Abstract
In survival analysis, models typically assess the impact of a single Time-Varying Exposure (TVE), where the exposure status can change over time. However, situations with multiple TVEs frequently arise, and adequate statistical handling remains an area of active research. To apply multiple time-varying approaches and to compare estimates derived from different models using an application with real-world data in the paediatric field. The Italian national paediatric database Pedianet was used to identify children aged between six months and 14 years at the beginning of the epidemiological season (October 1, 2017, to May 31, 2018). Influenza vaccine administrations and antibiotic prescriptions were modeled using both time-fixed and time-varying approaches. Cox proportional-hazard models with random intercept for the region of residence to address the association between antibiotic use, influenza vaccination, and the onset of influenza/influenza-like illness (ILI). Estimates for influenza vaccination remained relatively stable across the different modeling approaches, likely due to the relatively short length of the potential misspecification window. In contrast, estimates for antibiotic use varied significantly between the different scenarios, highlighting the need for careful evaluation and selection of the most appropriate statistical handling approach. It is of utmost importance to carefully evaluate the characteristics of each exposure included in the analysis. Statistical tools and techniques tailored for multiple TVEs need to be acknowledged.
Similar content being viewed by others
Introduction
The time-dependent exposures in observational cohort studies pose a substantial analytical challenge. Incorrectly, handling these exposures can introduce immortal time bias, leading to distorted and potentially misleading estimates of treatment effects1,2. Typically, while investigating a specific exposure, individuals are considered at risk from the beginning of the study. However, with time-dependent exposures, individuals only enter the at-risk period when the exposure begins, causing a shift in exposure status over time3. Considering patients who develop exposure during the follow-up as at-risk from study inception is methodologically incorrect. For example, consider a study examining whether statin use reduces the risk of a heart attack. In a time-fixed exposure model, individuals are classified as statin users if they were ever exposed to the medication at any point during follow-up, regardless of the timing of exposure. This means that a person is considered a user even if they started statins late in the follow-up period. In contrast, a time-varying exposure model updates each patient’s exposure status over time, allowing for a more accurate representation of the individual’s true exposure status. If a patient initiates statin therapy during follow-up, their status changes from non-user to user at the time of initiation, better capturing the dynamic nature of medication use and its temporal relationship with the outcome. These individuals, by definition, will survive until the exposure status changes, leading to the allocation of person-time to incorrect exposure groups and subsequent biased analysis results4. To prevent immortal time bias, it is essential to account for the time-varying nature of exposures employing a counting process framework5. This approach accurately reflects dynamic changes in exposure status over time, ensuring that individuals contribute person-time to the appropriate exposure category. While existing literature largely addresses the analysis of single TVE, with statistical software offering corresponding tools, there remains a gap in research focusing on clinical contexts involving multiple concurrent TVEs.
This study aimed to investigate appropriate statistical approaches for analyzing data with multiple TVEs, using a real-world pediatric dataset as an example. Specifically, we compared the results of a biased model, which treated both influenza vaccination and antibiotic treatment (i.e., exposures of interest) as time-fixed variables, with those of several other models that incorporated one or both exposures as time-varying. The outcome of interest was the incidence of influenza/influenza-like illness.
Methods
Study population and setting
The study cohort included Italian children aged 6 months to 14 years as of October 1, 2017, and who were followed throughout the 2017–2018 influenza season, from October 1, 2017, to May 31, 2018. The data were obtained from Pedianet (https://pedianet.it), a population-based Italian database that compiles patient-level information from family pediatricians participating in the Pedianet network. These data, collected during routine medical care, are anonymized in accordance with Italian privacy laws by assigning a unique numeric code to each patient record. The anonymized information is securely transmitted on a monthly basis to a central database in Padua, where it undergoes validation. Participation in the Pedianet system is entirely voluntary, and informed consent is obtained from parents or legal guardians to allow the storage and use of their child’s anonymized data for research purposes.
Only children followed by one of the family paediatricians from the Pedianet network adhering to the influenza vaccination program (i.e., who provided influenza vaccination to their patients during the influenza season in agreement with the National Health System) were included in the study, similarly to what was done in a previous study6. Study participants were considered at risk from October 1, 2017. Follow-up continued until the onset of influenza/ILI (i.e., the outcome of interest), the end of the study (i.e., May 31, 2018) or until participants dropped out due to unrelated reasons, whichever came first.
