistance (MDR), defined as resistance to three or more classes of antibiotics. This finding is consistent with other studies that reported high levels of MDR in Campylobacter32. In fact, the widespread prevalence of multidrug resistant Campylobacter in the food chain, human, animal or environment has a significant impact on both public health and veterinary in terms of the difficulty of treatment and the increased cost. Consequently, effective strategies to fight antibiotic resistance include responsible antibiotic use and the development of alternative treatments are vital13,54.
Our data revealed that the MAR indices of Campylobacter spp. isolated from animal, environmental and human samples were 0.393, 0.481, and 0.416, respectively. These findings are consistent with previous studies that reported mean MDR indices of 0.49 for animal isolates55, 0.4 for environmental isolates and 0.3–0.4 for human isolates56. Notably, the mean MDR index was highest for the environmental isolates, suggesting a relatively high level of antibiotic resistance among Campylobacter strains from the environment. Several factors might explain the high antibiotic resistance of environmental isolates, including exposure to antibiotics in the environment through pollution, horizontal gene transfer between bacteria, natural selection due to constant exposure to low levels of antibiotics, and the presence of diverse microbial communities that act as reservoirs for resistance genes14,57.
Conclusions
This study demonstrated a significantly high prevalence of MDR Campylobacter spp. in various samples. Phylogenetic analysis revealed a high degree of genetic similarity among isolates from different sources, including animal, environmental, and human samples. This finding underscores the potential for zoonotic transmission of Campylobacter spp. between animals, the environment, and humans. Future studies should focus on comparative genomics to identify zoonotic markers and assess the role of agricultural practices in the evolution of C. jejuni. Additionally, implementing measures to restrict the use of antibiotics as growth promoters and for prophylaxis on animal farms is crucial to combat antimicrobial resistance. Finally, mitigating the spread of diseases depends critically on a One Health strategy that combines environmental, human, and animal monitoring. Additionally, educating the public on safe food handling practices is essential to prevent human infection.
Data availability
The datasets generated during and/or analyzed during the current study are provided within the manuscript and supplementary information files.
References
Humphrey, T., O’Brien, S. & Madsen, M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 117, 237–257. https://doi.org/10.1016/j.ijfoodmicro.2007.01.006 (2007).
Mandel, T., Iulietto, M. F., Reinik, M. & Condoleo, R. Exploring frameworks for quantitative risk assessment of antimicrobial resistance along the food chain. EFSA J. 22, e221117. https://doi.org/10.2903/j.efsa.2024.e221117 (2024).
Chukwu, M. O., Abia, A. L., Ubomba-Jaswa, E., Obi, L. & Dewar, J. B. Characterization and phylogenetic analysis of campylobacter species isolated from paediatric stool and water samples in the Northwest Province, South Africa. Int. J. Environ. Res. Public Health https://doi.org/10.3390/ijerph16122205 (2019).
Yirgalem, M., Kemal, J., Wolkaro, T., Bekele, M. & Terefe, Y. Identification and antimicrobial susceptibility profiles of Campylobacter isolated from camel at municipal abattoirs in eastern Ethiopia. Sci. Rep. 14, 26335. https://doi.org/10.1038/s41598-024-76895-9 (2024).
Hoque, N. et al. Prevalence, risk factors, and molecular detection of campylobacter in farmed cattle of selected districts in Bangladesh. Pathogens https://doi.org/10.3390/pathogens10030313 (2021).
Ansarifar, E., Riahi, S. M., Tasara, T., Sadighara, P. & Zeinali, T. Campylobacter prevalence from food, animals, human and environmental samples in Iran: A systematic review and meta-analysis. BMC Microbiol. 23, 126. https://doi.org/10.1186/s12866-023-02879-w (2023).
Hansson, I., Olsson Engvall, E., Ferrari, S., Harbom, B. & Lahti, E. Detection of Campylobacter species in different types of samples from dairy farms. Vet. Rec. 186, 605–605. https://doi.org/10.1136/vr.105610 (2020).
Abd El-Aziz, N. K. et al. First report of aacC5-aadA7Delta4 gene cassette array and phage tail tape measure protein on class 1 integrons of campylobacter species isolated from animal and human sources in Egypt. Animals (Basel) https://doi.org/10.3390/ani10112067 (2020).
Kabir, S. M. Prevalence of Campylobacter species on human hands and its association with food handling. Int. J. Food Microbiol. 243, 1–6 (2017).
