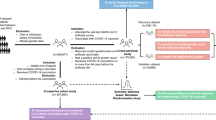
Abstract
The COVID-19 pandemic gave insight into how host genetics influence the ability to get infected and how severe the infection will be if contracted. Among these, the HLA polymorphisms and ABO blood groups are known to influence the immune system. This study examines the correlation between HLA-B genotypes, blood groups, and the severity of COVID-19. A total of 984 COVID-19 patient samples and control samples were collected from different regions of Pakistan. Throat and nasopharyngeal swabs were obtained for molecular detection of SARS-CoV-2, and blood samples were collected for HLA-B genotyping and ABO blood group analysis. Genomic DNA was extracted using the Phenol-Chloroform method, and HLA-B genotyping was performed using the OLERUP SSP HLA typing kit. The study identified a significant association between HLA-B07:02 and increased COVID-19 susceptibility (OR: 2.5, 95% CI: 1.6–3.9, p < 0.001), whereas HLA-B15:01 was associated with a protective effect (OR: 0.6, 95% CI: 0.4–0.9, p = 0.01). Blood group A was also linked to higher infection risk, while blood group O conferred protection. A strong correlation was observed between co-morbidities (hypertension, diabetes) and severe COVID-19 outcomes. This study provides convincing evidence that HLA-B polymorphisms and ABO blood groups play a significant role in COVID-19 susceptibility and severity.
Similar content being viewed by others
Introduction
The COVID-19 pandemic, caused by the novel SARS-CoV-2 virus, has resulted in over 90 million infections and nearly 2 million deaths globally by the beginning of 2021 (https://covid19.who.int/table). The severity and mortality of the infection differ among populations, with a fatality rate of 2.6% in certain regions. The infection’s severity varies from asymptomatic to potentially resulting in fatal acute respiratory syndrome1. South Asian nations, including India, were the second most impacted region globally, recording over 10 million cases and a fatality rate of approximately 1.3%, as reported in January 20212. Other South Asian nations, including Pakistan and Bangladesh, documented cases over 500,000 each, with a fatality rate of approximately 0.4%. The early spread of the virus in this region may have been facilitated by high population density and close interpersonal contact, combined with initial underestimation of airborne transmission routes. While surface contamination and hygiene practices were emphasized early in the pandemic, subsequent evidence indicated that respiratory droplets and aerosols were the primary modes of SARS-CoV-2 transmission3. Moreover, other factors, such as genetic variations, may potentially affect viral transmission and the severity of disease4. Although most individuals experience mild to moderate symptoms, a subset of patients develop severe respiratory distress, multi-organ inflammation, and life-threatening complications. These varying clinical outcomes have prompted investigations into the underlying factors contributing to disease susceptibility and severity5.
Multiple studies have identified host genetic factors as significant contributors to COVID-19 progression and immune response variability. Genes such as ACE2 and TMPRSS2, which are involved in viral entry, and IFNAR2, which plays a role in interferon signaling, have been linked to differing susceptibilities and disease outcomes6,7. However, one of the most extensively studied genetic systems in infectious disease immunology is the human leukocyte antigen (HLA) complex, which modulates antigen presentation and adaptive immune responses. The HLA system, particularly the highly polymorphic HLA class I genes (A, B, and C), is crucial in presenting viral peptides to cytotoxic T cells. Individual variations in HLA alleles can affect the binding affinity for viral antigens, thereby influencing the effectiveness of immune clearance8. A recent study in Italy indicated a heightened prevalence of B*27:07, DRB1*15:01, and DQB1*06:02 among severe COVID-19 patients in a sample of 99 Italians9. However, a study conducted in Sardinia, Italy, revealed an elevated prevalence of the HLA-DRB1*08:01 allele among hospitalized COVID‐19 patients compared to the control group10. A Spanish study demonstrated the significance of HLA‐A*11, C*01, and DQB1*04 concerning infection-related death in 72 severe patients10. Moreover, epidemiological and bioinformatics investigations from several groups have identified distinct HLA alleles as either conferring protection against or predisposing individuals to infection10,11,12. The disparities among the research can be attributed to the study methodology, sample size, and variances in ethnicity. Consequently, examining the correlation between HLA genotypes and COVID-19 across diverse global populations is imperative. In parallel, ABO blood group antigens have also emerged as potential biomarkers for COVID-19 susceptibility. Blood group A has been associated with an elevated risk of infection, while group O appears to confer some level of protection13. These effects may be due to differences in the expression of adhesion molecules or natural antibodies that interfere with viral entry mechanisms. ABO blood group influence has also been noted in previous viral infections, including SARS-CoV and norovirus outbreaks, making it a biologically plausible factor in COVID-19 outcomes14.
