Abstract
To characterize the hrHPV genotype distribution and attribution to cervical lesions in the cohort of women screened in Shanxi Province. Data from the records of cervical cancer screening programs for rural women in Shanxi Province were retrospectively collected. Women who underwent primary human papillomavirus (HPV) screening between January 2014 and December 2019 were included. The attribution proportion of specific hrHPV types for different grades of cervical lesions was calculated by using the type contribution weighting and the proportional attribution methods to estimate the potential impact of HPV vaccines on CIN II + lesions. CIN II + lesions were observed mainly with HPV16 (65.85%), HPV18 (10.20%), HPV58 (10.20%), HPV52 (7.98%), HPV31 (4.66%), HPV33 (4.43%), HPV51 (2.22%) and HPV56 (2.22%). A total of 97.42% of all CIN II + lesions were attributed to HPV16, HPV18, HPV52, HPV58, HPV31, HPV33 and HPV35. A total of 75.4% (95% CI, 71.4–79.3) of CIN II + lesions were attributable to HPV16/18, and 21.1% (95% CI, 17.4–25.1) were attributable to the 5 additional types (HPV31/33/45/52/58) covered by the 9-valent vaccine. The prevalence of hrHPV infection among women in Shanxi Province was high, and HPV16, HPV18, HPV58, HPV52, HPV31, HPV33 and HPV35 had the highest attributable proportion of CIN II + lesions. The type-specific HPV prevalence and attribution proportion of cervical precancerous lesions should be taken into consideration in both clinical management and the design of preventive strategies.
Similar content being viewed by others
Introduction
Cervical cancer is a major problem because of its high incidence and mortality in low-income and middle-income settings1. The cervical cancer incidence in countries and areas with a high or very high human development index was 11.3 per 100,000 women, which is markedly lower than that in those countries and areas with a low or medium human development index (18.8 per 100,000 women)2. This urban-rural gap is more pronounced in Shanxi Province, where mountainous terrain and fragmented healthcare access contribute to 3.7× higher late-stage diagnosis rates3. In 2009, the government-led cervical cancer screening pilot project was launched in 221 rural counties in China and national rollout female populations aged 35 to 64 in 20194. Currently, only 36.8% of women aged 35 to 64 have undergone cervical cancer screening5. The rural screening program is still constrained by limited healthcare facilities, cultural barriers, low HPV vaccine acceptance rates, and knowledge gaps. Recent studies suggest that if HPV screening and HPV vaccination programs expand to 80%−100% coverage over the next 50 years, successful elimination of cervical cancer will be possible by the end of the century6. Existing rural HPV epidemiology systems exhibit two evidence-based limitations: (1) Population-based genotype distribution datasets are absent in resource-limited regions, particularly for non-vaccine-targeted hrHPV types. (2) Prevailing vaccine efficacy models fail to incorporate rural-specific determinants, including fragmented healthcare access and culturally rooted vaccine hesitancy. Building on gaps above, three unresolved problems demand attention: Screening Interval Uncertainty: No evidence-based protocols exist to optimize resource-efficient screening intervals for populations with limited healthcare access. Vaccine-Reality Mismatch: Efficacy of current vaccines against regionally circulating recombinant HPV lineages remains unverified; (Cross-cutting) Algorithm Inflexibility: WHO risk-prediction algorithms lack adaptability to settings with < 5% cytology screening coverage. To overcome these interconnected gaps, our multi-year study aims to characterize the hrHPV genotype distribution and attribution to cervical lesions in the cohort of women screened in Shanxi Province.
Methods
Study population and ethics approval
Data from the records of cervical cancer screening programs for rural women in Shanxi Province were retrospectively collected. According to the “Urban Rural Classification Standards” (2020 edition) of the National Bureau of Statistics of China, the “rural areas” defined in this study refer to: administrative villages and natural village gathering areas; population density < 500 people/square kilometer; main industry is agriculture (accounting for > 60%)7. The research area covers 28 townships in 10 prefecture level cities in Shanxi Province. High-risk human papillomavirus (hrHPV) was defined as the 15 genotypes with established carcinogenicity (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68). Non-vaccine-targeted hrHPV genotypes were those not covered by locally available vaccines during the study period (2014–2019), which included the bivalent (16/18) and quadrivalent (6/11/16/18) formulations. Thus, HPV31/33/45/52/58 were classified as non-vaccine-targeted genotypes, despite inclusion in later 9-valent vaccines, due to the latter’s absence in the study region’s immunization programs until 2019.”
