Abstract
To address inherent limitations of standard diagnostic procedures for lung cancer, like long turn-around-time and the need for sufficient samples for analysis, we innovatively integrated traditional smear cytology (TSC) with quantitative polymerase chain reaction (qPCR) assays on micro cell samples (MCSs) for the diagnosis of lung cancer. All patients underwent TSC and qPCR assays targeting 11 genes based on different MCSs, including samples obtained by flushing needles used for endobronchial ultrasound biopsies and percutaneous aspiration biopsies of lung in 38 cases (G1) and 108 cases (G2), and lavage fluid samples obtained by bronchoalveolar lavage in 38 cases (G3). With clinical diagnosis and pathological biopsy as diagnostic gold standard, the diagnostic value of these MCSs were explored. In G1, G2 and G3, the diagnostic yield of MCSs-based TSC alone was 78.9%, 93.5% and 76.3%, respectively. Combination of TSC and genetic testing increased the diagnostic yield to 81.6%, 98.1% and 84.2% in G1, G2 and G3, respectively. In addition, the qPCR results of MCSs and paired tissue samples was all matched with a concordance rate of 100%, and the quantity of DNA extracted from our samples was significantly higher than that of tissue samples in G1 and G2. Notably, the TAT of our diagnostic method required only 24 h, which greatly improved the timeliness of diagnosis. Our study demonstrated that MCSs-based TSC combined with genetic testing could not only rapidly and reliably diagnose lung cancer, but also effectively detect gene targets, with potential for widespread application.
Similar content being viewed by others
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. The prognosis for lung cancer is relatively poor and more than 70% of patients are diagnosed at the advanced stage1,2. The insidious and complex nature of lung cancer leads to a time-consuming diagnostic process that prevents patients from receiving timely and effective treatment, which adversely affects clinical outcomes3. Therefore, rapid and precise diagnosis of lung cancer is crucial to the patient outcomes and well-being. To date, the standard diagnostic procedures, including imaging, pathology and molecular analyses, typically require a diagnostic period of 5-7days, resulting in long turn-around-time (TAT)4. Routine detection pathways prevent the patients from obtaining the treatment plan in a timely manner, or even makes them miss the optimal treatment time. Therefore, it is necessary to explore novel diagnostic pathways to shorten the TAT of test reports.
At present, tissue biopsy is the gold standard for the clinical diagnosis of lung cancer, encompassing techniques from large surgical resections to minimally invasive procedures5,6. While surgical resections yield sufficient samples for comprehensive analysis, most lung cancer cases are advanced at diagnosis, necessitating less invasive methods such as percutaneous aspiration biopsy (PAB) and endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA)7,8,9. These minimally invasive techniques often result in small biopsies, which have been reported to be insufficient for molecular profiling in approximately 30% of cases10. The challenge of obtaining adequate tissue samples persists, highlighting the potential utility of needle rinse solutions for genetic testing. Despite initial findings supporting their diagnostic value, further research is required to fully explore the potential of biopsy needle rinse solutions in lung cancer diagnosis and molecular analysis11,12.
Cytology biopsy is an acceptable alternative for patients with advanced-stage lung cancer or in poor physical condition whose tissue samples are difficult to obtain13. Currently, bronchoalveolar lavage (BAL) has received a great deal of attention in academic research for its easy availability and its potential to provide a definitive diagnosis and detect mutated cancer genes14. However, the cytology of BAL fluids (BALF) suffers a low detection sensitivity, and limited data have been published on the use of BALF for sequencing in lung cancer patients15. Meanwhile, pathologists have difficulty in making a definitive pathological diagnosis based on a small number of tumor cells. Therefore, it is necessary to investigate whether the use of cytology in combination with other diagnostic tools, such as genetic testing could improve the detection sensitivity of BALF, and to further explore the utility of BALF in genetic analysis.
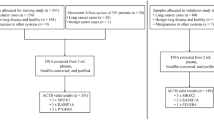
In this study, we aimed to determine the utility of the micro cell samples (MCSs), which were obtained from puncture needle rinse samples and BALF, for the diagnosis and genetic analysis of lung cancer. To this end, we first explored the diagnostic value of cytology based on MCSs in lung cancer, by considering pathological examination and clinical diagnosis as the diagnostic gold standard. Next, we further investigated the diagnostic performance of cytology combined with multi-gene testing by quantitative polymerase chain reaction (qPCR) assays based on MCSs in lung cancer and different histological types of lung cancer. Finally, we compared the quantity of DNA extracted from MCSs and paired tissue samples, and explored the consistency of MCSs and the paired tissue samples in detecting mutant genes. Figure 1 showed the experimental flowchart of this study.