The spread and impact of SARS-CoV-2 in South Asia have been significant, with countries such as India, Pakistan, and Bangladesh reporting millions of cases. Differences in population density, comorbidities, genetic makeup, and access to healthcare services may contribute to regional disparities in disease severity15. While earlier in the pandemic surface transmission and hygiene practices were emphasized, subsequent evidence established that airborne transmission through respiratory droplets and aerosols is the dominant route of viral spread16.
Although several international studies have explored the associations between HLA genotypes, blood groups, and COVID-19 outcomes, there remains a scarcity of data from South Asia, particularly Pakistan. To our knowledge, this is the first study to examine HLA-B gene polymorphisms and ABO blood groups in the context of COVID-19 susceptibility and severity in the Pakistani population. This study aims to fill a critical gap by investigating these genetic associations alongside demographic and clinical variables in a multi-ethnic cohort.
Methodology
Ethical approval
This hospital-based case-control study was conducted following ethical approval from the Research Ethics Committee of Kohat University of Science and Technology (Ref. No. KUST/Ethical Committee/976, dated 06-04-2023) and Saeed International Hospital (Biogene Labs and Diagnostics) (Approval Form No. 194). Written informed consent was obtained from all participants before sample and data collection. The study adhered to the Standards of Biomedical Research Ethics recommended by the WHO and other relevant authorities, ensuring the confidentiality and protection of participant identities and related information.
Study population and sample description
Participants were recruited from a pool of individuals presenting at hospitals and testing centers with suspected COVID-19 symptoms (e.g., fever, cough, respiratory distress). All participants underwent RT-PCR testing to confirm their infection status. A total of 984 participants were recruited between August 14, 2020, and January 3, 2021, from various hospitals and testing centers across multiple regions of Pakistan. Of these, 700 were PCR-confirmed COVID-19 patients, and 284 served as SARS-CoV-2 negative controls, verified through RT-PCR testing and absence of COVID-19 symptoms. A statistical power analysis was performed using G*Power software version 3.1.9.4, based on regional infection rates and published HLA allele frequencies from Pakistani population studies. Assuming an expected odds ratio of 2.0, an alpha level of 0.05, and 80% statistical power, the minimum required sample size was estimated to be at least 450 cases and 200 controls to detect significant differences in allele distribution between groups. Given our final sample of 700 cases and 284 controls, the study exceeds this requirement, ensuring sufficient statistical power to support robust subgroup analysis (e.g., by genotype, blood group, age, and sex).
Participants were divided into four groups based on WHO-defined clinical criteria:
-
Healthy controls: Asymptomatic individuals, PCR-negative, no known exposure.
-
Mild: Symptomatic without pneumonia or hypoxia.
-
Moderate: Clinical signs of pneumonia with SpO₂ ≥ 90% on room air.
-
Severe: SpO₂ < 90%, respiratory distress, or requiring ICU admission.
The inclusion criteria were:
-
Age ≥ 18 years.
Consent to participate:
-
For patients: positive SARS-CoV-2 PCR test.
-
For controls: negative PCR test and no COVID-19 symptoms.
Exclusion criteria:
-
Individuals with autoimmune or chronic immunodeficiency disorders.
-
Recent vaccination (within 4 weeks).
-
Incomplete demographic or clinical data.
A breakdown of participants by geographic region and group is provided in Supplementary Table S1.
Normality of data
Normality of continuous variables (e.g., age) was assessed using Shapiro-Wilk tests. Since age distribution did not deviate significantly from normality (p > 0.05), it was summarized using mean ± standard deviation. Other primary variables were categorical and analyzed using non-parametric tests (Chi-square), which do not require normality assumptions.
