Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease with heterogeneous clinical progression, reflecting distinct underlying pathological mechanisms. Early and accurate diagnosis and prognosis require reliable biomarkers to improve clinical management and therapeutic stratification. The present study explores the potential of platelet global metabolomics and fatty acid (FA) profiling as potential sources of diagnostic and prognostic biomarkers for ALS. We analysed platelets from 15 recently diagnosed ALS patients and 21 healthy controls (CTLs) using liquid chromatography-mass spectrometry (LC–MS) for metabolomics and gas chromatography-flame ionization detection (GC-FID) for FA profiling. ALS patients were classified as fast or slow progressors based on the median ALS Functional Rating Scale-Revised (ALSFRS-R) slope. While global metabolomic and FA profiles have shown limited potential for distinguishing ALS from CTL, preliminary molecular annotation based on mass and retention times disclosed specific metabolites with potential diagnostic value. Importantly, both global metabolomic and FA analyses demonstrated a marked capacity to differentiate fast progressors from slow progressors (receiver operating characteristic (ROC) curves of approximately 1), revealing distinct metabolic signatures associated with disease progression. Our findings demonstrate that platelet global metabolomics and FA profiling hold promise as prognostic biomarkers in ALS.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease and is characterized by the progressive degeneration of upper and lower motor neurons. This devastating disorder has a median survival of only 2–3 years after diagnosis1. The clinical presentation of ALS is highly heterogeneous, often resulting in a diagnostic delay of up to one year from the onset of symptoms1. Additionally, this fact places risk on the assessment of novel therapeutic approaches, as different pathogenies might require different approaches. Current ALS diagnosis relies on the clinical assessment of progressive motor neuron symptoms, alongside the exclusion of other potential causes through electrophysiological and neuroimaging studies2. Depending on the degree of clinical evidence, patients are categorized as clinically possible, probable, or definite ALS. This prolonged diagnostic process not only impacts clinical management but is also an independent predictor of reduced survival3. Therefore, the identification of robust and accessible biomarkers to support and accelerate the diagnostic process is a critical unmet need in ALS care.
In addition to being a diagnostic challenge, ALS is also characterized by significant heterogeneity in disease progression. The ALS Functional Rating Scale-Revised (ALSFRS-R) is a widely used standard for monitoring disability, evaluating a wide range of functions, including speech, swallowing, fine motor skills, mobility, and respiratory function4. The ALSFRS-R slope, which represents the rate of decline, is crucial for estimating disease progression and assessing treatment efficacy in randomized clinical trials (RCTs)5. Given this heterogeneity in clinical and progression, the identification of prognostic biomarkers could not only help predict disease trajectory but also aid in stratifying patients for clinical trials and personalized treatment approaches.
Emerging evidence suggests that differences in ALS progression might reflect distinct underlying disease mechanisms, potentially impacting treatment responsiveness. In particular, circulating lipids play key roles in disease heterogeneity. Circulating cholesterol and triglycerides have been associated with varying rates of disease progression6,7,8,9,10. Cholesterol and lipid metabolism are integral to maintaining cellular membrane fluidity and function, processes predominantly regulated by mitochondrial-associated membranes (MAMs). MAMs serve as crucial hubs for lipid synthesis and calcium signalling, and their disruption is a recognized feature of ALS pathogenesis11. In preclinical ALS models, MAM dysfunction leads to a metabolic shift from glucose metabolism to fatty acid metabolism, contributing to mitochondrial dysfunction and neuronal degeneration12. Our previous studies have reinforced this connection by demonstrating the interplay between impaired mitochondrial function, altered energy metabolism, and lipid biosynthesis in ALS models with TDP-43 loss of function13.
Metabolomics has emerged as a powerful tool for biomarker discovery, offering a comprehensive snapshot of metabolic alterations across diverse biochemical pathways, including lipid metabolism. Plasma metabolomics has been explored in ALS research since 200514. Several studies have independently validated15,16 and confirmed the utility of plasma metabolomics for ALS diagnosis and prognosis17 with platelets presenting an intriguing alternative for biomarker research. Unlike plasma, which reflects a complex mixture of systemic metabolites, platelets are cellular components with a high mitochondrial content18. Given the role of MAMs in lipid metabolism and the reported structural and functional alterations of mitochondria in ALS platelets19,20, we hypothesized that platelet metabolomics might better capture disease-specific changes in membrane composition and mitochondrial function.
Furthermore, the choice of platelets is supported by findings in primary fibroblast cultures from ALS patients, which exhibit metabolic shifts in both energetic and phospholipid pathways20. These alterations, independent of central nervous system (CNS) involvement, highlight the potential of peripheral tissues to reflect core ALS pathomechanisms. To complement the broad view provided by metabolomics, we integrated fatty acid (FA) profiling as a targeted approach to quantify specific lipid species and assess enzymatic activities involved in FA metabolism. Since MAMs influence FA synthesis and remodelling, this dual analytical strategy could provide deeper insights into the lipid dysregulation associated with ALS.