We adhered to the STROBE guidance for reporting on observational studies7.
This study adhered to the principles outlined in the Declaration of Helsinki and followed the methodological standards recommended by the European Network of Centres for Pharmacoepidemiology and Pharmacovigilance (ENCePP). Approval for the study and database access was granted by the Internal Scientific Committee of Società Servizi Telematici Srl, the legal entity responsible for Pedianet. In accordance with Italian legislation, retrospective, observational, and non-interventional studies are exempt from the requirement for ethics committee approval.
Definition of exposures
Vaccination status was defined based on the electronic health record from the Pedianet database and assumed to provide protection throughout the influenza season, consistent with evidence of influenza vaccine effectiveness8. Children under 6 months were excluded due to age-related ineligibility for influenza vaccination. Antibiotic exposure was identified using the prescription registry within the Pedianet database. Five exposure scenarios were considered to model antibiotic exposure: one assuming full-season coverage, and three others assuming coverage for five, seven, or ten days after each prescription. These timeframes reflect treatment durations commonly recommended by national and European pediatric guidelines for acute respiratory tract infections. Such infections, including pneumonia, acute otitis media, and bacterial pharyngitis, are typically treated with antibiotics for five to ten days. While these durations represent simplified assumptions and do not account for individual pharmacodynamics, they provide clinically grounded estimates for defining exposure windows. Our use of multiple windows also allowed us to assess the sensitivity of estimates to different definitions of exposure9,10,11,12,13,14.
Definition of outcome
The outcome of interest was influenza/influenza-like illness (ILI), identified using a machine-learning algorithm applied to electronic health records from the Pedianet database. The algorithm was trained to detect clinical diagnoses of influenza/ILI based on pediatricians’ ICD codes, clinical notes, and visit patterns. To ensure accurate case detection, a custom-developed and validated Natural Language Processing (NLP) algorithm trained on a comprehensive, gold-standard labeled dataset was employed. This algorithm demonstrated excellent performance, with an accuracy of approximately 99%15. Laboratory-confirmed influenza was not systematically available, as testing was only performed and reported at the discretion of the family pediatrician. Therefore, the outcome reflects clinically diagnosed influenza/influenza-like illness (ILI), rather than laboratory-confirmed influenza.
Statistical analysis
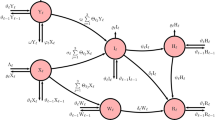
As a first step, the main characteristics of the study cohort were presented and summarized. Categorical variables are reported with numbers and proportions, while continuous variables are reported with median and quartiles. Cox regression mixed effects models with random intercept for the region of residence were fitted to estimate the hazard ratios (HRs) and 95% confidence intervals (CIs) for influenza/ILI associated with influenza vaccination and antibiotic use. Both time-fixed and time-dependent effects of these exposures were evaluated. When exposures were treated as time-fixed variables, the proportional hazards assumption was assessed using Schoenfeld residuals (see Appendix—Models Summaries section). Models were adjusted for age, sex, deprivation index16, comorbidity, and the number of visits and antibiotic prescriptions in the previous year (except for those under one year of age), as proxies for the clinical condition. Children were classified as having comorbidity if they had at least one disease-based healthcare exemption for chronic complex condition (i.e., cystic fibrosis, diabetes, chronic obstructive pulmonary disease, and asthma), immunodeficiency or immunosuppressive therapy, neurological and neurocognitive conditions (including Trisomy 21), prematurity (less than 37 weeks of gestation), renal failure, congenital cardiac disease (including heart failure), or chronic liver conditions15. The number of visits and antibiotic prescriptions were categorized as < 8 or ≥ 8, and as 0, 1–2, or ≥ 3, respectively, following the classification used in a previous study based on the same data source15. Bootstrap resampling was conducted to assess model stability (see Appendix, Model Fit and Stability sections). Six different model specifications were compared to assess the optimal approach (Fig. 1). Specifically:
Model 1: vaccine and antibiotic prescriptions as time-fixed
Model 2: vaccine as time-varying exposure (TV subscript), antibiotics as time-fixed
Model 3: vaccine and antibiotics as time-varying exposures (TV subscript)
Model 4: vaccine and antibiotics as time-varying exposures, coverage of the antibiotics prescribed ends after 10 days (TV10 subscript)
Model 5: vaccine and antibiotics as time-varying exposures, coverage of the antibiotics prescribed ends after 7 days (TV7 subscript)