Konkel, M. E., Gray, S. A., Kim, B. J., Garvis, S. G. & Yoon, J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J. Clin. Microbiol. 37, 510–517. https://doi.org/10.1128/jcm.37.3.510-517.1999 (1999).
Nayak, R., Stewart, T. M. & Nawaz, M. S. PCR identification of Campylobacter coli and Campylobacter jejuni by partial sequencing of virulence genes. Mol. Cell. Probes 19, 187–193. https://doi.org/10.1016/j.mcp.2004.11.005 (2005).
Sierra-Arguello, Y. M. et al. Identification of pathogenic genes in Campylobacter jejuni isolated from broiler carcasses and broiler slaughterhouses. Sci. Rep. 11, 4588. https://doi.org/10.1038/s41598-021-84149-1 (2021).
Du, Y. et al. Molecular identification of multidrug-resistant Campylobacter species from diarrheal patients and poultry meat in Shanghai China. Front. Microbiol. 9, 1642. https://doi.org/10.3389/fmicb.2018.01642 (2018).
Bunduruș, I. A. et al. Overview of virulence and antibiotic resistance in Campylobacter spp. livestock isolates. Antibiotics https://doi.org/10.3390/antibiotics12020402 (2023).
Portes, A. B., Panzenhagen, P., Pereira Dos Santos, A. M. & Junior, C. A. C. Antibiotic Resistance in Campylobacter: A systematic review of South American isolates. Antibiotics https://doi.org/10.3390/antibiotics12030548 (2023).
Roberts, D. & Greenwood, M. Practical Food Microbiology 3rd edn. (John Wiley & Sons, 2008).
Linton, D., Owen, R. & Stanley, J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147, 707–718. https://doi.org/10.1016/s0923-2508(97)85118-2 (1996).
Wang, G. et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbial. 40, 4744–4747. https://doi.org/10.1128/jcm.40.12.4744-4747.2002 (2002).
Shin, E.-J. & Lee, Y.-H. Comparison of three different methods for Campylobacter isolation from porcine intestines. J. Microbiol. Biotechnol. 19, 647–650 (2009).
Al Amri, A., Senok, A. C., Ismaeel, A. Y., Al-Mahmeed, A. E. & Botta, G. A. Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. J. Med. Microbiol. 56, 1350–1355. https://doi.org/10.1099/jmm.0.47220-0 (2007).
Lagacé, L., Pitre, M., Jacques, M. & Roy, D. Identification of the bacterial community of maple sap by using amplified ribosomal DNA (rDNA) restriction analysis and rDNA sequencing. Appl. Environ. Microbiol. 70, 2052–2060. https://doi.org/10.1128/AEM.70.4.2052-2060.2004 (2004).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
CLSI, I. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. CLSI Supplement VET08 (2018).
EUCAST. European Committee on Antimicrobial Susceptibility Testing (EUCAST): breakpoint tables for interpretation of MICs and zone diameters. (2021).
Ghasemian, S. O., Mahmoodipour, H. & Gholami-Ahangaran, M. Study on antibiotic resistance profile and multiple antibiotic resistance index (MAR Index) in the Campylobacter spp. isolates from domestic animals and water. Pathobiol. Res. 25, 6–14 (2022).
Steel, R. G., Torrie, J. H. & Dickey, D. A. Principles and Procedures of Statistics: a Biometrical Approach. (1997).
EFSA. The European Union One Health 2023 Zoonoses report. EFSA J. 22, e9106. https://doi.org/10.2903/j.efsa.2024.9106 (2024).
Guevremont, E., Lamoureux, L., Loubier, C. B., Villeneuve, S. & Dubuc, J. Detection and characterization of Campylobacter spp. from 40 dairy cattle herds in Quebec Canada. Foodborne Pathog Dis 11, 388–394. https://doi.org/10.1089/fpd.2013.1706 (2014).
Wanja, D. W., Mbuthia, P. G., Aboge, G. O. & Bebora, L. C. Seasonal prevalence and molecular identification of thermophilic Campylobacter from chicken, cattle, and respective drinking water in Kajiado county, Kenya. Int. J. Microbiol. 2022, 1526641. https://doi.org/10.1155/2022/1526641 (2022).
Khadr, A., Haggag, Y. & Khaleil, S. Prevalence of Campylobacter jejuni and Campylobacter coli in calves and lambs with and without diarrhea and their public health importance. Assiut Vet. Med. J. 52, 179–190. https://doi.org/10.21608/avmj.2006.177645 (2006).