In this study, we combined non-targeted LC–MS platelet global metabolomics with FA profiling to evaluate their potential as diagnostic and prognostic biomarkers for ALS. Our results reveal limited diagnostic utility of platelet global metabolomics but underscore its promising role in prognosis, particularly in distinguishing fast from slow progressors. These findings support the feasibility of using platelet-based biomarkers for ALS management and lay the groundwork for future validation in larger cohorts using targeted analytical methods.
Results
Characteristics of the population
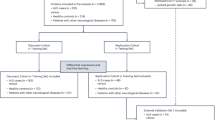
The cohort included 15 individuals who were diagnosed with the disease at a mean age of 60 ± 13 years (Table 1). Among them, six were male, and nine were female. Based on disease progression, eight individuals were classified as fast progressors, whereas seven were classified as slow progressors. With respect to onset type, 12 individuals presented with spinal onset, two presented with bulbar onset, and one presented with respiratory onset. The mean ALSFRS-R slope value was 0.53 ± 0.64.
Metabolomic analysis for ALS diagnosis
To explore the potential of non-targeted LC–MS platelet global platelet metabolomics as a diagnostic tool for ALS, we analysed platelet samples from ALS and CTL patients (Table 1). A total of 295 metabolites were detected, with only five showing statistically significant differences between the ALS and CTL groups (p < 0.05). The temptative identities of these metabolites were determined using the Human Metabolome Database (HMDB)21 and validated by matching m/z and retention times with internal standards from the lipid family (Table 2).
We employed unsupervised analyses, including principal component analysis (PCA) and hierarchical clustering, to visualize potential global metabolomic differences. However, PCA did not achieve a clear separation between the ALS and CTL groups (Fig. 1a), and hierarchical clustering of the top 25 differential metabolites also failed to differentiate between groups (Fig. 1b). These findings suggest that global metabolomic profiles in platelets are not sufficient for ALS diagnosis.
Multivariate analyses of platelet metabolome profile for ALS and CTL comparison. (a) PCA and (b) hierarchal clustering analyses of the 25 top differential metabolites show a limited effect of global metabolomic data for disease status prediction. (c) ROC analysis of a potential PI(34:0) and (d) a potential LysoSM(d18:0) shows an AUC > 0.7 and a p value < 0.05 for ALS diagnostic classification.
Given the lack of separation in unsupervised analyses, we performed receiver operating characteristic (ROC) analyses of individual metabolites. Five metabolites showed a p < 0.05, and two of them had an AUC > 0.7, including a potential phosphatidylinositol (PI) (34:0) (Fig. 1c) and lysosphingomyelin (LysoSM) (d18:0) (Fig. 1d), along with three unidentified metabolites with an AUC < 0.7 (Table 2). Despite these isolated significant hits, the overall global metabolomic profile demonstrated limited diagnostic value.
Fatty acid (FA) profiling for ALS diagnosis
Recent studies suggest a link between ALS pathology and disruptions in MAMs, which play a key role in regulating FA composition, with a pivotal role in energy metabolism11,12. Therefore, we conducted complementary FA profiling of platelets to determine whether FA homeostasis could provide additional diagnostic insights complementary to the metabolites found in the global metabolomic analysis.
The FA analysis included individual FA species, estimations of desaturase and elongase activities, and the peroxidability index22. Similar to global metabolomic results, unsupervised analyses did not reveal significant diagnostic potential. PCA of the FA profile did not distinguish ALS from CTL individuals (Fig. 2a), nor did hierarchical clustering of the top 25 FA species (Fig. 2b). Moreover, no statistically significant FA biomarkers were identified in the ROC analyses for diagnostic purposes (data not shown).
Metabolomic analysis for ALS prognosis
We next focused on evaluating platelet global metabolomics for prognostic purposes. Despite the small size of our cohort, heterogeneity in progression still existed (Table 1). ALS patients were categorized into fast (F) (≥ median ALSFRS-R slope) and slow (S) (< median ALSFRS-R slope) progression groups. Interestingly, 15 metabolites were significantly different between these groups, a notable increase compared with the three significant metabolites in the diagnostic approach (Table 3).
Unsupervised analyses confirmed the prognostic potential of platelet global metabolomics: although PCA did not show a clear separation between patients with fast and slow progressions (Fig. 3a), hierarchical clustering of the top 25 differential metabolites demonstrated strong discrimination between these prognostic groups (Fig. 3b).
Multivariate analyses of the platelet metabolome for slow (S) and fast (F) progressors. (a) PCA did not result in good group separation. However, (b) hierarchal clustering analyses of the 25 top differential analyses show a good separation of both populations. (c) Three metabolites correlated with the ALSFRS-R slope according to Pearson correlation analysis. (d) Potential DG(28:2), (e) PC(38:3), (d) P-Nme(42:7), and (f) unidentified metabolites with an AUC > 0.9 and a p value > 0.05 for ALS prognosis. *p value < 0.05, **p value < 0.01.