Method
Patients
A total of 184 patients with suspected lung cancer who undergone EBUS-TBNA, percutaneous aspiration biopsy of lung (PABL), or BAL, for diagnosis from Henan Cancer Hospital Affiliated Cancer Hospital of Zhengzhou University between July 2023 and February 2024 were enrolled in this study. Clinic data including the age, gender, stage, and histology type were collected from the medical records. Informed consents were obtained from all patients and this study was carried out in compliance with the Declaration of Helsinki and approved by the ethics committee of Henan Cancer Hospital Affiliated Cancer Hospital of Zhengzhou University (2023-KY-0065).
Specimen collection
The sampling procedures was performed by an experienced oncologist with the help of a radiologist. Histologic specimens of all patients were obtained by PABL. Of the 184 patients, MCSs were obtained by flushing needles used for EBUS-TBNA in 38 cases (G1), by flushing needles used for computed tomography (CT)-guided PABL in 108 cases (G2), and by the BAL in 38 cases (G3). Meanwhile, 37 and 35 sufficient cell samples for liquid-based cytology (LBC) were obtained by the EBUS-TBNA in G1 and G3, respectively.
The sampling procedure for the EBUS-TBNA was as follows: the routine bronchoscopy was performed and then BF-UC260F-OL8 (Olympus Ltd, Tokyo, Japan) was brought into contact with the airway wall and moved in all directions to identify the lesions for sampling; after identification of the target lymph node by EBUS, a 21-gauge EBUS-TBNA needle (NA-201SX-4021, Olympus Ltd, Tokyo, Japan) was used to puncture into the lesion, and then moved back and forth within the lesion for sampling. After pushing the tissue from the puncture needle onto the plain glass with a needle core, the sufficient cell sample for LBC was obtained by rinsing the residue of the needle with PreservCyt Solution (ThinPrep PreservCyt Solution, Hologic Inc., MA, USA). The MCSs for G1 were obtained by washing the residue of the needle again in a 5-mL tubes containing 5 mL of PreservCyt Solution.
The sampling procedure for PABL was as follows: the CT scans of the area of interest were performed to locate the puncture site and an 18-gauge end-cut full core biopsy needle (CORCA1820SB, Merit Medical Systems Inc., Utah, USA) was used to obtain tissue specimens for pathological diagnosis. After placing the tissue specimens in saline, the MCSs for G2 were obtained by rinsing the needle several times with cell storage solution.
The sampling procedure for BAL was as follows: the flexible bronchoscope was “wedged” into the target bronchial segment, and 10–20 mL of saline was injected into the alveolar cavity via the bronchoscope, followed by negative pressure suction to recover perfusion. The recollected fluids were used as the MCSs for G3.
Histological and cytological tests
The tissue sample obtained by PABL was coated on the slide, and the visible tissue was put into a vial containing 10% formalin for examination. After treatment, the tissue was embedded in paraffin, sliced into 5-µm-thick sections, stained with hematoxylin and eosin (HE), and observed under a light microscope (CX31, Olympus Corporation).
All the liquid samples were sent for cytological analysis, with sufficient cell samples examined by LBC and MCSs examined by traditional smear cytology (TSC). The procedure for LBC was as follows: the sample was centrifuged at a radius of 10 cm at 1500 r/min for 5 min and the supernatant was discarded; 25 ml of cleaning fluid was added and then oscillated prior to centrifugation at 1500 r/min for 5 min; the supernatant was discarded again, and the sediment was transferred into a Thinprep liquid then oscillated and mixed. After 15 min, an ultrathin cell smear was made by a TCT microcomputer processing system, fixed with 95% ethanol for 15 min, stained with HE, sealed, and observed under the microscope. The MCSs were centrifuged (3,000 rpm) and included in blocks, followed by HE staining and microscopic examination. Positive samples were defined by the presence of malignant cells on HE staining. The diagnoses of biopsy tissue samples and corresponding MCSs were conducted separately by experienced pathologists, respectively.