Sample collection
Approximately 5–10 mL of peripheral blood was collected from each participant into EDTA-coated vacutainer tubes by trained phlebotomists. Nasopharyngeal and oropharyngeal swabs were also obtained from suspected COVID-19 cases for PCR-based confirmation. Following WHO-approved protocols, SARS-CoV-2 infection status was determined using real-time reverse transcription-polymerase chain reaction (RT-PCR). Only individuals with a positive PCR test were included in the patient group, while those with negative PCR results and no symptoms or exposure history were included as controls. Demographic data, clinical history, comorbidities, and symptom onset were recorded using a structured questionnaire following WHO surveillance guidelines.
Extraction of genomic DNA
Genomic DNA was extracted using the Phenol-Chloroform method, following standard protocols. DNA was stored at 4 °C until further analysis. Concentration and purity were assessed using a Nano drop spectrophotometer (Thermo Fisher Scientific). Only samples with an absorbance ratio (260/280) near 1.8 were selected as suitable for HLA genotyping, as this range signifies low protein contamination and ensures dependable DNA integrity for subsequent genetic analyses.
HLA-B genotyping
HLA-B genotyping was conducted using the OLERUP SSP HLA Typing Kit, targeting polymorphic exons of the MHC class I gene locus. Each PCR reaction included positive and negative controls to ensure genotyping accuracy. The PCR products were analyzed via agarose gel electrophoresis, and allele-specific typing was determined using kit software. The HLA-B gene was specifically selected for this study due to its critical immunological function and polymorphic nature, which make it a key determinant in host-pathogen interactions. As part of the HLA class I complex, HLA-B encodes molecules that present endogenous viral peptides to CD8+ cytotoxic T lymphocytes, initiating targeted immune responses. Among HLA class I loci (A, B, and C), HLA-B is the most polymorphic, and has shown strong associations with viral disease progression, including HIV, HBV, and SARS-CoV, making it a compelling candidate for SARS-CoV-2 susceptibility research.
Allele frequency estimation and clustering
Allele frequencies were calculated using the formula:
Frequency = (Number of observed alleles)/(2 × total number of individuals).
Hardy-Weinberg equilibrium (HWE) was tested using the Arlequin v3.5 software, applying the exact test by Guo and Thompson for each allele. To explore genetic structure and allele distribution patterns among different severity groups and controls, Maximum Likelihood (ML) methods implemented in Arlequin were used to assess HLA-B genotype clustering. These analyses were limited to human HLA-B polymorphism data; no viral genomic or clade data were analyzed in this study. The ML-based clustering aimed to identify any allele-based groupings or population stratification that might correlate with COVID-19 severity or susceptibility in the Pakistani population.
Statistical analysis
Statistical analyses were performed using SPSS v25 and R v4.1.0. Categorical variables, including HLA-B genotypes, blood groups, and comorbidities, were evaluated using Chi-square tests to identify COVID-19 susceptibility and severity associations. Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to quantify the strength of these associations. Subgroup-specific effects were explored via stratified analysis by age and sex. A p-value < 0.05 was considered statistically significant. Descriptive statistics (means, standard deviations, percentages) were used to summarize participant characteristics.
Results
A total of 984 participants were included in the study, comprising 700 COVID-19 PCR-positive cases and 284 PCR-negative healthy controls. Participants were recruited from various regions of Pakistan and classified based on WHO criteria into mild, moderate, or severe disease categories. The study cohort consisted of individuals from diverse age groups, ethnic backgrounds, and genders. Basic demographic characteristics are summarized in Table 1.
Ethnicity and COVID-19 status
Participants were stratified based on their ethnic backgrounds. The distribution of COVID-19 status across different ethnic groups (Fig. 1) revealed that certain ethnicities exhibited a higher prevalence of infection.
Co-morbidities and COVID-19 susceptibility
The prevalence of co-morbidities among participants is illustrated in Fig. 2. Hypertension and diabetes were the most frequently reported conditions, with a notable correlation between the presence of co-morbidities and severe COVID-19 outcomes. Their presence correlated significantly with disease severity (Chi-square test, χ² = 36.71, df = 2, p < 0.001).