We further analysed the correlations between metabolite levels and the ALSFRS-R slope, a measure of functional decline. Three unidentified metabolites were significantly correlated with functional deterioration (Fig. 3c). These findings suggest that specific metabolic alterations may reflect ALS progression dynamics.
To further assess the clinical relevance of these metabolites, we performed ROC analyses. Potential biomarkers with an AUC > 0.9 and p < 0.05 included diacylglycerol (DG) (28:2) (Fig. 3d), phosphatidylcholine (PC) (38:3) (Fig. 3e), phosphatidyl-N-monomethylethanolamine (P-Nme) (42:7) (Fig. 3f), and unidentified metabolites (Fig. 3g). Additional significant metabolites are listed in Table 3.
Fatty acid (FA) profiling for ALS prognosis
In parallel, we analysed the potential of FA profiling for predicting ALS prognosis. PCA did not result in a clear separation (Fig. 4a), but the top 25 hierarchical clusters (Fig. 4b) of the FA profile demonstrated effective separation of patients with fast and slow progression, indicating a stronger prognostic capacity than that of diagnostic assessments.
Multivariate analyses of the platelet metabolome for slow (S) and fast (F) progressors. (a) Although PCA did not yield good separation, (b) hierarchal clustering analyses of the 25 top differential metabolites revealed good separation of both populations. (c) The n-6 pathway and f22:5n6 were positively correlated with the ALSFRS-R slope according to Pearson correlation analysis, whereas the f18:2n-6 pathway was negatively correlated. ROC analysis of the FA analysis revealed an AUC > 0.9 and p value < 0.05 for the following measurements: (d) Δ8 n-6, (e) the n-6 pathway, (f) f22:5n6 fatty acid, and (g) f20:2n6 fatty acid. * p value < 0.05, ** p value < 0.01.
Several FA species and ratios were significantly correlated with the ALSFRS-R slope. Positive correlations were observed for the n-6 pathway and the f22:5n6 fatty acid, whereas the f18:2n6 fatty acid was negatively correlated (Fig. 4c).
ROC analysis further validated the prognostic potential of FA profiling. Notable biomarkers with an AUC > 0.9 and p < 0.05 included Δ8 n-6 (Fig. 4d), the n-6 pathway (Fig. 4e), f22:5n6 fatty acid (Fig. 4f), and f20:2n6 fatty acid (Fig. 4g). These findings align with the global metabolomic data, reinforcing the complementary value of FA analysis in identifying ALS prognostic biomarkers.
To investigate potential sex-related differences, we stratified the metabolomics and fatty acid data by sex within both the control and ALS groups. In the controls, hierarchical clustering of the top 25 most variable metabolites suggested partial separation by sex, with modest separation also observed in the PCA (Supplementary Fig. S1a–b). However, no clear grouping was observed in the fatty acid composition data (Supplementary Figs. S2a–b). In the ALS group, neither PCA nor clustering analysis revealed strong separation by sex for either dataset (Supplementary Figs. S1c–d and S2c–d). Significant metabolites associated with sex differences are listed in Supplementary Tables S1–S4.
To further support the PCA and hierarchical clustering analyses, volcano plots were generated for key group comparisons (Supplementary Fig. S3). This allowed visualization of both the statistical significance and magnitude of change for each feature. In the comparison between the control and ALS samples, several metabolites and fatty acids were significantly altered (Fig. S3a–b). Additional volcano plots comparing fast and slow progressors revealed fewer but distinct metabolic differences (Fig. S3c–d), supporting the findings observed in the clustering analyses.
Discussion
In previous studies, we demonstrated the potential of plasma metabolomics for both the diagnosis and prognosis of ALS14. Despite these promising findings, the use of platelets as a source of biomarkers in ALS research remains largely unexplored. This gap is noteworthy considering that platelets exhibit disturbances in membrane distribution and mitochondrial structure, which could be linked to the energetic and FA metabolism dysregulations commonly observed in ALS19,18,23.
Our current results suggest a limited diagnostic utility for platelet global metabolomic biomarkers in ALS, but due to the small sample size, we cannot exclude this possibility. Among the metabolites analysed, certain phospholipids presented the highest area under the curve (AUC) in the ROC analysis. Importantly, the identification of five statistically significant metabolites (p < 0.05) among the 295 metabolites tested may include findings that occurred by chance. Given the relatively small sample size and the lack of correction for multiple testing, the possibility of false positives cannot be excluded. These results should therefore be interpreted as exploratory, and validation in independent and larger cohorts will be essential to confirm the potential of these metabolites as biomarkers. Phospholipids are integral to platelet activation24, and interestingly, previous studies have also identified differentially expressed phospholipids in cerebrospinal fluid (CSF) and the MAM fraction of fibroblasts from ALS patients20,25. Our findings align with this evidence, extending the alterations in phospholipids to peripheral samples and reinforcing their potential as ALS biomarkers14. However, when the platelet metabolome is considered, the overall metabolic profile of ALS patients is remarkably similar to that of controls, suggesting that early diagnostic changes are restricted to specific phospholipid species. Similarly, platelet FA profiling does not provide a clear diagnostic advantage, although the use of metabolomics cannot be discarded.