Genetic testing
DNA/RNA extraction and genetic testing were performed by an experienced medical technologist blinded to the pathological diagnoses. Total DNA/RNA was isolated from the tissue samples and MCSs using QIAamp DNA Micro Kit (QIAGEN, Germany) and RNeasy Micro Kit (QIAGEN, Germany) according to the manufacture’s protocol. Nucleic acids were quantified using the Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and NanoDrop2000 (Thermo Fisher Scientific, Wilmington, DE, USA). The NanoDrop measured the single strand of DNA, and the Qubit measured the double strand of DNA. qPCR was performed on an Mx3000P PCR instrument (Agilent, Santa Clara, CA, USA) according to the instructions of Lung Cancer 11Gene Mutations Detection Kit (ACCB Biotech Ltd., Beijing, China). The assay detected lung cancer-related genes including epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1), proto-oncogene tyrosine-protein kinase receptor Ret (RET), hepatocyte growth factor receptor (MET), neurotrophic receptor tyrosine kinase 1 (NTRK1), NTRK2, NTRK3, human epidermal growth factor receptor 2 (HER2), kirsten rat sarcoma viral (KRAS), b-raf proto-oncogene (BRAF). The genetic variants tested in the assay included mutations in the EGFR, HER2, MET, KRAS and BRAF genes, as well as fusions in the ALK, ROS1, RET, NTRK1, NTRK2, and NTRK3 genes. The specific genetic testing information is displayed in supplementary Tables 1–2. For cases of inconsistent results of genetic testing between tissue samples and MCSs, we resequenced these samples, and the accuracy of the genetic test results was ultimately assessed by the effectiveness of the clinical medications. In all group, samples were diagnosed as positive when the results of either TSC and genetic testing were determined to be positive.
Statistical analysis
Statistical analysis was performed using R package (version 4.1.2). Quantitative data was compared by t-test, Mann-Whitney U test, or Kruskal-Wallis test. Categorical variables were evaluated using the Fisher’s exact test followed by Bonferroni correction. All tests were two-tailed, P < 0.05 was considered to be significantly different. Graphs were made in Prism 8, v8.2.0 (GraphPad Software Inc.) and Adobe Illustrator 2021 (Adobe Inc.).
Results
Baseline characteristics
A total of 184 patients with lung cancer were enrolled in this study. Patients’ ages at diagnosis ranged from 27 to 90 years with a median age of 64 years. The majority of patients were male (62.0%) and diagnosed at an advanced stage of IV (68.6%). 170 cases were diagnosed as lung cancer by pathological and clinical diagnosis, including adenocarcinoma (ADC) in 100 (54.3%) cases, squamous cell carcinoma (SCC) in 30 (16.3%) cases, adenosquamous carcinoma (ASC) in 2 (1.1%) cases, small cell lung cancer (SCLC) in 12 (6.5%) cases, neuroendocrine neoplasms (NEN) in 4 (2.2%) and unable to divide cancer cell types in 22 (12.0%) cases. A summary of the clinical characteristics of 184 patients was presented in Table 1. No significant difference in age (P = 0.496), gender (P = 0.439), and stage (P = 0.479) was present between G1, G2 and G3, while there was statistically significant difference in histology types between these groups (P = 0.034).
The diagnostic value of traditional smear cytology based on different micro cell samples
The results of the diagnostic performance of TSC based on different MCSs and the comparison of the diagnostic performance between TSC and LBC were displayed in Table 2. Of 184 patients, 32, 104 and 34 patients were diagnosed with lung cancer by pathological examination and clinical diagnosis in G1, G2 and G3, respectively. The accuracy/sensitivity/specificity of LBC in G1 and G3 for diagnosing lung cancer were 73%/68.8%/100% and 80%/78.1/100%, respectively. The diagnostic concordance between LBC and pathological examination in G1 (k = 0.37) and G3 (k = 0.38) was fair. The sensitivities/specificity for TSC based on MCSs in G1, G2 and G3 were 75%/100%, 93.3/100%, and 73.5%/100%, respectively, to identify a patient with lung cancer. The discrimination of lung cancer patients from patients with non-tumor lung pathology in G1, G2 and G3 reached an accuracy of 78.9%, 93.5%, and 76.3%, respectively. The diagnostic agreement between TSC and biopsy was moderate in G1 (k = 0.49) and G2 (k = 0.51), and fair in G3 (k = 0.37). With pathological and clinical diagnosis as the diagnostic gold standard, no significant difference in diagnostic accuracy was found between TSC and LBC in G1 (P = 0.597) and G3 (P = 0.782).