Inclusion Criteria: (1) Women aged 35–64 who participated in the rural cervical cancer screening project in Shanxi Province from 2014 to 2019;(2) Complete HPV genotyping for 15 high-risk types of human papillomavirus (hrHPV). Exclusion criteria: (1) Specimens without hrHPV genotyping (N = 10,590);(2) Incomplete genotyping results (N = 493); (3) Loss to follow-up (N = 6). The 2014–2019 timeframe was strategically selected because: (1) To capture complete pre-vaccine baseline data (9-valent HPV vaccine introduced in 2019);(2) To capture Coronavirus Disease 2019 (COVID-19) pandemic data distortions. This study was approved by the Ethics Committee of Shanxi Maternal and Child Health Hospital (approval number: IRB-KYYN-2021-001(5)). All data were anonymized and analyzed under the ethical exemption for retrospective studies, as informed consent had been obtained from all participants during the original data collection phase of the Shanxi Province Cervical Cancer Screening Program (2014–2019). We confirm that all methods were performed according to the relevant guidelines and regulations. We followed the Strengthening the Reporting of Observational Studies for Epidemiology (STROBE) guidelines for cross-sectional studies8.
Screening procedures
Basically, the screening procedure was performed according to the released American Colposcopy and Cervical Pathology Association interim (ASCCP) guidelines9. The data were collected through the Shanxi Maternal and Child Health Information Platform, including: (1) Exposure variable: HPV genotyping results; (2) Outcome variables: CIN II + diagnosed by histopathology (dual-pathologist blinded); (3) Covariates: age (from census records), education level (last educational record), screening history. The participants underwent hrHPV genotyping as the primary test, and those who tested positive were further triaged by cytology and/or colposcopy. The management procedures for hrHPV-positive women were as follows: ① An immediate colposcopy referral was recommended for women with clinically suspicious cervical cancer or HPV16/18 infection. ② Other hrHPV-positive and HPV16/18-positive participants underwent cervical cytology or visual inspection with acetic acid and lugol’s lodine (VIA/VILI) If cervical cytology showed atypical squamous cells of undetermined significance (ASC-US), the patients underwent colposcopy. Those suspected by colposcopy or cervical smear underwent cervical biopsy and were referred for pathological examination. Pathological diagnostic reports of cervical precancerous lesions were based on the traditional classification, i.e., cervical intraepithelial neoplasia (CIN) I, CIN II, and CIN III, or a dichotomy, i.e., low-grade squamous intraepithelial lesions (LSIL) and high-grade squamous intraepithelial lesions (HSIL).
Statistical analyses
The detection rate of hrHPV was defined as a comparison of the number of positive cases of hrHPV to the total number of hrHPV cases that had complete genotyping results (six women were excluded because they received no further testing and dropped out from the screening program). To calculate the detection rate of hrHPV type-specific, GraphPad Prism 8.0.1.244 (GraphPad Software, San Diego, CA, USA) was used. To analyze subtype specific carcinogenic risk, the “ adjusted OR” with 95% confidence intervals (CIs) related to individual hrHPV types in the subcohort of patients with a single infection were analysed using logistic regression analysis. SPSS 22.0 (IBM, Armonk, NY, USA) was used to analyse the data.
The calculations of attributable proportions of lesions caused by specific hrHPV types have been described previously10,11. Regarding the Attribution proportion Formula: for example, there are 6 single-type HPV16 cases and 4 single-type HPV18 for a CIN II case, the derivation of the attributable proportion of each genotype for the CIN II lesions positive for HPV16 and 18 in a study is as follows: 6/(6 + 4) = 0.6 of the multi-type infected lesions would be attributed to HPV16, and 4/(6 + 4) = 0.4 would be attributed to HPV18. We calculated genotype-specific attributable proportions (AP) using the principle of proportional attribution. For cases with single-type hrHPV infections, the causative agent was fully attributed to the detected genotype. In multi-type infections (e.g., HPV16/18 co-infection in CINII + lesions), we assigned proportional attribution based on population-level single-type prevalence: Formulation: APₖ= P(k)/∑P(i). Where: APₖ= Proportion attributed to genotype; P(k) = Prevalence of single-type infections caused by genotyp, ∑P(i) = Summed prevalence of all detected genotypes in co-infection. Example application (HPV16/18 co-infection): If population data show: 6 CINII + cases with HPV16 single-infection, 4 CINII + cases with HPV18 single-infection. Then for HPV16/18 co-infected lesions: AP₁₆= 6/(6 + 4) = 0.60 (60% attributed to HPV16), AP₁₈= 4/(6 + 4) = 0.40 (40% attributed to HPV18).