HLA-B allele distribution in the study cohort
To further understand the genetic landscape of the study population, we analyzed the distribution of HLA-B alleles among all participants. Table 1 shows the frequency of each allele detected. The most prevalent alleles were HLA-B*55 (3.97%), B*15 (3.82%), and B*40 (3.82%), followed by B*44 (3.54%) and B*51 (2.83%). The presence of these alleles in varying proportions underscores the genetic diversity within the Pakistani population and highlights potential polymorphisms associated with COVID-19 susceptibility or resistance. These frequency data provide essential context for the genotype-phenotype associations reported in this study and may serve as a reference for future immune genetic investigations in similar populations (Table 2).
Association of HLA-B genotypes with COVID-19 susceptibility
The analysis of HLA-B genotypes and COVID-19 susceptibility (Fig. 3) revealed significant genetic predispositions. Individuals carrying the HLA-B07:02 allele exhibited a markedly increased susceptibility to COVID-19 (OR: 2.5, 95% CI: 1.6–3.9, p < 0.001), whereas those with the HLA-B15:01 allele had a lower susceptibility (OR: 0.6, 95% CI: 0.4–0.9, p = 0.01). The findings suggest that specific HLA-B variants may influence host immune response mechanisms.
COVID-19 Susceptibility by HLA-B Genotype: This stacked bar chart illustrates the distribution of COVID-19-positive cases across different HLA-B genotypes. The blue bars represent participants with PCR-confirmed COVID-19 infections, while the red bars indicate cases where PCR results were not recorded.
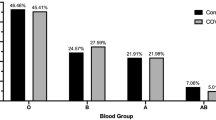
ABO blood groups and COVID-19 susceptibility
The analysis of blood group distribution and its correlation with COVID-19 susceptibility (Fig. 4) revealed that persons with blood group A exhibited a heightened risk of infection. In contrast blood group O was linked to a diminished risk Chi-square = 9.72, df = 3, p = 0.02.
HLA-B genotypes and COVID-19 severity
The direct association between HLA-B genotypes and COVID-19 severity was further examined (Fig. 5). The data indicate that several HLA-B genotypes exhibit distinct distributions across severity levels. The HLA-B14 and HLA-B55 genotypes were notably overrepresented among individuals experiencing severe COVID-19, whereas HLA-B08 and HLA-B15 showed lower prevalence in severe cases. These results suggest a genetic predisposition where specific HLA-B alleles may contribute to increased disease severity, while others confer a potential protective effect. These insights further reinforce the role of host genetic variability in COVID-19 outcomes.
Interaction between HLA-B genotypes and blood groups
The relationship between HLA-B genotypes and blood groups for susceptibility to and severity of COVID-19 was further examined. Figure 6 displays a heatmap depicting the prevalence of various HLA-B genotypes among blood groups. Our findings indicate that specific HLA-B alleles exhibit differential prevalence among blood group subtypes, with certain genotype-blood group combinations correlating with increased disease severity. Notably, HLA-B14 demonstrated a strong association with blood group A + ve and a higher number of COVID-19 cases, whereas HLA-B40 showed a higher prevalence in individuals with blood group B + ve. These results highlight the complex interplay between host genetics and blood group antigens in modulating disease outcomes.
Interaction Between HLA-B Genotypes and Blood Groups: This heatmap visualizes the distribution of HLA-B genotypes across different blood groups, illustrating potential interactions between genetic polymorphisms and blood group antigens with COVID-19 susceptibility and severity. The color intensity represents the number of participants, with red shades indicating higher frequencies and blue shades indicating lower frequencies.
Interaction between HLA-B genotypes and blood groups in COVID-19 severity
The combined effect of HLA-B genotypes and blood groups on COVID-19 severity is presented in Table 3. The data revealed that individuals with the HLA-B07:02 genotype and blood group A had a significantly higher risk of severe disease (OR: 2.5, 95% CI: 1.6–3.9, p < 0.001). Conversely, carriers of HLA-B15:01 with blood group O demonstrated reduced severity (OR: 0.6, 95% CI: 0.4–0.9, p = 0.01). Additionally, HLA-B35:01 carriers with blood group B had an increased risk of severe disease (OR: 2.1, 95% CI: 1.4–3.2, p = 0.001). These findings suggest a potential synergistic effect between HLA polymorphisms and blood group antigens in determining COVID-19 severity.