The diagnostic value of traditional smear cytology complemented with genetic testing based on different micro cell samples
Next, we explored the diagnostic value of TSC complemented with genetic testing based on different MCSs. The sensitivity/specificity of TSC combined with qPCR assays targeting 11-genes in distinguishing malignant tumors from benign lesions in G1, G2 and G3 was 78.1%/100%, 98.1%/100%, and 82.4%/100%, respectively. The AUC values of the diagnostic procedure of TCS combined with genetic testing were higher than that of the diagnostic procedure of TSC alone (G1, 0.875 vs. 0.891; G2, 0.966 vs. 0.990; G3, 0.868 vs. 0.911) (Fig. 2A-C), suggesting that the diagnostic performance of TSC in combination with genetic testing for lung cancer was dramatically improved compared to TSC alone. The TAT of our diagnostic method mostly required only 24 h in all groups, whereas the median TAT of standard diagnostic procedures needed 5 d (range 1–14), 3 d (range 1–11), and 5 d (range 1–12) in G1, G2 and G3, respectively. The TAT of our diagnostic method was significantly lower than that of standard diagnostic procedures (G1, P < 0.001; G2, P < 0.001; G3, P < 0.001, Fig. 2D-F).
Diagnostic performance and turn-around-time of TSC combined with genetic testing based on micro cell samples for lung cancer. With pathological and clinical diagnosis as the diagnostic gold standard, the diagnostic value of TSC combined with genetic testing in G1 (A), G2 (B) and G3 (C) was estimated by receiver operating characteristic (ROC) curve analysis. Comparison of turn-around-time between standard diagnostic procedures and our novel diagnostic procedures. Comparisons of round-around-time between our diagnostic method and standard diagnostic procedures in G1 (D), G2 (E) and G3 (F).
Moreover, we investigated the diagnostic value of TSC complemented with genetic testing based on different MCSs in different histology types of lung cancer. G1 exhibited higher diagnostic sensitivity in ADC (94.4%) patients, and lower diagnostic sensitivity in SCC (40%), SCLC (75%) and undefined (75%) patients. G2 demonstrated excellent diagnostic performance in all histological subtypes of lung cancer, and, especially, had a diagnostic sensitivity of 100% for patients with ADC, ASC, SCLC and NEN. G3 exhibited higher diagnostic sensitivity in SCLC (92.9%) and ADC (100%) patients, and lower diagnostic sensitivity in SCC (66.7%) and undefined (66.7%) patients (Table 3).
Comparison of genetic testing of different micro cell samples and the paired tissue samples
Finally, we compared the distribution of the quantity of DNA extracted from MCSs and paired tissue samples, and the concordance of gene variations in MCSs with paired tissue samples. In G1, G2 and G3, a total of 14, 50 and 10 MCSs and paired tissue samples had available DNA concentrations measured by Nanodrop and Qubit. In G1, the concentration of DNA extracted from MCSs was significantly higher than that extracted from paired tissue samples, regardless of the method of DNA concentration measurement (Nanodrop, P = 0.036; Qubit, P = 0.001, Fig. 3A-B). In contrast, in G3, there was no significant difference between the concentration of DNA extracted from MCSs and that extracted from paired tissue samples, regardless of the DNA concentration measurement method used (Nanodrop, P = 0.964; Qubit, 0.137, Fig. 3E-F). In G2, the concentration of DNA extracted from MCSs was significantly higher than that extracted from paired tissue samples when DNA concentration was measured using the Qubit (P < 0.001 Fig. 3D), whereas there was no significant difference between the two types of samples when measured using the Nanodrop (P = 0.063, Fig. 3C).
In addition, a total of 17, 61 and 11 paired MCSs and tissue samples in G1, G2 and G3, respectively, underwent the detection of 11 genes by qPCR. The concordance of detectable variations between MCSs and paired tissue samples was 100% in all groups. In G1, 11 genetic aberrations were identified in 52.9% (9/17) of cases. The mutation rates of EGFR L858R, EGFR 19del, EGFR T790M, BRAF V600E and KRAS G12C were 29.41%, 11.76%, 11.76%, 5.88% and 5.88% (Fig. 4A). In G2, a total of 41 genetic aberrations were detected in 60.7% (37/61) of cases, among which the most frequently genetic aberrations was EGFR L858R (22.95%), followed by EGFR 19del (13.11%), EGFR T790M (8.20%), ALK fusion (4.92%), EGFR S768I (4.92%), EGFR G719 × (3.28%), KRAS G12C (3.28%), BRAF V600E (1.64%), EGFR ins20 (1.64%), KRAS G13C (1.64%), and RET fusion (1.64%) (Fig. 4B). In G3, 6 genetic variations including EGFR 19del, EGFR T790M, BRAF V600E, KRAS G12X/G13X, ALK fusion, and ROS1 fusion were detected in 45.5% (5/11) of case (Fig. 4C).