In contrast, the prognostic potential of platelet global metabolomic biomarkers appears more promising. Hierarchical clustering based on the top 25 metabolites almost perfectly distinguishes patients with fast progression from those with slow progression. This distinct global metabolomic fingerprint indicates that these subgroups may not only exhibit different disease mechanisms but may also benefit from tailored therapeutic approaches. A plausible explanation is the hypermetabolic state observed in fast progression, which is consistent with previous plasma and CSF metabolomic analyses14. The identification of robust prognostic biomarkers is critical for stratifying patients in clinical trials and improving therapeutic outcomes. For example, the efficacy of edaravone has been shown to depend heavily on the rate of disease progression26.
Our study’s unique design—sampling at the diagnostic stage and correlating with longitudinal follow-up—adds a predictive value to the identified biomarkers. While we classified patients into fast and slow progressors, ALS is characterized by a highly variable functional decline. Notably, several metabolites were strongly associated with this decline, suggesting their potential inclusion in predictive algorithms for functional trajectory. Moreover, platelet FA profiling reinforced these findings, demonstrating a high discriminative capacity between progression groups. The complementary nature of these two techniques is particularly valuable, while global metabolomics offers a broad metabolic overview, FA profiling enables precise molecular identification and additional insights into enzymatic activity and lipid peroxidability indices.
Our data specifically highlight that an increased n-6 pathway is associated with greater functional decay (ALSFRS-R slope). This finding underscores the role of peripheral inflammation in ALS progression, echoing previous studies showing n-6 polyunsaturated fatty acid (PUFA) dysregulation in human central nervous system samples27,28. Notably, modulating these pathways through dietary interventions in preclinical models has shown potential to influence disease progression and lifespan28. Among the prognostic biomarkers, FADS2 Δ8-desaturase activity demonstrated exceptional performance, with an AUC of 1, accurately classifying 100% of fast progressors in our cohort. This enzyme is upregulated in the spinal cord of sporadic ALS patients and is linked to excitotoxicity in vitro29, further validating the use of peripheral tissues as reporters of disease-specific mechanisms, including lipid metabolism dysregulation.
While sex-based differences in metabolism are well documented, our subgroup analysis revealed that in ALS patients, sex did not account for major variance in either global metabolomic or fatty acid profiles. In contrast, controls exhibited some degree of sex-associated clustering in the metabolomics data but not in fatty acid composition. These findings may indicate that disease-driven metabolic alterations in ALS are more dominant than sex-specific metabolic signatures.
This study is not without limitations. We analysed whole platelet lysates to capture a broad metabolic profile. However, this approach combines both cytosolic and membrane-associated metabolites, which may obscure more localized metabolic changes. Future studies could benefit from subcellular fractionation of platelets (e.g., via differential centrifugation) to analyse the cytosolic and membrane fractions separately. This strategy could enhance detection sensitivity and help identify more compartment-specific biomarkers for ALS. Moreover, the use of a nontargeted metabolomics approach restricts our ability to confidently identify specific metabolites, as potential identities are inferred based on m/z and retention time properties in the Human Metabolome Database (HMDB)21, which provides only moderate confidence. Another technical limitation is found in unsupervised analysis: the marked separation of samples into two distinct groups observed in the PCA score plot appears to be driven primarily by technical variability. Specifically, the samples were processed and analysed in two different analytical batches, which we believe contributed to the large variance captured in PC1. While batch effects are a recognized challenge in metabolomics studies, we acknowledge that in our dataset, this may have overshadowed more subtle biological differences. Potential confounding factors that may have influenced the observed metabolomic profiles cannot be fully excluded in this study. Although all samples were collected following a standardized protocol, variability in pre-analytical conditions (e.g., storage time, temperature fluctuations, or platelet count), individual dietary habits, or concomitant medications could have impacted the metabolome. Future studies will require stricter control of batch processing, inclusion of technical replicates, and statistical correction for batch effects to ensure robust interpretation of metabolomic variation. Importantly, ALS progression is heterogeneous, and the sampling time point used in this study, although informative, may not fully capture the dynamic trajectory of metabolic alterations over time. Future studies will benefit from a prospective longitudinal design incorporating at least three or more time points per patient at regular intervals (e.g., every 3–4 months). Such an approach would allow for better modelling of disease progression and strengthen the identification of reliable prognostic biomarkers in ALS. Additionally, the relatively small sample size and regional homogeneity (all patients were from Catalonia) may limit the generalizability of our findings. Nevertheless, as a pilot study, our primary objective was to assess whether global platelet metabolomics and FA profiling warrant further investigation. Our promising results should be validated through targeted analytical methods and larger, prospective and more diverse cohorts.