Model 6: vaccine and antibiotics as time-varying exposures, coverage of the antibiotics prescribed ends after 5 days (TV5 subscript)
A time-fixed baseline model approach was adopted in Model 1 (Fig. 1.A), considering both exposures as time-fixed. This implied that all children receiving vaccination, antibiotics or both were considered exposed from the beginning of the follow-up. Model 2 (Fig. 1.B) evaluated influenza vaccination as time-varying exposure, utilizing a counting process structure that duplicated observations for vaccinated individuals. Conversely, antibiotic exposure remained time-fixed. Model 3 considered both antibiotics and influenza vaccination as time-varying exposures. This represents an extension of the conventional approach, where typically, only one variable is treated as time-varying, mainly because restructuring the dataset to such a format might be overly complex. This issue is addressed by the mtvc R package (version 1.1.0)17(see Appendix—Multiple Time-Varying Covariates (MTVC) in R—survival package section). This tool accepts one or more time-varying variables, along with their corresponding exposure change dates. It generates a restructured dataset where each individual has a row representing their exact exposures during each time window (Fig. 1.C). In addition, as it might not be appropriate to consider that exposure to antibiotics lasts for several months, three different models were developed (Fig. 1.D), in which each individual who received a prescription for antibiotics would be considered no more exposed after a coverage period of five, seven, or ten days.
The analyses were conducted using R version 4.4 (R Foundation for Statistical Computing, Vienna, Austria), with packages mtvc, survival, coxme, dplyr and forestplot. The code to reproduce this analysis is publicly available on GitHub18.
Results
A total of 56,069 children aged 6 months to 14 years (median age: 7.3 [IQR: 4.4–10.3], Table 1) were identified. Of these, 4,572 (8.2%) received influenza vaccination, and 13,975 (25%) had at least one antibiotic prescription during the follow-up. Most patients resided in Veneto (62%) or Campania (12%). In total, 20,466 had a low deprivation index (37%), whereas 8,396 (15%) did not report their socio-economic status. The remaining 27,207 (49%) had a high deprivation index. The number of patients with at least one comorbidity was 2,434 (4.3%).
Figure 2 summarizes the estimated impact of antibiotic and vaccination exposures on the risk of influenza/ILI for each model. The time-fixed baseline model , which treated both influenza vaccination and antibiotic exposure as time-fixed, estimated HRs of 0.47 (95% CI: 0.38;0.58) for influenza vaccination and 0.57 (95% CI: 0.53;0.62) for antibiotic exposure. These results would suggest a protective effect of both exposures against influenza/ILI infections. A similar estimate (HR = 0.47 [95% CI: 0.38;0.59]) was obtained in the second model, where influenza vaccination was considered time-varying. Indeed, this was explained by the fact that vaccines are dispensed to patients a few months after the study begins, meaning that the amount of person-time misallocated did not significantly affect the estimate. Conversely, the observed association for antibiotics remained unchanged, indicating a consistently protective effect against influenza/ILI. This finding is unexpected, given that antibiotics primarily target bacterial infections, while influenza/ILI is caused by a virus. This unforeseen result may indicate potential misspecification in the initial models, particularly those that treated antibiotic exposure as time-fixed.
In contrast, the third model, which considered both exposures as time-varying, returned a different estimate for the effect of antibiotic treatment, with an HR of 1.18 (95% CI: 1.10; 1.28), indicating a positive association with influenza/ILI.
The last three models, which used different antibiotic coverage protection periods (5, 7, and 10 days), reported an even stronger positive relationship between antibiotic use and influenza/ILI, with increased risks ranging from 1.58 to 2.99, respectively, for 10 to 5 days of coverage.
Discussion
This study investigated the impact of multiple time-varying exposures on clinical outcomes by comparing different statistical approaches, including treating exposures as time-fixed or as single- or multiple-time-varying variables. Our findings underscore the novel importance of accounting for the dynamic interplay of multiple time-varying exposures when conducting cohort studies, highlighting the need for researchers to properly consider the impact of misclassification of exposures.
This study used real-world data as an application to assess the association between antibiotic treatments and influenza vaccination on influenza/ILI infections. The first two models, which treated antibiotic use as a time-fixed variable, paradoxically suggest a protective effect against influenza/ILI. However, this is not reasonable and in contrast with medical knowledge18. In contrast, subsequent models, which more accurately captured the time-varying nature of antibiotic exposure, revealed a positive association between antibiotic use and the risk of influenza/ILI.