Discussion
Lung cancer is the deadliest cancer worldwide. Timely and accurate diagnosis is critical to improving patient prognosis16. In our study, we innovatively integrate TSC with genetic tests based on MCSs for the diagnosis of lung cancer. This approach holds the potential to improve patient outcomes by enabling timely diagnosis and treatment.
Tissue samples are the gold standard for pathological diagnosis, and the methods commonly used for tissue sampling are PABL, EBUS-TNBA, etc17. In the past, the needles used in aspiration biopsies were usually discarded after the procedure was finished. Of note, a few recent studies have found that the puncture needle rinse solution have the potential to detect malignant tumors and their associated genetic mutations11,12,18. However, data regarding the diagnostic value of the needle rinse solution in the lung cancer is limited. Our study found that the TSC of needle rinse solution obtained by flushing the needle used for EBUS-TBNA and CT guided PABL correctly recognized 75% and 93.3% of the lung cancer, respectively, suggesting that needle rinse solution may be valuable diagnostic material in the lung cancer. For patients in poor physical condition and in advanced stages who are not eligible for invasive sampling procedure. BALF has emerged as an alternative liquid biopsy source19. Previous studies have found that the sensitivity of BALF cytology alone was modest, ranging from 28% − 87%, but the specificity is exceptionally high, ranging from 90% − 100%7,20. In our study, the sensitivity and the specificity of BALF for diagnosing lung cancer were 73.5% and 100%, respectively, which was in line with previous findings. Of note, the various groups exhibited inconsistent diagnostic performance, which may be attributed to differences in sampling methods. Specifically, the sample volume obtained by PABL is generally larger than that obtained by EBUS-TBNA and BAL21,22,23. Meanwhile, PABL is primarily performed for diagnosing peripheral lung cancers, mainly lung adenocarcinoma, which has a relatively higher incidence of variation in the 11 genes detected in our study24. These reasons may explain why G2 displayed the highest diagnostic yield compared to the other two groups.
Since the complex nature of lung cancer can lead to misdiagnosis, repeat diagnosis is necessary for accurate diagnosis25. In our lab, LBC of EBUS-TBNA cytology samples is commonly used as a complementary diagnostic tool. LBC is a novel method of cytology production that has emerged in recent decades. It can eliminate the effect of blood and mucous fluid, making the background and the cell structures clearer26,27. However, our study found the diagnostic yield of the LBC method was similar to that of the TSC method in EBUS-TBNA samples of G1, with no significant difference (P = 0.597), which was accordance with the previous studies report by Hou et al.28 and Gauchotte et al.29. This result suggested that the LBC method may not be superior to the TSC method for lung diagnosis, which may be caused by the inherent drawbacks of the LBC method, which sometimes loses background material such as necrosis, mucus, etc., creating a diagnostic dilemma in confirming the malignant nature of the lesion, and thus resulting in a false-negative diagnosis.