Multiple hrHPV infections were defined as testing positive for two or more different types of hrHPV. To evaluate the contribution proportion of each genotype for individuals with single or multiple infections, the standard was set as the proportion of hrHPV genotypes that caused a single infection in the population with the same histological grade12. To estimate the potential protective effects of the 9-valent HPV vaccine on CIN II + lesions, hrHPV classification data were divided into three categories: HPV16/18, HPV31/33/45/52/58, and other high-risk types (HPV35/39/51//53/56/59/66/68). The 95% confidence interval (CI) was determined using the Wilson scoring method. When describing the sociodemographic characteristics of the participants, we used three age groups: 35–40 years, 41–50 years and 51–64 years. We classified educational level as junior high school and below, high school and technical secondary school or above. Whether the participants had a history of cervical cancer screening was also evaluated. To assess the association between hrHPV type and demographic characteristics, we used the Pearson X22test. A p value < 0.05 was considered statistically significant. we performed sensitivity analyses under a worst-case scenario, in the complete HPV genotyping population, where all 6 excluded cases (due to loss to follow-up) were assumed to have developed CIN II + lesions. The primary analysis yielded a CIN II + incidence of 9.99% (451/4,516) in the study cohort. After incorporating the excluded cases with maximum risk inflation (absolute risk overestimation: 0.12%), the adjusted incidence remained minimally changed at 10.11% (457/4,522). This marginal difference demonstrates that the selection bias due to exclusion had non-significant change on our core findings. Selection Bias Assessment: We performed inverse probability weighting (IPW) analysis to assess potential selection bias. Participation probabilities were estimated using logistic regression with age, region, and educational level as predictors. Weighted analyses were conducted to evaluate the robustness of our findings.
Results
Descriptive analysis
Screening results of the study population
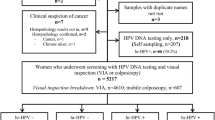
In total, 111,353 women underwent HPV primary screening between January 2014 and December 2019 according to the registry records. Among them, 15,605 (1,5605/111,353, 14.01%) participants were hrHPV positive, including 4,522 (4,522/1,5605, 28.97%) women who had complete genotyping results. Six women were excluded because they received no further testing and dropped out from the screening program. Finally, 4,516 (4,516/1,5605, 28.94%) patients with a median age of 47.89 years (range, 47.67–48.11 years) were included in the present study, and 1,431 (1,431/4,516, 31.69%) of them underwent a cervical biopsy. According to the pathological examination results, 451 (451/4,516, 9.99%) women had CIN II + lesions, including four with CIN II, three with CIN III, 403 (403/4,516, 8.92%) with HSIL, four with adenocarcinoma in situ (AIS), and thirty-seven with cervical cancer; 396 (396/4,516, 8.77%) women had CIN I or LSIL (Fig. 1).
HPV genotype distribution in cervical lesions
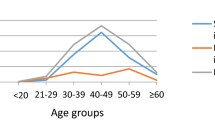
Of the 4,516 women who were positive for hrHPV revealed by complete genotyping tests, 4,071 (4,071/4,516, 90.15%) had single infections, and 445 (445/4,516, 9.85%) had multiple infections. The proportions of multiple infections in women with no detected lesions, CIN I or LSIL, and CIN II + were 9.27% (340/3,669), 13.38% (53/396), and 11.53% (52/451), respectively. In the entire cohort, HPV16 27.81% (1,256/4,516), HPV52 16.54% (747/4,516), HPV58 12.11% (547/4,516), HPV18 8.79% (397/4,516), and HPV53 6.36% (287/4,516), were the most common genotypes. The distribution pattern differed between different subgroups. Notably, HPV16 was the only genotype that showed an upward trend in infection prevalence from negative lesions 20.99% (770/3,669) to CIN I or LSIL 47.73% (189/396), then to CIN II + 65.85% (297/451), while the other hrHPV types only showed modest differences between groups or even the lowest prevalence in the CIN II + subgroup. In the CIN II + subgroup (N = 451), HPV16 was the predominant type, with a positive rate of 65.85%(297/451), followed by HPV18 10.20%(46/451), HPV58 10.20%(46/451), HPV52 7.98%(36/451) and HPV31 4.66%(21/451). These prevalent HPV subtypes were predominantly identified as single infections in CIN II + patients, while less prevalent subtypes, such as HPV51, HPV53, HPV68, HPV59, HPV45, and HPV39, were mainly found in women with multiple infections (Fig. 2).