-
Odds Ratio (OR): measures the association between the combination of HLA-B genotype and blood group, and COVID-19 susceptibility and severity.
-
95% Confidence Interval (CI): range of values within which the true OR is likely to lie.
-
p-value: measures the statistical significance of the association.
-
Susceptibility and Severity: indicates whether the combination of HLA-B genotype and blood group is associated with increased or reduced susceptibility and severity of COVID-19.
This table shows the interactions between specific HLA-B genotypes and blood groups, and their associations with COVID-19 susceptibility and severity.
Stratified analysis by age and sex
A stratified analysis based on age and sex (Table 4) further elucidated the differential impact of HLA-B genotypes on COVID-19 severity. Older adults (≥ 65 years) carrying HLA-B07:02 were at a significantly higher risk of severe disease (OR: 4.2, 95% CI: 2.5–7.1, p < 0.001) compared to younger adults (< 65 years) (OR: 2.1, 95% CI: 1.3–3.4, p = 0.01). Similarly, the protective effect of HLA-B15:01 was more pronounced in females (OR: 0.4, 95% CI: 0.2–0.7, p = 0.01) compared to males (OR: 0.8, 95% CI: 0.5–1.2, p = 0.05). These findings underscore the importance of demographic variables in genetic susceptibility to infectious diseases.
-
Odds Ratio (OR): measures the association between HLA-B genotype and COVID-19 susceptibility and severity in each subgroup.
-
95% Confidence Interval (CI): range of values within which the true OR is likely to lie.
-
p-value: measures the statistical significance of the association.
-
Susceptibility and Severity: indicates whether the HLA-B genotype is associated with increased or reduced susceptibility and severity of COVID-19 in each subgroup.
This table indicates the original results categorized by age and sex, where there seemed to be a difference in the relationship between HLA-B genotype and COVID-19 risks and severities of different subgroups.
Discussion
This study is the first of its kind to investigate the association between HLA-B gene polymorphisms and ABO blood groups with COVID-19 susceptibility and severity in the Pakistani population, a demographically and genetically diverse region that has been underrepresented in global immune genetic research. By integrating genetic, demographic, and clinical data, the study provides novel insights into how specific HLA-B alleles (e.g., B07:02 and B15:01) and blood group antigens interact with host immunity and disease progression. These findings have important implications for personalized risk prediction, targeted intervention strategies, and contribute to the growing global dataset needed for cross-ethnic genomic comparisons. Furthermore, this work establishes a foundation for future population-specific GWAS and HLA-based vaccine response studies in South Asia. The COVID-19 pandemic has shown the significance of host genetic variables in vulnerability, severity, and diversity of immunological response. HLA polymorphisms influence varied disease outcomes, particularly in their capacity to alter antigen presentation and immunological activation17,18. A recent study indicates that ABO blood groups influence the efficacy of viral entry into human bodies19. Medical study indicates that comorbidity such as hypertension, diabetes, and cardiovascular diseases worsen the severity of coronavirus disease20. Our research investigates the association between HLA-B blood group genes and the severity of COVID-19, including patient demographics and medical history. Our integrated genomic and immunological research, including population data, enhances our methodologies for examining virus-host interactions in medical contexts.
Specific HLA-B genotypes are significantly correlated with susceptibility and severity of COVID-19, alongside other genetic factors influencing infectious diseases. As previously established with SARS-CoV and MERS-CoV, we demonstrate a correlation between the HLA-B07:02 alleles and heightened susceptibility to Coronavirus COVID-19 (OR: 2.5, p < 0.001). This is supported by the research that a HLA-B07:02 carrier showed the weaker T cell responses that might result in higher levels of viral loads and acute manifestations of disease21,22. On the other hand, our study identified HLA-B15:01 as a protective allele, conferring a significantly reduced risk of infection (OR: 0.6, p = 0.01). This align with the result reported by Augusto, et al.23, where HLA-B15:01 boosted CD8 + T cell-mediated immune response, leading to faster viral clearance.