Molecular testing plays an important role in the diagnosis and targeted therapy of lung cancer, and has become a standard of clinical practice30,31. Our study showed that combining TSC and genetic testing, increased the diagnostic yield versus TSC alone, suggesting that genetic testing had the potential to improve the diagnostic yield of lung cancer. Moreover, the detection of EGFR, BRAF and MET mutations as well as the analysis of ALK, ROS1, RET and NTRK translocations have already been incorporated in the lung cancer diagnostic standards, and the inhibitors of these kinases are in routine clinical use32. Our genetic testing panel incorporated those common targetable oncogenic driver alterations, ensuring that clinicians can make timely and precise decisions for treatment strategies33,34. Specifically, EGFR was the most frequently detected mutation in our cohort. For patients with EGFR L858R and EGFR exon 19 deletion, first-generation EGFR-TKIs (e.g. gefitinib and erlotinib) and second generation EGFR-TKIs (e.g. afatinib and dacomitinib) are recommended as targeted therapy options. And, for patients harboring EGFR T790M mutations, third-generation EGFR-TKIs (e.g. osimertinib) represent an effective therapeutic strategy35. In our study, the concordance rate between MCSs- and tissue-based genetic testing in G1, G2 and G3 was 100%, and the TAT of the genetic assessment only required 24 h, which suggested that MCSs-based genetic testing via qPCR assay is a rapid and efficient method for assessing genetic variation in the lung patients. Notably, Pisapia et al.36 found that next-generation sequencing (NGS) can identify more actionable EGFR mutations compared to RT-qPCR, thereby offering more patients opportunities for targeted EGFR therapy. This finding implied that NGS may offer more comprehensive molecular mutation profiling than PCR-based methods. With the advancement of NGS technology, the TAT of NGS can be shortened to 3 days by choosing the appropriate platforms and panels. Therefore, the widespread application of NGS in the clinical management of lung cancer will be our future research direction. In addition, higher DNA concentrations were observed in MCSs of G1 and G2 than in paired tissue samples, highlighting the puncture needle rinse as a rich source of tumor cell DNA. Tissue-based gene mutation testing involves the paraffin fixation, which is not only time-consuming but also limited by DNA fragmentation, chemical cross-linking and artificial mutations introduced by formalin fixation37. In addition, the concentration of DNA in the MCSs was lower than in the paired tissue samples in G3, suggesting that the BALF contained fewer tumor cells. Our data demonstrated that routinely discarded puncture needle rinses are an adequate source of material for molecular diagnostic testing.
Moreover, previous studies reported medians diagnosis-to-treatment time ranging from 6 to 45 days, and improved survival when the waiting time was shorter38,39. In our institution, the TAT of routine diagnostic procedure is approximately 7 days, whereas the TAT of the TSC combined with genetic testing by qPCR assays based on MCSs in this study took only 24 h. The novel diagnostic pathway we explored dramatically decrease the TAT for laboratory reports, potentially helping to accelerate treatment initiation and improve patient prognosis. Moreover, the new diagnostic pathway has comparable costs to existing methods during disease diagnosis. However, it is worth noting that our novel method significantly shortens the diagnostic TAT, which may shorten the patient’s hospital stay and consequently decrease the overall expenses throughout the entire treatment cycle. In terms of clinical applicability, our novel diagnostic method offers the following advantages: (1) demonstrating strong diagnostic accuracy for lung cancer; (2) addressing the clinical challenge of sample inadequacy; (3) significantly reducing diagnostic TAT; and (4) showing potential to reduce total patient cost. Collectively, these strengths ensure the applicability of this approach in routine clinical workflows.
There are some limitations in our study. To begin with, the size of our study is relatively small. Second, as different sampling methods are applied to different locations of lung cancer, this could lead to a potential bias in patient selection in our study. Third, our study lacked the exploration of the diagnostic performance of non-invasive methods (e.g., sputum). Finally, the gene panel used in this study did not include clinically relevant P53 genes because of the lack of methodological validation of PCR-based P53 mutation detection. We look forward to larger cohorts or more data that can be accumulated at our center in the future to verify the generalizability of our study. Moreover, growing evidence suggests that other approaches (e.g. long non-coding RNAs and reinforcement learning) are being used to improve the diagnosis and treatment of lung cancer40,41,42, which is also a potential direction for our future research.
Collectively, we demonstrated that the novel diagnostic pathway, i.e. MCSs-based cytology and genetic testing was highly reliable in the diagnosis of lung cancer and identification of genetic variations. This diagnostic pathway has the advantage of not requiring sufficient tissue samples and long TAT for analysis, which may benefit patients with advanced lung cancer who do not have access to sufficient tissue samples and urgently need diagnostic results.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Wadowska, K. et al. Genetic markers in lung cancer diagnosis: A review. Int. J. Mol. Sci. 21, 13 (2020).
Nasim, F., Sabath, B. F. & Eapen, G. A. Lung cancer. Med. Clin. North. Am. 103 (3), 463–473 (2019).
Leiro-Fernández, V. et al. Predicting delays in lung cancer diagnosis and staging. Thorac. Cancer. 10 (2), 296–303 (2019).
Shi, C. et al. Implementation of a pathological diagnosis and treatment pathway May improve the molecular detection of lung cancer. Ann. Transl Med. 10 (2), 45 (2022).