Predictive modeling
Subtype-specific carcinogenic risk quantification
To estimate the hrHPV type-specific risk of CIN II+, we analysed the adjust OR related to individual hrHPV types in the subcohort of patients with single infections (N = 4,071). There were 399 (399/4071, 9.80%) CIN II + cases in this subcohort. HPV39, HPV45, and HPV68 were not included because they did not cause any single infections. Among the 12 hrHPV subtypes analysed, as expected, HPV16 (crude OR = 70.59, 95% CI: 9.85-505.76; aOR = 70.58, 95% CI: 9.85-505.72), and HPV18 (crude OR = 29.69, 95% CI: 4.05-217.88; aOR = 29.32, 95% CI: 3.99-215.22) showed the highest risks for CIN II+, followed by HPV31 (crude OR = 18.20, 95% CI: 2.32-142.52; aOR = 18.46, 95% CI 2.36-144.57), HPV33 (crude OR = 17.09, 95% CI: 2.20-132.76; aOR = 17.31, 95% CI 2.23-134.52), and HPV58 (crude OR = 15.00, 95% CI: 2.03-110.83; aOR = 15.22, 95% CI 2.06-112.45). In contrast, HPV51 (crude OR = 2.85, 95% CI: 0.26–31.68; aOR = 2.79, 95% CI 0.25–31.06), HPV59 (crude OR = 2.27, 95% CI: 0.14–36.70; aOR = 2.27, 95% CI 0.14–36.68) were ranked as the three types with the lowest risks (Fig. 3).
The attributable proportions of different grades of cervical lesions in HrHPV
The prevalence of lesions attributed to different hrHPV types was low when weighting multi-infection lesions. A total of 46.94% of CIN I cases and 65.44% of CIN II + cases were attributed to HPV16. In total, HPV16, HPV18, HPV52, and HPV58 combined caused 77.70% of all CIN I lesions. A total of 97.42% of all CIN II + lesions were attributed to HPV16, HPV18, HPV52, HPV58, HPV31, HPV33 and HPV35 combined (Table 1).
Using the proportional attribution method to estimate the potential impact of the 9-valent HPV vaccine on CIN II + lesions in this study 75.4% of CIN II + lesions were attributable to HPV16/18 21.1% to the 5 additional types (HPV31/33/45/52/58) covered by the candidate 9-valent vaccine, and 3.5% of CIN II + lesions were attributable to hrHPV types not covered by the 9-valent vaccine (Fig. 4).
Sensitivity assessment
HPV16/18 was responsible for the largest percentage of CIN II + lesions across all age groups (Table 2). Among 35-40-year-olds, HPV16/18 attribution was (OR = 81.6, 95% CI 72.5–88.7). The attribution was lowest (OR = 72.2, 95% CI 65.5–78.2) in the age group of 41-50-year-olds. Conversely, the proportion of CIN II + lesions attributable to HPV31/33/45/52/58 was notably greater among women aged 41–50 years in comparison to younger age groups. There was no significant difference in the distribution of HPV16/18 or HPV31/33/45/52/58 types based on educational level among women with CIN II+. In individuals without a history of cervical cancer screening, HPV16/18 was responsible for the majority of CIN II + lesions. Nonetheless, the proportion of CIN II + lesions attributable to HPV31/33/45/52/58 and other high-risk types was significantly higher among women with a history of cervical cancer screening than among those without. Participation Rate Analysis: Among 111,353 women screened, 4,516 (4.06%) underwent complete HPV genotyping. Six women were excluded because they received no further testing and dropped out from the screening program. Regional participation rates varied significantly, ranging from 0.96% in Jincheng to 17.13% in Lvliang (p < 0.001). Baseline Characteristics Comparison: Women who received complete genotyping were significantly older (mean age 47.9 vs. 46.9 years, p < 0.001), prevalence increases significantly with age (peak in 51–64 age group), higher education (high school+) correlates with higher hrHPV rates(Table S1).