Indeed, antibiotic treatment has been previously associated with an increased risk of viral infections19. The relationship may be explained by the disruption of the gut microbiome balance, which plays a critical role in immune system regulation, induced by antibiotic exposure, which has been identified as a predisposing factor to viral infections, including influenza/ILI20. Moreover, antibiotic treatment may be considered as a proxy for acute infections that can transiently impair the immune system, thereby increasing susceptibility to subsequent infections, including influenza/ILI. This hypothesis could account for the positive association observed in this study between antibiotic exposure and influenza/ILI risk when using time-varying models, which are widely recognized as the appropriate methodological choice for such analyses.
As already known, the impact of considering a time-varying exposure as time-fixed may change considerably depending on the nature of that variable. In our application, considering the vaccination as time-fixed does not significantly impact the estimates, meaning that even a more straightforward approach, considering only antibiotics therapy as time-varying, would have returned reasonable estimates. Vaccination was administered to patients only a few weeks after the beginning of the study, and its coverage lasted until the end of the study. Consequently, the impact of misclassification is minimal for this exposure.
On the other hand, this is not true for antibiotics, which could be prescribed at any time and lasted only for a few days. Indeed, classifying such exposure as fixed in time, or for longer periods than actual, would significantly bias the respective HR.
This study shows that the bias generated by wrongly defining the exposure window cannot be underestimated, especially when multiple time-varying exposures are considered. More complex approaches that accurately allocate person-time to the correct exposure categories are necessary for valid conclusions.
When it comes to interpreting our results from a clinical perspective, it is important to note that this study has several limitations. First, antibiotics were not further classified based on their spectrum of activity or diagnosis, leading to a potential misinterpretation of the results, with children presenting acute respiratory infections being at higher risk of developing subsequent influenza/ILI. Furthermore, misclassification bias might be introduced by using electronic health records, with the assumption of full compliance with prescribed antibiotics regimens. Moreover, the decision to model antibiotic exposure for five, seven, or ten days may oversimplify complex biological processes. However, the selected durations are supported by national and European pediatric clinical guidelines and consensus, recommending 5–7 days of treatment for community-acquired pneumonia and up to 10 days for acute otitis and pharyngitis in younger or high-risk children. These evidence-based choices strengthen the rationale for the exposure windows tested in our models and allow for a structured sensitivity analysis of the effect estimates. Additionally, as our outcome was defined using a machine-learning algorithm applied to clinical records, and laboratory-confirmed influenza data were not systematically available, as testing was only performed and reported at the discretion of the family pediatrician, our results refer to clinically diagnosed influenza/ILI. This may have introduced some degree of misclassification, which we acknowledge as a limitation. Lastly, unmeasured confounders, such as healthcare-seeking behaviour or concurrent bacterial infections, could also influence the associations between antibiotic use and influenza/ILI risk.
Conclusions
The overall estimates can be substantially influenced by the statistical approach adopted, particularly when the exposure under investigation is subject to immortal time bias. The impact of misspecification is particularly pronounced for exposures with a dynamic, time-varying nature.
In this study, influenza vaccination was administered a few weeks after the beginning of the study, with coverage lasting until the end of the study, resulting in minimal misclassification impact for this exposure. On the other hand, antibiotic treatment could be prescribed at any time and lasted for only a few days. Misclassifying such exposure as time-fixed or extending the duration beyond its actual periods would have led to substantial bias in the HR estimates.
Data availability
The data supporting this study’s findings are available on request from the corresponding author (AC). The data are not publicly available due to restrictions (containing information that could compromise the privacy of research participants). De-identified data could be shared upon reasonable request to the corresponding author, subject to approval by the Internal Scientific Committee of Società Servizi Telematici Srl, the legal owner of Pedianet.
Abbreviations
- TVE:
-
Time-varying exposure
- ILI:
-
Influenza/influenza-like illness
- HRs:
-
Hazard ratios
- Cis:
-
Confidence intervals
References
Suissa, S. Immortal time bias in pharmacoepidemiology. Am. J. Epidemiol. 167(4), 492–499 (2008).
Targownik, L. E. & Suissa, S. Understanding and avoiding immortal-time bias in gastrointestinal observational research. ACG. 110(12), 1647–1650 (2015).