In conclusion, our findings indicate that while platelet global metabolomic and FA profiling offer limited diagnostic value for ALS, they hold significant promise as prognostic biomarkers. Several metabolites and FAs identified in this study could contribute to predicting functional decline, offering a potential tool for better patient stratification and therapeutic decision making in clinical settings.
Methods
Human samples
The study followed the guidelines of the relevant Spanish legislation (Real Decreto 1716/2011) and was approved by the Institutional Ethics Committee of Bellvitge University Hospital. Accordingly, informed consent was obtained from all participants. Platelets were obtained from 15 ALS patients at Bellvitge University Hospital between 2015 and 2016 and from 21 age-matched non-ALS individuals (CTLs). The ALS patients included 9 women and 6 men (12 spinal, 2 bulbar and 1 respiratory) and age-matched controls (15 women and 6 men) (Table 1). Biological samples were obtained between 10 and 24 months after the beginning of symptoms, and ALS was diagnosed according to the El Escorial criteria for ALS. The ALSFR was calculated and validated by two independent neurologists (MP and RD). The progression rate was calculated (at baseline or at the last visit) as 48 minus the ALS Functional Rating Scale-Revised score, divided by the disease duration from the onset of symptoms (Lu et al., 2015). We defined the ALSFR slope of the median value (0.5) as the cut-off point for classifying progression rates as fast progression (F) or slow progression (S). Blood was obtained in the morning after overnight fasting.
Platelet isolation
Platelets were isolated from peripheral blood using Histopaque-1077 (10771, Sigma). Briefly, 3 ml of Ficoll was added to the bottom of a centrifuge tube. Carefully, 3 ml of blood mixed with 3 ml of PBS was layered onto Ficoll without mixing the phases. The tubes were centrifuged at 400×g for 30 min at room temperature with the lowest acceleration and brake program. The upper layer and the opaque interphase were collected into a new tube, and then this phase was washed with PBS and centrifuged at 300×g. The supernatant contained the platelet fraction and the pelleted PBMCs. The platelets were centrifuged at 360×g, and the pellets (containing PBMCs) were discarded. The supernatant was collected and centrifuged at 800×g. The resulting pellet was washed with PBS and centrifuged at 800×g to obtain the platelets.
Metabolomic analysis
A total of 100 μL of PBS was added to the platelet pellet, and the protein concentration was quantified using the Bradford method, following the manufacturer’s instructions (#5000006, Bio-Rad). An equal amount of protein (150 μg) was used for metabolite extraction. The samples were diluted to a final volume of 100 μL in PBS, and 300 μL of ice-cold acetone was added. The samples were vortexed for 10 s and incubated for 30 min at 4 °C. The samples were then centrifuged at 4 °C for 10 min at 1000×g. The supernatant was separated into a clean glass tube and evaporated in Speed Vac. A total of 250 μL of methanol and 500 μL of chloroform were added together with the standard mixture. The samples were vortexed for 10 s, 200 μl of 0.7% KCl was added, the mixture was vortexed for 10 s, and the mixture was centrifuged at 4 °C for 10 min at 1000 ×g. The upper and bottom phases were separated. The bottom phase contained chloroform-soluble metabolites (lipids), and the upper phase contained methanol-soluble metabolites (employed for the metabolomic analysis).
For the metabolomic study, an Agilent 1290 liquid chromatography system coupled to an ESI-Q-TOF MS/MS 6520 instrument (Agilent Technologies, Santa Clara, CA, US) was used. In all cases, 2 μL of extracted sample was applied onto a reversed-phase column (Zorbax SB-Aq 1.8 μm 2.1 × 50 mm; Agilent Technologies) equipped with a precolumn (Zorbax-SB-C8 Rapid Resolution Cartridge 2.1 × 30 mm 3.5 μm; Agilent Technologies) with a column temperature of 60 °C. The flow rate was 0.6 mL/min. Solvent A was composed of water containing 0.2% acetic acid, and solvent B was composed of 0.2% acetic acid. The gradient started at 2% B, increased to 98% B in 13 min and was held at 98% B for 6 min. Post-time was established in 5 min.