Discussion
Key findings
This study described the distribution of hrHPV types among different grades of cervical lesions and their attribution proportions among women in Shanxi Province, China. In our study, CIN II + lesions occurred mainly with HPV16 (65.85%), HPV18 (10.20%), HPV58 (10.20%), HPV52 (7.98%), HPV31 (4.66%), HPV33 (4.43%), HPV51 (2.22%) and HPV56 (2.22%). HPV58, HPV52, HPV31, and HPV33 accounted for 27.27% of CIN II + lesions. Consistent with previous studies13,14,15HPV16, HPV52, HPV58, HPV33, HPV31 and HPV18 were the main genotypes in HSIL + patients. A European study16 analysed the difference in the prevalence of HPV types between HSIL and invasive cervical cancer, which showed that the most common types of HPV in women with HSIL were HPV16/33/31 and those in women with cervical cancer were HPV16/18/45. In our study, the two with the highest rate of CIN II + lesions were also HPV16 and HPV18. however, HPV45 demonstrated minimal involvement. This dual pattern highlights the necessity for locally adapted cervical cancer screening and vaccination strategies.
Risk stratification insights
The risk of progression and disease contribution varied by individual HPV genotype17. To exclude the influence of the varied prevalence of specific HPV genotypes, we estimated the hrHPV type-specific risk of CIN II+. Our findings revealed that HPV16, HPV18, HPV31, HPV33, HPV58, HPV35, HPV52, HPV56, and HPV66 often carried a high immediate risk for CIN II+ (≥ 4%). This is consistent with previous research18 that reported the risk ratio for individuals at high risk of progressing to CIN III; seven HPV types (HPV16, HPV18, HPV31, HPV33, HPV35, HPV52 and HPV58) showed a high risk of progression. The absolute metrics highlight both individual-level risk magnitude and population-level prevention priorities.
Considering the incidence and carcinogenic potential of different genotypes, we used attribution to explore the genotype distribution of hrHPV. The prevalence of lesions attributed to different hrHPV types was low when weighting multi-infection lesions. A global19 study on HPV genotypes and hrHPV DNA-based screening tests and protocols focused on HPV16, HPV18, and HPV45. In our study, the two with the highest attribution rate of CIN II + lesions were also HPV16 and HPV18. However, HPV52, HPV58, HPV31, HPV33 and HPV35 were often detected more frequently than HPV45 in CIN II + lesions. Consistent with previous conclusions20HPV16, HPV52, HPV58, HPV31 and HPV33 were more common in women with cervical lesions in eastern China. A study21 showed that adding HPV35 to the vaccine can prevent a small subset of CIN III and SCC, with a greater potential impact on CIN III + in black women. Similarly, based on our cross-sectional study, HPV35 may deserve special attention in addition to HPV16, HPV18, HPV52, HPV58, HPV31 and HPV33.
Vaccine implementation implications
The distribution of specific HPV genotypes in the general population and in patients with cervical lesions is critical to developing precise CC prevention strategies10,22. In a 20-year nationwide study23the incidence of severe cervical precancerous lesions (CIN III and AIS) as well as cervical cancer (squamous cell types) decreased after the implementation of the national multicohort HPV vaccination program in Denmark. In our data, we analysed the potential impact on CIN II + lesions from candidates for the 9-valent HPV vaccine. HPV16/18 accounted for 75.4% of CIN II + lesions. The additional 5 HPV types in the 9-valent vaccine, HPV31/33/45/52/58, contributed to 21.1% of CIN II + cases. Similar to a global study24the 9-valent HPV vaccine can prevent most CIN II + cases in Shanxi Province. The non-negligible attributable fraction of HPV35 underscores potential benefits from expanded valency vaccines.