Chala, G., Eguale, T., Abunna, F., Asrat, D. & Stringer, A. Identification and characterization of Campylobacter species in livestock, humans, and water in livestock owning households of Peri-urban Addis Ababa, Ethiopia: a one health approach. Front. Public Health 9, 750551. https://doi.org/10.3389/fpubh.2021.750551 (2021).
Hanlon, K. E. et al. Presence of Salmonella and Escherichia coli O157 on the hide, and presence of Salmonella, Escherichia coli O157 and Campylobacter in feces from small-ruminant (goat and lamb) samples collected in the United States, Bahamas and Mexico. Meat. Sci. 135, 1–5. https://doi.org/10.1016/j.meatsci.2017.08.003 (2018).
Oporto, B., Esteban, J., Aduriz, G., Juste, R. & Hurtado, A. Prevalence and strain diversity of thermophilic campylobacters in cattle, sheep and swine farms. J. Appl. Microbiol. 103, 977–984. https://doi.org/10.1111/j.1365-2672.2007.03328.x (2007).
Serraino, A. et al. Presence of Campylobacter and Arcobacter species in in-line milk filters of farms authorized to produce and sell raw milk and of a water buffalo dairy farm in Italy. J. Dairy Sci. 96, 2801–2807. https://doi.org/10.3168/jds.2012-6249 (2013).
Almashhadany, D. A. Isolation, biotyping and antimicrobial susceptibility of Campylobacter isolates from raw milk in Erbil city, Iraq. Ital. J. Food Saf. https://doi.org/10.4081/ijfs.2021.8589 (2021).
Sotohy, S. A., Emam, S. M., Diab, M. S. & Ewida, R. M. Prevalence of Campylobacter Spp. in marketable milk and some milk products in New Valley Governorate, Egypt. J. Adv. Vet. Res. 13, 833–836 (2023).
Haghi, F. et al. Detection of major food-borne pathogens in raw milk samples from dairy bovine and ovine herds in Iran. Small Rumin. Res. 131, 136–140. https://doi.org/10.1016/j.smallrumres.2015.08.005 (2015).
Abd El Tawab, A. A., Ammar, A. A., Ahmed, H. A. & Hefny, A. A. Bacteriological and molecular identification of some Campylobacter species in broilers and their macrolide resistance profile. Benha Vet. Med. J. 34, 374–391. https://doi.org/10.21608/bvmj.2018.54483 (2018).
El-Naenaeey, E.-S., Abd El-Hamid, M. & Khalifa, E. Prevalence and antibiotic resistanc patterns of Campylobacter species isolated from different sources in Eygpt. J. Microbiol. Biotechnol. Food Sci. https://doi.org/10.15414/jmbfs.3723 (2021).
Debelo, M., Mohammed, N., Tiruneh, A. & Tolosa, T. Isolation, identification and antibiotic resistance profile of thermophilic Campylobacter species from Bovine, Knives and personnel at Jimma Town Abattoir, Ethiopia. PLoS ONE 17, e0276625. https://doi.org/10.1371/journal.pone.0276625 (2022).
Bai, Y. et al. Quantification of cross-contamination of Campylobacter jejuni during food preparation in a model kitchen in China. J. Food Prot. 84, 850–856. https://doi.org/10.4315/JFP-20-280 (2021).
Gwida, M., Zakaria, A., El-Sherbiny, H., Elkenany, R. & Elsayed, M. Prevalence of Campylobacter, Enterococcus and Staphylococcus aureus in slaughtered camels. Vet. Med. 64, 521–530. https://doi.org/10.17221/104/2019-VETMED (2019).
Yamasaki, S. et al. Isolation, molecular identification and antimicrobial resistance patterns of Campylobacter species of dairy origin: First report from Bangladesh. Vet. Sci. Dev. https://doi.org/10.4081/vsd.2018.7838 (2019).
Aung, W. W. et al. Occurrence of Campylobacter in dairy and beef cattle and their farm environment in Malaysia. Pak. Vet. J 35, 470–473 (2015).
Agunos, A., Waddell, L., Leger, D. & Taboada, E. A systematic review characterizing on-farm sources of Campylobacter spp. for broiler chickens. PLoS ONE 9, e104905. https://doi.org/10.1371/journal.pone.0104905 (2014).