Our study also emphasizes the importance of ABO blood groups in determining the susceptibility to SARS-CoV-2 infection; individuals with ABO blood group A have increased infection risk (p < 0.05) as opposed to those with ABO blood group O. The result agrees with a meta-analysis by Liu, et al.24, which reports that blood group A is associated with increased ACE2 receptor expression, facilitating viral entry, while group O subjects have higher levels of anti-A antibodies, possibly offering some protection. The findings of genotype frequency association between HLA-B and blood groups have provided enough evidence for further research to study immunogenetics25.
Looking at the co-morbidities, hypertension and diabetes were identified as the most prevalent co-morbidities in patient conditions, followed by chronic neurological conditions, cardiovascular disease, and renal co-morbidities among others, and the presence of co-morbid conditions made patient outcome severe COVID-19 related (p < 0.001). These findings corroborate with other epidemiological Scenario studies done in the past, including in this study by26, in which Hypertension, diabetes, and cardiovascular disease were the most important and significant risk factors leading to severe Covid disease manifestation. This indicates that apart from the co-morbidities, genetic factors might also play a dual role in determining the risk of developing more severe forms of the disease that require a specific approach to managing these conditions27.
Our results confirm the hypothesis concerning different disease severity depending on the HLA-B allele. In particular, COVID-19 severe patients were more frequently carriers of HLA-B14 and HLA-B55, while HLA-B08 and HLA-B15 were identified in the patients with mild symptoms. Earlier research concerning viral infections has linked the molecule HLA-B14 to compromised antigen presentation and, thus, weak immune response, and severe disease outcome28. As for the HLA-B15, individuals carrying this allele were found to have cross-reactive T-cell immunity, which could explain the protective effect detected in the given population. These results align with Tavasolian, et al.8, who stated that polymorphism of HLA-B affects cytokine storm, which directly affects clinical features. Studies across diverse populations have highlighted the role of HLA-DRB1 alleles in modulating susceptibility and severity of COVID-19. For instance, HLA-DRB1*01 has been associated with increased susceptibility in a large cohort from 28 Mexican states29. Similarly, DRB1*08 was linked to higher infection risk in Saudi Arabia, while DRB1*09:01 in Japan and DRB1*15:01 in Italy showed comparable associations with disease susceptibility. In contrast, DRB1*04 has been implicated in severe disease outcomes in the Iranian population, and DRB1*08 has been associated with elevated mortality rates in Italian patients. Conversely, DRB1*01:01 and DRB1*04:01 confer a protective effect against COVID-19 in individuals from Northeast England, and similar protective associations with DRB1*04 were reported in the Saudi population30.
The other notable discovery identified by the current study is the genotyping of HLA-B alleles and blood groups on the severity of COVID-19. Patients with HLA-B07:02 and blood factor A possessed the highest OR, 2.5, at a significance level < 0.001, while HLA-B15:01 and blood factor O had an OR, 0.6 at a significance level of 0.01. Thus, genetic predisposition and ABO blood group antigens may have a combined effect that may be associated with immune response effectiveness and inflammatory processes. This supports the research by13, where they concluded that genetic variations near the ABO genes and HLA areas are related to COVID-19 severity.
The stratification results confirmed the significant differences of HLA-B genotypes in response to age and sex. The severe disease in the current study was significantly higher in older adults (≥ 65 years) who were HLA-B07:02 carriers compared to younger adults (< 65 years), with an OR of 4.2, p < 0.001 for older and 2.1, p = 0.01 for younger adults. This emphasis arises because genetic susceptibility is an age-related factor and has relationships with immune senescence and chronic inflammation. In the same way, HLA-B15:01 had a fairly higher protective effect on females; OR: 0.4; p = 0.01 and less on males; OR: 0.8; p = 0.05, suggesting that male sex hormones might interact with immune mediators. This supports other studies that have also noted that estrogen helps to boost antiviral immune response and that testosterone has immune depressive properties on human leukocytes31,32.
Clinically, these findings have significant implications for personalized medicine and risk stratification. Understanding an individual’s HLA-B genotype and blood group profile could assist healthcare providers in predicting disease progression, prioritizing high-risk patients for early intervention, and informing vaccine response expectations. From a public health perspective, identifying genetically vulnerable subgroups could enable more targeted resource allocation and preventive strategies, particularly in resource-constrained settings like Pakistan.