Strengths and limitations
This study addresses critical knowledge gaps in cervical cancer prevention by (1) Establishing the first population-based HPV genotype distribution for rural Shanxi, China, where cervical cancer incidence significantly higher than exceeds national averages; (2) Quantifying the carcinogenic progression risks for understudied HPV types by estimating the hrHPV type-specific risk of CIN II+; (3) Formulating evidence-based recommendations for vaccine formulation optimization, particularly highlighting the necessity to include HPV35 in next-generation vaccines. These findings provide essential epidemiological evidence to recalibrate prevention strategies in resource-limited regions. However, this study has some limitations. The data were acquired from an official records system, and the original data cannot be backtracked. Single-province sampling constrain the extrapolation to other rural populations. In our study, only 4% of the population underwent full genotyping, and more prospective studies with larger sample sizes are needed to determine the specific hrHPV genotypes that cause CIN II+. In the work of Baay M.F.D2513% of cancers were HPV negative. The analysis of the incidence of CIN II + attributed to individual HPV types was novel, but the contribution of hrHPV may have been overestimated. These limitations are partially addressed by our large sample size (N = 111,353) and standardized pathology review. Future multicenter studies that incorporate viral load quantification could further refine risk prediction models. Our study’s 4.06% genotyping rate raises concerns about external validity. Quantitative bias analysis revealed that original genotype prevalence estimates may be overestimated by 1.5% due to selection toward higher-risk women. However, sensitivity analyses using IPW showed that core findings regarding HPV16/18 dominance remained robust (aOR difference < 10%). Rural-urban generalizability is limited, as our sample predominantly represents rural women with healthcare access.
Conclusions
In summary, HPV16, HPV18, HPV52, and HPV58 were the dominant high-risk genotypes in this population and accounted for 89.71% of all CIN II + lesions, with HPV31/33/35 contributing an additional 7.71%. The 9-valent vaccine covers 96.21% of CIN II + causes in rural Shanxi, but the 1.21% protection gap attributable to HPV35 requires revision of existing vaccine standards. This study provides three paradigm shifts for cervical cancer prevention and control in rural areas of developing countries: (1): Shifting from “high-risk pan screening” to “regional adaptive subtyping testing”; (2): Shifting from “unified review interval” to “type-dependent dynamic monitoring”; (3): Transitioning from the “gold standard of histology” to “molecular subtyping guided predictive medicine”.
Data availability
The datasets generated during the current study are not yet publicly available due to privacy concerns and ongoing additional research. Data can be made available for peer review upon reasonable request by contacting the corresponding author.
Abbreviations
- CIN II+:
-
Cervical intraepithelial neoplasia grade II and above
- HPV:
-
Human papillomavirus
- hrHPV:
-
High-risk human papillomavirus
- ACS:
-
American Cancer Society
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- ASCCP:
-
American Society for Colposcopy and Cervical Pathology
- ASCP:
-
American Society for Clinical Pathology
- ICC:
-
Invasive cervical carcinoma
- AIS:
-
Adenocarcinoma in situ
- VIA/VILI:
-
visual inspection with acetic acid and lugol’s lodine
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 68 (6), 394–424 (2018).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249 (2021).
Xing, N. Z. W. J. & Zhang, Y. Z. Shanxi Provincial Cancer Registry Annual Report. 2022. (2021).
Di, J., Rutherford, S. & Chu, C. Review of the cervical cancer burden and Population-Based cervical cancer screening in China. Asian Pac. J. Cancer Prev. 16 (17), 7401–7407 (2015).
Zhang, M. et al. Cervical cancer screening Coverage - China, 2018–2019. China CDC Wkly. 4 (48), 1077–1082 (2022).
Simms, K. T. et al. Impact of scaled up human papillomavirus vaccination and cervical screening and the potential for global elimination of cervical cancer in 181 countries, 2020–99: a modelling study. Lancet Oncol. 20 (3), 394–407 (2019).
China NBoSo. Statistical classification criteria for urban and rural areas. (2020).
Vandenbroucke, J. P. et al. Strengthening the reporting of observational studies in epidemiology (STROBE): explanation and elaboration. Int. J. Surg. 12 (12), 1500–1524 (2014).
Huh, W. K. et al. Use of primary High-Risk human papillomavirus testing for cervical cancer screening. J. Lower Genit. Tract Dis. 19 (2), 91–96 (2015).
Zhao XL, Hu SY, Zhang Q, Dong L, Feng RM, Han R, Zhao FH. High-risk human papillomavirus genotype distribution and attribution to cervical cancer and precancerous lesions in a rural Chinese population. J Gynecol Oncol. 28(4), e30 (2017).