Han, Y. & Li, J. Sample types applied for molecular diagnosis of therapeutic management of advanced non-small cell lung cancer in the precision medicine. Clin. Chem. Lab. Med. 55 (12), 1817–1833 (2017).
Griffin, J. P. et al. Diagnosis of lung cancer: a bronchoscopist’s perspective. J. Bronchol. Interv Pulmonol. 19 (1), 12–18 (2012).
Zhang, H. et al. Bronchoalveolar lavage fluid assessment facilitates precision medicine for lung cancer. Cancer Biol. Med. 21 (3), 230–251 (2023).
Nooreldeen, R. & Bach, H. Current and future development in lung cancer diagnosis. Int. J. Mol. Sci. 22 (16), 8661 (2021).
Sardi, A. H. & Islam, S. Early lung cancer detection, mucosal, and alveolar imaging. Curr. Opin. Pulm Med. 22 (3), 271–280 (2016).
Brainard, J. & Farver, C. The diagnosis of non-small cell lung cancer in the molecular era. Mod. Pathol. 32 (Suppl 1), 16–26 (2019).
Boonsarngsuk, V., Saengsri, S. & Santanirand, P. Endobronchial ultrasound-guided transbronchial needle aspiration rinse fluid polymerase chain reaction in the diagnosis of intrathoracic tuberculous lymphadenitis. Infect. Dis. (Lond). 49 (3), 193–199 (2017).
Lan, Z. et al. Utility of liquid-based cytology on residual needle rinses collected from core needle biopsy for lung nodule diagnosis. Cancer Med. 10 (12), 3919–3927 (2021).
Li, T. et al. A rapid liquid biopsy of lung cancer by separation and detection of exfoliated tumor cells from Bronchoalveolar lavage fluid with a dual-layer PERFECT filter system. Theranostics 10 (14), 6517–6529 (2020).
Andreasen, C. B. Bronchoalveolar lavage. Vet. Clin. North. Am. Small Anim. Pract. 33 (1), 69–88 (2003).
Domagala-Kulawik, J. The relevance of Bronchoalveolar lavage fluid analysis for lung cancer patients. Expert Rev. Respir Med. 14 (3), 329–337 (2020).
Kadara, H. et al. Early diagnosis and screening for lung cancer. Cold Spring Harb Perspect. Med. 11 (9): a037994 (2021).
Galvin, J. R. & Franks, T. J. Lung cancer diagnosis: radiologic imaging, histology, and genetics. Radiology 268 (1), 9–11 (2013).
Fuller, M. Y. et al. Next-Generation sequencing identifies gene mutations that are predictive of malignancy in residual needle rinses collected from Fine-Needle aspirations of thyroid nodules. Arch. Pathol. Lab. Med. 142 (2), 178–183 (2018).
Casagrande, G. M. S. et al. Liquid biopsy for lung cancer: Up-to-Date and perspectives for screening programs. Int. J. Mol. Sci. 24 (3), 2505 (2023).
Wongsurakiat, P. et al. Diagnostic value of Bronchoalveolar lavage and postbronchoscopic sputum cytology in peripheral lung cancer. Respirology 3 (2), 131–137 (1998).
Roncarati, R. et al. Molecular testing on bronchial washings for the diagnosis and predictive assessment of lung cancer. Mol. Oncol. 14 (9), 2163–2175 (2020).
Azour, L. et al. Percutaneous Transthoracic Lung Biopsy: Optimizing Yield and Mitigating Risk. J. Comput. Assist. Tomogr 2021 Sep-Oct 01(5), 765–775 (2021).
Medford, A. R. et al. Endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA): applications in chest disease. Respirology 15 (1), 71–79 (2010).
Ding, Y. et al. Comparative study on the mutational profile of adenocarcinoma and squamous cell carcinoma predominant histologic subtypes in Chinese non-small cell lung cancer patients. Thorac. Cancer. 11 (1), 103–112 (2020).
Han, S., Yang, W. & Li, H. A study of the application of fiberoptic bronchoscopy combined with liquid-based cytology test in the early diagnosis of lung cancer. Oncol. Lett. 16 (5), 5807–5812 (2018).
Nguyen-Dang, K. et al. The role and associated factors of Liquid-Based cytology of Bronchoalveolar lavage fluid in lung cancer diagnosis: A prospective study. Cureus 15 (11), e48483 (2023).