Conclusion
This study provides convincing evidence of the genetic and immunological factors influencing COVID-19 susceptibility and severity. Our findings indicate that HLA-B07:02 is associated with increased susceptibility, whereas HLA-B15:01 exhibits a protective effect, reinforcing the role of host genetic variation in disease outcomes. The notable correlation between ABO blood groups and COVID-19 risk, especially the increased vulnerability in those with blood group A and the comparative protection in blood group O, reinforces the immune genetic foundation of disease susceptibility. Additionally, the strong correlation between comorbidities such as hypertension and diabetes with severe COVID-19 outcomes highlights the need for targeted clinical interventions in high-risk populations. The demographic analysis highlighted that older individuals and males exhibit a greater risk of severe disease, suggesting that genetic predisposition, in conjunction with demographic and clinical factors, plays a crucial role in determining COVID-19 outcomes. Despite these insights, the study’s scope was limited by the sample size, the focus on a single gene locus (HLA-B), and the absence of longitudinal follow-up to assess long-term outcomes. These findings lay the groundwork for future large-scale, multi-locus immune genetic studies in diverse populations. Expanding this work to include other HLA loci, cytokine gene interactions, and vaccine response profiling could further advance personalized medicine and improve pandemic preparedness strategies.
Data availability
All data generated or analysed during this study are included in this published article.
References
Rabaan, A. A. et al. SARS-CoV-2/COVID-19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic. Ann. Clin. Microbiol. Antimicrob. 19 (1), 40 (2020).
Malik, Y. S. et al. Responses to COVID-19 in South Asian association for regional Cooperation (SAARC) countries in 2020, a data analysis during a world of crises. Chaos Solitons Fractals. 152, 111311 (2021).
Rabaan, A. A. et al. Airborne transmission of SARS-CoV-2 is the dominant route of transmission: droplets and aerosols, (2022).
Guhathakurata, S., Saha, S., Kundu, S., Chakraborty, A. & Banerjee, J. S. South Asian countries are less fatal concerning COVID-19: a hybrid approach using machine learning and M-AHP. In Computational Intelligence Techniques for combating COVID- Vol. 19 1–26 (Springer, 2021).
Larsen, J. R., Martin, M. R., Martin, J. D., Kuhn, P. & Hicks, J. B. Modeling the onset of symptoms of COVID-19. Front. Public. Health. 8, 551338 (2020).
Dieter, C. et al. Polymorphisms in ACE1, TMPRSS2, IFIH1, IFNAR2, and TYK2 genes are associated with worse clinical outcomes in COVID-19. Genes 14(1):29 (2022).
Pecoraro, V., Cuccorese, M. & Trenti, T. Genetic polymorphisms of ACE1, ACE2, IFTM3, TMPRSS2 and TNFα genes associated with susceptibility and severity of SARS-CoV-2 infection: a systematic review and meta-analysis. Clin. Experimental Med. 23 (7), 3251–3264 (2023).
Tavasolian, F. et al. HLA, immune response, and susceptibility to COVID-19. Front. Immunol. 11, 601886 (2021).
Novelli, A. et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. Hla 96(5), 610–614 (2020).
Littera, R. et al. Human leukocyte antigen complex and other Immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. The Sardinian experience. Front. Immunol. 11, 605688 (2020).
Pisanti, S. et al. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid-19. J. Translational Med. 18, 1–16 (2020).
Nguyen, A. et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 94, 13. https://doi.org/10.1128/jvi.00510-20 (2020).
Dai, X. ABO blood group predisposes to COVID-19 severity and cardiovascular diseases. Eur. J. Prev. Cardiol. 27 (13), 1436–1437 (2020).
Hoiland, R. L. et al. The association of ABO blood group with indices of disease severity and multiorgan dysfunction in COVID-19. Blood Adv. 4 (20), 4981–4989 (2020).
Salman, H. M. et al. An epidemiological, strategic and response analysis of the COVID-19 pandemic in South asia: a population-based observational study. BMC Public. Health. 22 (1), 457 (2022).