Data were collected in positive electrospray mode in full-scan mode at 100–3000 m/z in an extended dynamic range (2 GHz), with N2 used as the nebulizer gas (5 L/min, 350 °C). The capillary voltage was 3500 V with a scan rate of 1 scan/s. The ESI source used a separate nebulizer for the continuous, low-level (10 L/min) introduction of reference mass compounds: 121.050873, 922.009798 (positive ion mode) and 119.036320, 966.000725 (negative ion mode), which were used for continuous, online mass calibration. MassHunter Data Analysis Software (Agilent Technologies) was used to collect the results, and MassHunter Qualitative Analysis Software (Agilent Technologies) was used to obtain the molecular features of the samples, which represent different, co-migrating ionic species of a given molecular entity using the Molecular Feature Extractor algorithm (Agilent Technologies), as described previously (Jove et al., 2015).
Fatty acid analysis
Total lipids from platelets were extracted using a chloroform/methanol mixture (2:1, v/v)30 in the presence of 0.01% butylated hydroxytoluene to prevent oxidation. The extraction also included 1,2-dinonadecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, ref. 850,367) as an internal standard. The chloroform phase containing the lipids was collected and evaporated under a nitrogen stream, and the resulting fatty acids were converted into their methyl ester derivatives (FAMEs) for analysis via gas chromatography (GC), following previously established protocols30. A total of 23 species were analysed (Table S5).
Separation was achieved using a DB-WAX capillary column (30 m × 0.25 mm × 0.20 μm) on a 7890A GC system equipped with a 7683B Series automatic injector and a flame ionization detector (FID) (Agilent Technologies, Barcelona, Spain). Fatty acid methyl esters were identified by comparing their retention times with those of authenticated standards from Larodan Fine Chemicals (Malmö, Sweden). The results are reported as mol%.
Additionally, the following fatty acyl indices were calculated: saturated fatty acids (SFAs); unsaturated fatty acids (UFAs); monounsaturated fatty acids (MUFAs); polyunsaturated fatty acids (PUFAs) from the n-3 and n-6 series (PUFAn-3 and PUFAn-6, respectively); and average chain length, ACL = [(Σ%Total14 × 14) + (Σ% Total16 × 16) + (Σ%Total18 × 18) + (Σ%Total20 × 20) + (Σ% Total22 × 22) + (Σ% Total24 × 24)]/100. The density of the double bonds in the membrane was calculated with the double bond index, DBI = [(1 × Σmol% monoenoic) + (2 × Σmol% dienoic) + (3 × Σmol% trienoic) + (4 × Σmol% tetraenoic) + (5 × Σmol% pentaenoic) + (6 × Σmol% hexaenoic)]. Membrane susceptibility to peroxidation was calculated with the peroxidisability index, PI = [(0.025 × Σmol% monoenoic) + (1 × Σmol% dienoic) + (2 × Σmol% trienoic) + (4 × Σmol% tetraenoic) + (6 × Σmol% pentaenoic) + (8 × Σmol% hexaenoic)]31, and PI = [(0.015 × Σmol% monoenoic) + (1 × Σmol% dienoic) + (2 × Σmol% trienoic) + (3 × Σmol% tetraenoic) + (4 × Σmol% pentaenoic) + (5 × Σmol% hexaenoic)]31,32.
Elongase and desaturase activity was estimated from specific product/substrate ratios33: Δ9(n-7) = 16:1n-9/16:0; Δ9(n-9) = 18:1n-9/18:0; Δ5(n-6) = 20:4n-6/20:3n-6; Δ6(n-3) = 18:4n-3/18:3n-3; Δ6(n-3) = 24:6n-3/24:5n-3; Elovl3(n-9) = 20:1n-9/18:1n-9; Elovl6 = 18:0/16:0; Elovl1-3-7a = 20:0/18:0; Elovl1-3-7b = 22:0/20:0; Elovl1-7c = 24:0/22:0; Elovl5(n-6) = 20:2n-6/18:2n-6; Elovl2-5 (n-6) = 22:4n-6/20:4n-6; Elovl 2–5(n-3) = 22:5n-3/20:5n-3, and Elovl 2(n-3) = 24:5n-3/22:5n-3. Finally, the n-3 and n-6 pathways were estimated according to ratios of 22:6n-3/18:3n-3 and 22:5n-6/18:2n-6, respectively.
Data analysis
All investigations were performed in a double-blinded manner. After the data were decoded, molecular changes were extracted using MassHunter Qualitative Analysis (Agilent Technologies, Barcelona, Spain), and the molecular profiles were obtained using the MassHunter Profiler Professional (Agilent Technologies, Barcelona, Spain), as previously described34. Molecules within a 0.1% ± 0.25 min retention time window and 30 ppm ± 2 mDA mass window were considered the same. Only common features (those found in at least 50% of the samples of the same condition) were considered to minimize individual bias. The peak intensities were relativised by internal standard peak intensity. TTESTs (normalized by the autoscale option), Spearman correlations, multivariate analyses (principal component analysis (PCA) and hierarchical clustering) and ROC analyses were performed in Metaboanalyst.ca.