Saraiya M, Unger ER, Thompson TD, Lynch CF, Hernandez BY, Lyu CW, Steinau M, Watson M, Wilkinson EJ, Hopenhayn C, Copeland G, Cozen W, Peters ES, Huang Y, Saber MS, Altekruse S, Goodman MT; HPV Typing of Cancers Workgroup. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst. 107(6), djv086 (2015).
Julious, S. A. Two-sided confidence intervals for the single proportion: comparison of seven methods by Robert G. Newcombe,Statistics in Medicine. ;17:857–872. Statistics in Medicine. 2005;24(21):3383–3384. (1998).
Nygård, M. et al. Human papillomavirus genotype-specific risks for cervical intraepithelial lesions. Hum. Vaccines Immunotherapeutics. 17 (4), 972–981 (2020).
Sundström, K. & Dillner, J. How many human papillomavirus types do we need to screen for?? J. Infect. Dis. 223 (9), 1510–1511 (2021).
Song F, Yan P, Huang X, Wang C, Du H, Qu X, Wu R. Roles of extended human papillomavirus genotyping and multiple infections in early detection of cervical precancer and cancer and HPV vaccination. BMC Cancer. 22(1), 42 (2022).
Tjalma, W. A. et al. Differences in human papillomavirus type distribution in high-grade cervical intraepithelial neoplasia and invasive cervical cancer in Europe. Int. J. Cancer. 132 (4), 854–867 (2012).
Sakamoto, J. et al. Single type infection of human papillomavirus as a cause for high-grade cervical intraepithelial neoplasia and invasive cancer in Japan. Papillomavirus Res. 6, 46–51 (2018).
So KA, Lee IH, Lee KH, Hong SR, Kim YJ, Seo HH, Kim TJ. Human papillomavirus genotype-specific risk in cervical carcinogenesis. J Gynecol Oncol. 30(4), e52 (2019).
De Sanjose, S. et al. Human Papillomavirus Genotype Attribution in Invasive Cervical Cancer: a Retrospective cross-sectional Worldwide Study (LANCET ONCOLOGY., 2010).
Zhang L, Bi Q, Deng H, Xu J, Chen J, Zhang M, Mu X. Human papillomavirus infections among women with cervical lesions and cervical cancer in Eastern China: genotype-specific prevalence and attribution. BMC Infect Dis. 17(1), 107 (2017).
Mix, J. et al. Cervical precancers and cancers attributed to HPV types by race and ethnicity: implications for vaccination, screening, and management. J. Natl. Cancer Inst. 114 (6), 845–853 (2022).
Molina-Pineda, A. et al. High frequency of HPV genotypes 59, 66, 52, 51, 39 and 56 in women from Western Mexico. BMC Infect. Dis. 20 (1), 889 (2020).
Ring, L. L. et al. Incidence of cervical precancerous lesions and cervical cancer in Denmark from 2000 to 2019: population impact of multi-cohort vaccination against human papillomavirus infection. Int. J. Cancer. 152 (7), 1320–1327 (2023).
Serrano, B. et al. Potential impact of a nine-valent vaccine in human papillomavirus related cervical disease. Infect. Agent Cancer. 7 (1), 38 (2012).
Baay, M. F. et al. Human papillomavirus infection in the female population of antwerp, belgium: prevalence in healthy women, women with premalignant lesions and cervical cancer. Eur. J. Gynaecol. Oncol. 22 (3), 204–208 (2001).
Funding
The National Natural Science Foundation of China (82072891). The Project of the Central Government Guiding Local in Shanxi Province (YDZJSX2022B012). Basic Research Program of Shanxi Province (202103021224352). Shanxi Maternal and Child Health Hospital (2021009).
Author information
Authors and Affiliations
Contributions
RS and JC conceived and designed the study, having full access to all of the data in the study and taking responsibility for the content of the manuscript. RS and ZHW analyzed the data, took responsibility for the accuracy of the data analysis and wrote the first draft of the manuscript. ZHW and MZ reviewed the paper. RS, WJQ, JC, ZHW, and MZ contributed to the interpretation of the data and clinical inputs. All authors were involved in the revision of the manuscript for important intellectual content and approved the final version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, R., Qi, W., Wang, Z. et al. High-risk human papillomavirus genotype distribution and attribution to cervical lesions in a Shanxi Province screening population. Sci Rep 15, 28217 (2025). https://doi.org/10.1038/s41598-025-14228-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-14228-0