Dekker, F. W., De Mutsert, R., Van Dijk, P. C., Zoccali, C. & Jager, K. J. Survival analysis: time-dependent effects and time-varying risk factors. Kidney Int. 74(8), 994–997 (2008).
Platt, R. W., Hutcheon, J. A. & Suissa, S. Immortal time bias in epidemiology. Curr. Epidemiol. Rep. 6, 23–27 (2019).
Fleming, T. R. & Harrington, D. P. Counting Processes and Survival Analysis (Wiley, 2013).
Cantarutti, A. et al. Influenza vaccination effectiveness in paediatric “healthy” patients: a population-based study in Italy. Vaccines (Basel). 10, 4 (2022).
Cuschieri, S. The STROBE guidelines. Saudi J. Anaesth. 13(Suppl 1), S31–S34 (2019).
Osterholm, M. T., Kelley, N. S., Sommer, A. & Belongia, E. A. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet. Infect. Dis 12(1), 36–44 (2012).
Donà, D. et al. Treatment of mild to moderate community-acquired pneumonia in previously healthy children: an Italian intersociety consensus (SIPPS-SIP-SITIP-FIMP-SIAIP-SIMRI-FIMMG-SIMG). Ital. J. Pediatr. 50(1), 217 (2024).
Mathur, S., Fuchs, A., Bielicki, J., Van Den Anker, J. & Sharland, M. Antibiotic use for community-acquired pneumonia in neonates and children: WHO evidence review. Paediatr. Int. Child Health. 38(sup1), S66–S75 (2018).
Marchisio, P. et al. Updated guidelines for the management of acute otitis media in children by the italian society of pediatrics: treatment. Pediatr. Infect. Dis. J. 38, 12 (2019).
Castelli Gattinara, G. et al. Antibiotic treatment of acute and recurrent otitis media in children: an Italian intersociety Consensus. Ital. J. Pediatr. 51(1), 50 (2025).
Suzuki, H. G., Dewez, J. E., Nijman, R. G. & Yeung, S. Clinical practice guidelines for acute otitis media in children: a systematic review and appraisal of European national guidelines. BMJ Open 10(5), e035343 (2020).
Chiappini, E. et al. Treatment of acute pharyngitis in children: an Italian intersociety consensus (SIPPS-SIP-SITIP-FIMP-SIAIP-SIMRI-FIMMG). Ital. J. Pediatr. 50(1), 235. https://doi.org/10.1186/s13052-024-01789-5 (2024).
Rigamonti, V. et al. Real-world effectiveness of influenza vaccination in preventing influenza and influenza-like illness in children. Vaccine 53, 126946 (2025).
Batzella, E. et al. The association between pediatric COVID-19 vaccination and socioeconomic position: nested case-control study from the Pedianet Veneto Cohort. JMIR Public Health Surveill. 9, e44234 (2023).
Elia Gonzato LA. Multiple Time varying covariate (2024). https://cran.r-project.org/web/packages/mtvc/index.html.
Elia Gonzato LA. Time varying exposure (2024). https://github.com/egonzato/TimeVaryingExposure.
Sutton, S. S., Magagnoli, J., Cummings, T. & Hardin, J. Association between the use of antibiotics, antivirals, and hospitalizations among patients with laboratory-confirmed influenza. Clin. Infect. Dis. 72(4), 566–573 (2020).
Luo, C. et al. Influenza and the gut microbiota: a hidden therapeutic link. Heliyon 2024, 562 (2024).