Molecules showing statistically significant expression (with a p value < 0.05 in differential analyses) were annotated by comparing their exact mass and retention time and isotopic distribution with a specific database (hmdb.ca) to obtain potential identities.
The data were analysed using the MetaboAnalyst platform. A t test was employed for comparison. ROC analysis was employed for biomarker potential assessment. Pearson correlation was used to quantify association levels. Principal component analysis (PCA) was performed using Euclidean distance as the distance measure. The separation between groups observed in the PCA was statistically assessed using PERMANOVA (Permutational Multivariate Analysis of Variance), as implemented within MetaboAnalyst.
Hierarchical clustering was performed using Ward’s linkage method and Euclidean distance. Heatmaps were generated using the 25 most variable metabolites across samples (top 25 features), which were automatically selected based on their variance. This approach enhances visualisation and improves separation in exploratory clustering. Prior to clustering, features were filtered using univariate statistical tests (t test or ANOVA, depending on the comparison) provided by the software. Volcano plots were generated using the MetaboAnalyst 6.0 platform. Differential metabolites were identified based on a fold change threshold greater than 1 (FC > 1) and a p value < 0.05; p < 0.05 was used as the cut-off for significance. Unless specified, the annotation of metabolite is: calculated exact mass (in mass units)_retention time (min).
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author, Dr. Manuel Portero-Otín ( [manuel.portero@udl.cat](mailto: manuel.portero@udl.cat) ) or Dr Pascual Torres ( [pascual.torres@udl.cat](mailto: pascual.torres@udl.cat) ) , on reasonable request.
References
van Es, M. A. et al. Amyotrophic lateral sclerosis. Lancet 390, 2084–2098 (2017).
Brooks, B. R., Miller, R. G., Swash, M. & El Munsat, T. L. Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 1, 293–299 (2000).
Ganesalingam, J. et al. Latent cluster analysis of ALS phenotypes identifies prognostically differing groups. PLoS ONE 4, e7107 (2009).
Cedarbaum, J. M. et al. The ALSFRS-R: A revised ALS functional rating scale that incorporates assessments of respiratory function. J. Neurol. Sci. 169, 13–21 (1999).
Castrillo-Viguera, C., Grasso, D. L., Simpson, E., Shefner, J. & Cudkowicz, M. E. Clinical significance in the change of decline in ALSFRS-R. Amyotroph. Lateral Scler. 11, 178–180 (2010).
Ingre, C. et al. Lipids, apolipoproteins, and prognosis of amyotrophic lateral sclerosis. Neurology 94, E1835–E1844 (2020).
Esteban-García, N. et al. Body complexion and circulating lipids in the risk of TDP-43 related disorders. Front. Aging Neurosci. 14, 838141 (2022).
Mariosa, D. et al. Blood biomarkers of carbohydrate, lipid, and apolipoprotein metabolisms and risk of amyotrophic lateral sclerosis: A more than 20-year follow-up of the Swedish AMORIS cohort. Ann. Neurol. 81, 718–728 (2017).
Michels, S. et al. Association of blood lipids with onset and prognosis of amyotrophic lateral sclerosis: Results from the ALS Swabia registry. J. Neurol. 270, 3082–3090 (2023).
Nakamura, R. et al. Investigation of the prognostic predictive value of serum lipid profiles in amyotrophic lateral sclerosis: Roles of sex and hypermetabolism. Sci. Rep. 12, 1–10 (2022).
Bernal, A. F. et al. MAM-Tata: Investigating the role of mitochondrial-associated membranes in ALS. Biochim. Biophys. Acta Mol. Basis Dis. 1869, 166716 (2023).
Larrea, D. et al. Altered mitochondria-associated ER membrane (MAM) function shifts mitochondrial metabolism in amyotrophic lateral sclerosis (ALS). Nat. Commun. 16, 379 (2025).
Ceron-Codorniu, M. et al. TDP-43 dysfunction leads to bioenergetic failure and lipid metabolic rewiring in human cells. Redox Biol. 75, 103301 (2024).
Sol, J. et al. Lipidomic traits of plasma and cerebrospinal fluid in amyotrophic lateral sclerosis correlate with disease progression. Brain Commun. 3, fcab143 (2021).
Lanznaster, D. et al. Metabolomics: A Tool to Understand the Impact of Genetic Mutations in Amyotrophic Lateral Sclerosis. Genes (Basel) 11, 537 (2020).
Zharikov, S. & Shiva, S. Platelet mitochondrial function: From regulation of thrombosis to biomarker of disease. In Biochemical Society Transactions vol. 41 118–123 (Biochem Soc Trans, 2013).
Lanznaster, D., de Assis, D. R., Corcia, P., Pradat, P. F. & Blasco, H. Metabolomics biomarkers: A strategy toward therapeutics improvement in ALS. Front. Neurol. 9, 1126 (2018).
Chacko, B. K. et al. Mitochondria in precision medicine: Linking bioenergetics and metabolomics in platelets. Redox Biol. 22, 101165 (2019).