Klompas, M., Milton, D. K., Rhee, C., Baker, M. A. & Leekha, S. Current insights into respiratory virus transmission and potential implications for infection control programs: a narrative review. Ann. Intern. Med. 174 (12), 1710–1718 (2021).
van der Made, C. I., Netea, M. G., van der Veerdonk, F. L. & Hoischen, A. Clinical implications of host genetic variation and susceptibility to severe or critical COVID-19. Genome Med. 14 (1), 96 (2022).
Lorente, L. et al. HLA genetic polymorphisms and prognosis of patients with COVID-19. Med. Intensiva. 45 (2), 96–103 (2021).
Muñiz-Diaz, E. et al. Relationship between the ABO blood group and COVID-19 susceptibility, severity and mortality in two cohorts of patients. Blood Transfus. 19 (1), 54 (2020).
Sanyaolu, A. et al. Comorbidity and its impact on patients with COVID-19. SN Compr. Clin. Med. 2, 1069–1076 (2020).
Naranbhai, V. et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell 185(6), 1041–1051 (2022).
Pretti, M. A. M. et al. Class I HLA allele predicted restricted antigenic coverages for Spike and nucleocapsid proteins are associated with deaths related to COVID-19. Front. Immunol. 11, 565730 (2020).
Augusto, D. G. et al. A common allele of HLA is associated with asymptomatic SARS-CoV-2 infection. Nature 620 (7972), 128–136 (2023).
Liu, N. et al. The impact of ABO blood group on COVID-19 infection risk and mortality: A systematic review and meta-analysis. Blood Rev. 48, 100785 (2021).
Pojero, F. et al. The role of immunogenetics in COVID-19. Int. J. Mol. Sci. 22 (5), 2636 (2021).
Biswas, M., Rahaman, S., Biswas, T. K., Haque, Z. & Ibrahim, B. Association of sex, age, and comorbidities with mortality in COVID-19 patients: a systematic review and meta-analysis. Intervirology 64(1), 36–47 (2021).
Dolan, M. E. et al. Investigation of COVID-19 comorbidities reveals genes and pathways coincident with the SARS-CoV-2 viral disease. Sci. Rep. 10 (1), 20848 (2020).
Mosaad, Y. M. Clinical role of human leukocyte antigen in health and disease. Scand. J. Immunol. 82 (4), 283–306 (2015).
Garcia-Silva, R. et al. Mayan alleles of the HLA-DRB1 major histocompatibility complex might contribute to the genetic susceptibility to systemic lupus erythematosus in Mexican patients from Tapachula, Chiapas. Clin. Rheumatol. 40(8), 3095–3103 (2021).
Dhaouadi, T., Riahi, A., Ben Abdallah, T., Gorgi, Y. & Sfar, I. Association of HLA-DR, HLA-DQ, and HLA-B alleles with inclusion body myositis risk: A systematic review, a meta-analysis, a meta-regression and a trial sequential analysis. Int. J. ImmunoPathol Pharmacol. 39, 03946320251321747 (2025).
Harding, A. T. & Heaton, N. S. The impact of estrogens and their receptors on immunity and inflammation during infection. Cancers 14(4), 909 (2022).
Statsenko, Y. et al. Impact of age and sex on COVID-19 severity assessed from radiologic and clinical findings. Front. Cell. Infect. Microbiol. 11, 777070 (2022).
Acknowledgements
We express our sincere gratitude to Saeed International Hospital for funding this research project on “Impact of Human Leukocyte Antigen-B genes polymorphism on the severity of Covid − 19 in Pakistani population”. Their generous support has been instrumental in facilitating this study.
Author information
Authors and Affiliations
Contributions
Muhammad Noaman Saeed1, 2, Maha Rehman1, Muhammad Nughman Khattak2, Niamat Khan1, Faiyaz Ahmed, Saad Ullah1, Muzaffar Shojonov3, Bekzod Madaminov4, Rehan Naeem1*, Abdela Befa Kinki-all the authors take part equally.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Saeed, M.N., Rehman, M., Nughman, M. et al. Association of HLA-B gene polymorphism and blood groups with COVID-19 susceptibility and severity. Sci Rep 15, 31775 (2025). https://doi.org/10.1038/s41598-025-15455-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-15455-1