Acknowledgements
This work is supported by PNRR 2022-NAZ-0524 — PRIN 2022 under the National Recovery and Resilience Plan (PNRR), Mission 4, Component 2, Investment 1.1 – Call 1409/22: Covid-19 and Acute Respiratory Infections: the Clinical and Epidemiological Changes in the Pediatric Population (the CARICE project); CUP: H53D23007460001. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The authors gratefully acknowledge Professor Francesca Ieva and Dr. Vittorio Torri from the Polytechnic University of Milan for implementing the machine learning algorithm used to identify the outcome of interest (i.e., influenza/influenza-like illness), the head of Pedianet, Dr. Luigi Cantarutti, and the contributions of all the family pediatricians participating in Pedianet: Accardo Fabiana, Alberti Arturo, Alboresi Stefano, Alfieri Eva, Alfiero Bordigato Michela, Alongi Angelo, Amato Denise, Amoroso Biagio, Ancarola Rosaria, Andreola Barbara, Andretta Maria Luisa, Anese Giampaolo, Angelini Roberta, Apostolo Maria Grazia, Arcangeli Bruno, Argo Giovanna, Assirelli Valentina, Avarello Giovanni, Azzoni Lucia, Bacciarini Marta, Balliana Franco, Barbazza Maria Carolina, Barberi Frandanisa Maria, Barbieri Patrizia, Bardella Davide, Barone Roberto, Bellavere Donatella, Belluzzi Gabriele, Benetti Eleonora, Berti Fabio, Bezzi Roberto, Biasci Filippo, Biondi Claudio, Biserni Giovanni Battista, Boe Franca, Bollettini Stefano, Bonaiuto Francesco, Bonfigli Emanuela, Bontempelli Anna Maria, Bonza Matteo, Borsari Gloria, Boselli Lucia, Bozzetto Sara, Britta Rosa, Bruna Andrea, Brusaterra Ivana, Brusoni Guido, Budassi Roberto, Caccini Massimo, Calì Laura, Cammarata Maria Grazia, Camposilvan Sonia, Cantalupi Laura, Caprio Luigia, Carbogno Simone, Cardarelli Chiara, Carli Giovanna, Carnazza Sylvia, Casalboni Rita, Castaldo Massimo, Castelli Stefano, Castronuovo Serenella, Cavedagni Monica, Censini Stefania, Cera Giuseppe Egidio, Chillemi Chiara, Cichello Francesca, Cicione Giuseppe, Ciliani Niccolò, Cimatti Anna Giulia, Ciscato Carla, Clerici Schoeller Mariangela, Cocchiola Samuele, Coletta Maurizio, Collacciani Giuseppe, Comaita Fabrizio, Conte Ugo Alfredo, Conte Valeria, Corchia Matteo, Corrò Roberta, Costagliola Rosaria, Costanzo Nicola, Cozzani Sandra, Cuboni Giancarlo, Curia Giorgia, Curti Valentino, Curto Salvatore, D'Alia Caterina, Dalla Casa Chiara, D'Amanti Vito Francesco, D'Avino Antonio, De Angelis Rita, De Clara Roberto, De Giovanni Lorenzo, De Marchi Annamaria, De Nicolò Emanuele, De Polo Nicoletta, Del Bono Gian Piero, Del Ponte Gigliola, Dell’Antonia Fabio, Di Giampietro Tiziana, Di Mauro Francesco, Di Mauro Giuseppe, Di Palma Salvatore, Di Renzo Anna Paola, Di Santo Giuseppe, Di Saverio Piero, Dieli Mattea, Dolci Marco, Doria Mattia, Drago Stefano, El Mazloum Dania, Elio Giuseppe, Fadda Maria Carmen, Faedi Clara Maria, Falco Pietro, Falcon Elena, Fama Mario, Faraci Marco, Farina Maria Immacolata, Favali Alessio, Favilli Tania, Federico Mariagrazia, Felice Michele, Ferraiuolo Maurizio, Ferrara Enrico, Ferrarese Marta, Ferretti Mauro Gabriele, Ferretti Michele, Forcina Paolo, Foti Patrizia, Frattini Claudio Paolo, Freo Luisa, Frison Ezio, Fusco Fabrizio, Fusco Teresa, Gabutti Alessandra, Gallo Giovanni, Gallo Roberto, Galvagno Andrea, Gentili Alberta, Gentilucci Pierfrancesco, Giacomelli Erica, Giampaolo Giuliana, Giancola Giuseppe, Gianfredi Francesco, Giaretta Letizia, Girotto Silvia, Giuseppin Isabella, Gnesi Laura, Gobbi Costantino, Granzon Renza, Grelloni Mauro, Grugnetti Mirco, Hamarneh Marwan, Isca Antonina, Lagrasta Urania Elisabetta, Lanci Maurizio, Landi Massimo, Lazzari Maura, Letta Maria Rosaria, Levi Della Vida Francesca, Lietti Giuseppe, Ligas Marianna, Lista Cinzia, Lorusso Giuseppe, Lucantonio Ricciardo, Lucchi Elide, Luise Francesco, Luotti Diego, Macropodio Nadia, Malusa Tommaso, Manzali Elisabetta, Marano Enrico, Marine Francesca, Mariniello Lorenzo, Marostica Gabriella, Marzetti Valentina, Masotti Sergio, Mauri Laura, Mazzini Franco, Meneghetti Stefano, Milani Massimo, Milone Stella