Hänninen, M. L. et al. Campylobacter spp. prevalence on farm surfaces, including walls, in poultry facilities. Int. J. Food Microbiol. 82, 105–112 (2003).
Falodun, O. I. & Waleola, O. A. Antibiotic resistance and virulence genes in Campylobacter species from pig and cattle samples in Ibadan, Nigeria. Eur. J. Biol. Res. 14, 33–44. https://doi.org/10.5281/zenodo.10909573 (2024).
Bang, D. D. et al. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J. Appl. Microbiol. 94, 1003–1014. https://doi.org/10.1046/j.1365-2672.2003.01926.x (2003).
Zhang, M. J., Gu, Y. X., Ran, L. & Zhang, J. Z. Multi-PCR identification and virulence genes detection of Campylobacter jejuni isolated from China. Zhonghua Liu Xing Bing Xue Za Zhi 28, 377–380 (2007).
Zhang, Q., Beyi, A. F. & Yin, Y. Zoonotic and antibiotic-resistant Campylobacter: A view through the One Health lens. One Health Adv. https://doi.org/10.1186/s44280-023-00003-1 (2023).
Veronese, P. & Dodi, I. Campylobacter jejuni/coli infection: Is it still a concern?. Microorganisms https://doi.org/10.3390/microorganisms12122669 (2024).
Liao, Y.-S. et al. Antimicrobial resistance in Campylobacter coli and Campylobacter jejuni from human campylobacteriosis in Taiwan, 2016 to 2019. Antimicrob. Agents Chemother. 66, e01736-e11721. https://doi.org/10.1128/aac.01736-21 (2022).
Mohan, V. et al. Antimicrobial resistance in Campylobacter spp. focussing on C. jejuni and C. coli–A narrative review. J. Glob. Antimicrob. Resist. https://doi.org/10.1016/j.jgar.2025.05.008 (2025).
Banisharif Dehkordi, G., Marhamatizadeh, M. H. & Momtaz, H. Phenotypic and genotypic identification of antimicrobial resistance amongst the Campylobacter jejuni and Campylobacter coli strains isolated from raw milk of animal species. J. Food Qual. 2024, 6980374. https://doi.org/10.1155/2024/6980374 (2024).
Ghoneim, N. H., Sabry, M. A., Ahmed, Z. S. & Elshafiee, E. A. Campylobacter species isolated from chickens in Egypt: Molecular epidemiology and antimicrobial resistance. Pak. J. Zool. 52, 917. https://doi.org/10.17582/journal.pjz/20190324080346 (2020).
Chibwe, M., Odume, O. N. & Nnadozie, C. F. Emerging Pollutants: Protecting Water Quality for the Health of People and the Environment. In Sarantuyaa Zandaryaa (eds Fares, A. & Eckstein, G.) (Springer Nature Switzerland, 2025).
Acknowledgements
The authors also thank all members of the Department of Animal Hygiene and Zoonoses, Faculty of Veterinary Medicine at New Valley University for their valuable assistance in this study.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). No funding has been provided.
Author information
Authors and Affiliations
Contributions
MSD, MK, MGS, AF and NKA proposed and designed the research idea. ARE, RME, MSD, and DAA outlined the research and designed the methodology. MGS and DAA carried out the sample collection, performed the laboratory work, and analyzed and interpreted the data. MGS, AF and MK prepared the first draft of the manuscript for publication. MSD, NKA, and ARE reviewed the final version of the manuscript. DAA was involved in providing laboratory consumables. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Human or animal rights
All experimental protocols in this study were approved by Institutional Review Board of New Valley University, Egypt (Approval Number: NVREC 0213-20249). All the farm owners included in this study were informed of all the study procedures and aims of the study. Additionally, Informed consent was obtained from the human participants and/or their legal guardian. The study was conducted in accordance with the Declaration of Helsinki for medical research involving human subjects. All experimental protocols in this study were approved by Institutional Review Board of New Valley University, Egypt (Approval Number: NVREC 0213-20249.All methods were performed in accordance with the ARRIVE guidelines for the reporting of animal experiments (https://arriveguidelines.org). All methods were carried out in accordance with relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kholeif, M., Sayed, M.G., Fotouh, A. et al. Prevalence and molecular characterization of multidrug resistant Campylobacter isolated from animals and humans as a one health approach. Sci Rep 15, 30262 (2025). https://doi.org/10.1038/s41598-025-13120-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13120-1