Wang, H. et al. Clinical values of different specimen Preparation methods for the diagnosis of lung cancer by EBUS-TBNA. Diagn. Pathol. 19 (1), 61 (2024).
Hou, G. et al. Clinical impact of liquid-based cytology test on diagnostic yields from transbronchial needle aspiration. Respirology 17 (8), 1225–1228 (2012).
Gauchotte, G. et al. A combination of smears and cell block preparations provides high diagnostic accuracy for endobronchial ultrasound-guided transbronchial needle aspiration. Virchows Arch. 461 (5), 505–512 (2012).
Ettinger, D. S. et al. Non-Small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 20 (5), 497–530 (2022).
Dingemans, A. C. et al. Small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up(☆). Ann. Oncol. 32 (7), 839–853 (2021).
Imyanitov, E. N., Iyevleva, A. G. & Levchenko, E. V. Molecular testing and targeted therapy for non-small cell lung cancer: current status and perspectives. Crit. Rev. Oncol. Hematol. 157, 103194 (2021).
Castellanos, E., Feld, E. & Horn, L. Driven by mutations: the predictive value of mutation subtype in EGFR-Mutated Non-Small cell lung cancer. J. Thorac. Oncol. 12 (4), 612–623 (2017).
Hirsch, F. R. et al. Lung cancer: current therapies and new targeted treatments. Lancet 389 (10066), 299–311 (2017).
Su, P. L. et al. Recent advances in therapeutic strategies for non-small cell lung cancer. J. Hematol. Oncol. 18 (1), 35 (2025).
Pisapia, P. et al. The relevance of the reference range for EGFR testing in non-small cell lung cancer patients. Lung Cancer. 198, 108002 (2024).
Sugimoto, A. et al. A Large-Scale prospective concordance study of Plasma- and Tissue-Based Next-Generation targeted sequencing for advanced Non-Small cell lung cancer (LC-SCRUM-Liquid). Clin. Cancer Res. 29 (8), 1506–1514 (2023).
Jacobsen, M. M. et al. Timeliness of access to lung cancer diagnosis and treatment: A scoping literature review. Lung Cancer. 112, 156–164 (2017).
Jiang, T. et al. The changing diagnostic pathway for lung cancer patients in shanghai, China. Eur. J. Cancer. 84, 168–172 (2017).
Ghahramani Almanghadim, H. et al. New insights into the importance of long Non-Coding RNAs in lung cancer: future clinical approaches. DNA Cell Biol. 40 (12), 1476–1494 (2021).
Ghorbian, M. & Ghorbian, S. Comprehensive review of reinforcement learning in lung cancer diagnosis and treatment: taxonomy, challenges and recommendations. Comput. Biol. Med. 183, 109326 (2024).
Khosravi, N. et al. Sarcoidosis and Non-small cell lung cancer: expression of miR-145, miR-301, and miR-449. Biomedical Biotechnol. Res. J. (BBRJ). 8 (4), 517–523 (2024).
Acknowledgements
We thank Shanghai Rightongene Biotechnology Co., Ltd. (Shanghai, China) for analysis of the data.
Funding
This study was supported by the grants from Major Science and Technology Project of Henan Provincial (No.221100310100) and Technological and scientific projects in Henan Province (No.232102311026).
Author information
Authors and Affiliations
Contributions
Ke Yang: Conceptualization, Investigation, Data curation, Writing-original draft; Jiuzhou Zhao: Investigation, Data curation, Writing-original draft; Yanping Hu: Investigation, Writing-review & editing; Pengfei Ren: Data curation, Writing-review & editing; Xinxin Wu, Dongqing Wang: Validation, Visualization; Dongxu Chen, Juan Yu, Jun Zhang, Zhizhong Wang, Rui Sun, Chengzhi Zhao: Methodology, Validation; Zhenzhen Liu, Jie Ma, Yongjun Guo; Bing Wei: Conceptualization, Writing-review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
The study was carried out based on the Declaration of Helsinki and approved by the Ethics Committee of Henan Cancer Hospital Affiliated Cancer Hospital of Zhengzhou University (2023-KY-0065).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, K., Zhao, J., Hu, Y. et al. Innovative diagnostic approaches for lung cancer: integrating traditional cytology with qPCR for rapid and reliable results. Sci Rep 15, 29085 (2025). https://doi.org/10.1038/s41598-025-13743-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13743-4