Vittoria, Minutoli Antonella, Moggia Donatella, Monolo Annalisa, Montagnani Enrico, Monteleone Angela Maria, Morresi Giulia Maria, Mortillaro Angela, Mussinu Pierangela, Muzzolini Carmen, Naccari Anna, Naso Immacolata, Nicoletti Laura, Nicoloso Flavia, Nitsch Monika, Novarini Cristina, Olimpi Laura Maria, Ongaro Riccardo, Palma Maria Maddalena, Pandolfini Vittorio, Pasinato Angela, Passarella Andrea, Pata Davide, Pazzola Pasquale, Perin Monica, Perone Vanessa, Perrera Cristina, Perri Danilo, Perrone Alberina, Pescosolido Silvana Rosa, Petrazzuoli Giovanni, Petrotto Giuseppe, Piazza Vanna, Picco Patrizia, Pinelli Elvira, Pirola Ambrogina, Pisanello Lorena, Pittarello Daniele, Polidoro Eleonora, Ponti Roberto, Porro Elena, Porto Adolfo Francesco, Prandoni Alfiero, Profumo Elisabetta, Puma Antonino, Puocci Maria Paola, Quitadamo Anna Lucia, Raffeiner Ruth, Ragazzon Ferdinando, Reghelin Giulia, Regini Paolo, Righetti Andrea, Rizzari Rosaria, Rizzi Maria Oliva, Rosafio Cristiano, Rosas Paolo, Rosignoli Rino, Rossitto Mariella, Ruffato Bruno, Ruggieri Lucia, Ruscitti Annamaria, Russo Annarita, Salamone Pietro, Sambugaro Daniela, Sanfilippo Francesco Emilio, Saretta Luigi, Sarno Vittoria, Sasso Marcella, Savastano Renato, Savio Valentina, Scarcella Antonio, Sciolla Nico Maria, Semenzato Flavio, Semenzato Rossella, Senesi Paolo, Silenzi Romina, Silvan Carla, Soldà Giorgia, Spanevello Valter, Speciale Sergio Maria.
Funding
This work is supported by PNRR 2022-NAZ-0524 — PRIN 2022 under the National Recovery and Resilience Plan (PNRR), Mission 4, Component 2, Investment 1.1 – Call 1409/22: Covid-19 and Acute Respiratory Infections: the Clinical and Epidemiological Changes in the Pediatric Population (the CARICE project); CUP: H53D23007460001. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Data extraction and management: V.R.; Statistical analysis plan: E.G., L.A., A.C., D.V.; Data analysis: E.G., L.A.; Draft of manuscript and revisions: E.G., L.A., A.C., C.DC. and D.V.. All authors provided approval of the final manuscript and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This is an observational, retrospective, noninterventional study. According to a bylaw on the classification and implementation of observational drug-related research, as issued by the Italian National Drug Agency (an entity belonging to the Italian Ministry of Health), this study does not require approval by an ethics committee in Italy (Italian Drug Agency note on August 3, 2007). This study was conducted in accordance with the tenets of the Declaration of Helsinki and was compliant with the European Network of Centers for Pharmacoepidemiology and Pharmacovigilance’s Guide on Methodological Standards in Pharmacoepidemiology. The Internal Scientific Committee of Società Servizi Telematici Srl, the legal owner of Pedianet approved ethical approval of the study and access to the database.
Consent for publication
This is an observational, retrospective, noninterventional study. According to a bylaw on the classification and implementation of observational drug-related research, as issued by the Italian National Drug Agency (an entity belonging to the Italian Ministry of Health), this study does not require approval by an ethics committee in Italy (Italian Drug Agency note on August 3, 2007). This study was conducted in accordance with the tenets of the Declaration of Helsinki and was compliant with the European Network of Centers for Pharmacoepidemiology and Pharmacovigilance’s Guide on Methodological Standards in Pharmacoepidemiology. The Internal Scientific Committee of Società Servizi Telematici Srl, the legal owner of Pedianet approved the study and access to the database.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gonzato, E., Annicchiarico, L., Cantarutti, A. et al. Handling multiple time-varying exposures in survival analysis using real-world pediatric data from the pedianet database. Sci Rep 15, 30827 (2025). https://doi.org/10.1038/s41598-025-14849-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-14849-5