Shrivastava, M., Das, T. K., Behari, M., Pati, U. & Vivekanandhan, S. Ultrastructural variations in platelets and platelet mitochondria: A novel feature in amyotrophic lateral sclerosis. Ultrastruct. Pathol. 35, 52–59 (2011).
Veyrat-Durebex, C. et al. Metabo-lipidomics of fibroblasts and mitochondrial-endoplasmic reticulum extracts from ALS patients shows alterations in purine, pyrimidine, energetic, and phospholipid metabolisms. Mol. Neurobiol. 56, 5780–5791 (2019).
Wishart, D. S. et al. HMDB: The human metabolome database. Nucl. Acids Res. 35, D521 (2007).
Ferrer, I. et al. Region-specific vulnerability to lipid peroxidation and evidence of neuronal mechanisms for polyunsaturated fatty acid biosynthesis in the healthy adult human central nervous system. Biochim. Biophys. Acta Mol. Cell Biol. Lip. 1862, 485–495 (2017).
Dupuis, L. et al. Platelet serotonin level predicts survival in amyotrophic lateral sclerosis. PLoS ONE 5, e13346 (2010).
Koseoglu, S. et al. Analytical characterization of the role of phospholipids in platelet adhesion and secretion. Anal. Chem. 87, 413–421 (2015).
Blasco, H. et al. 1H-NMR-Based metabolomic profiling of CSF in early amyotrophic lateral sclerosis. PLoS ONE 5, e13223 (2010).
Abe, K. et al. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet Neurol. 16, 505–512 (2017).
Ilieva, E. V. et al. Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain 130, 3111–3123 (2007).
Torres, P. et al. Gender-specific beneficial effects of docosahexaenoic acid dietary supplementation in G93A-SOD1 amyotrophic lateral sclerosis mice. Neurotherapeutics 17, 269–281 (2020).
Cacabelos, D. et al. Interplay between TDP-43 and docosahexaenoic acid-related processes in amyotrophic lateral sclerosis. Neurobiol. Dis. 88, 148–160 (2016).
Pamplona, R. et al. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 280, 21522–21530 (2005).
Yin, H., Xu, L. & Porter, N. A. Free radical lipid peroxidation: Mechanisms and analysis. Chem. Rev. 111, 5944–5972 (2011).
Holman, R. T. Autoxidation of fats and related substances. Prog. Chem. Fats Other Lip. 2, 51–98 (1954).
Guillou, H., Zadravec, D., Martin, P. G. P. & Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49, 186–199 (2010).
Servià, L. et al. A prospective pilot study using metabolomics discloses specific fatty acid, catecholamine and tryptophan metabolic pathways as possible predictors for a negative outcome after severe trauma. Scand. J. Trauma Resusc. Emerg. Med. https://doi.org/10.1186/s13049-019-0631-5 (2019).
Acknowledgements
We acknowledge funding from the Spanish Ministry of Economy and Competitiveness, Institute of Health Carlos III (PI20/0155; PI23/00176) to MP-O; from the Spanish Ministry of Science, Innovation and Universities (RTI2018-099200-B-I00), and the Generalitat of Catalonia (Agency for Management of University and Research Grants (2017SGR696) and Department of Health (SLT002/16/00250)) to RP. Support was also received in the form of a FUNDELA Grant, RedELA-Plataforma Investigación, and the Fundació Miquel Valls (Jack Van den Hoek donation). M.J, is a professor under the Serra Hunter program (Generalitat de Catalunya). This study has been co-financed by FEDER funds from the European Union (“A way to build Europe”) and funded by the PIRS program by the “Diputació de Lleida” and by the “la Caixa” Foundation. IRBLleida is funded by a CERCA Programme/Generalitat of Catalonia. P.T. received a Margarita Salas postdoctoral fellowship from the Spanish Ministry of Universities (Spanish Government), which was supported by NextGenerationEU. P.T. received a Beatriu de Pinós postdoctoral fellowship from the Departament de Recerca i Universitats de la Generalitat de Catalunya (2023 BP 00144).
Author information
Authors and Affiliations
Contributions
M.P-O. conceived and supervised the study. P.T., I.P., A.F.B., M.P., R.D., M.J., C.G-M., V.A., and I.F. performed the experiments and data collection. P.T., M.J., M.P-O., and I.P. analyzed and interpreted the results. I.F. and R.P. provided critical insights into the study design and manuscript preparation. M.P-O. and P.T. wrote the manuscript, with contributions from all co-authors. All authors reviewed, edited, and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Torres, P., Pradas, I., Fernàndez-Bernal, A. et al. Exploring platelet metabolomics and fatty acid profiles for ALS prognosis and diagnosis. Sci Rep 15, 34236 (2025). https://doi.org/10.1038/s41598-025-16220-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-16220-0