Abstract
Previously reported HLA class I correlated with the outcome of early-stage non-small cell lung cancer (NSCLC) in the Japanese population. The binding affinity capability of the EGFR mutation peptide and various HLA-A subtypes could explain this. We conducted a prospective cohort study to explore advanced EGFR-mutated NSCLC patients who received EGFR TKIs in various HLA-A subtypes, the outcomes of treatment, and tumor immune microenvironment (TIME). Eighty-four advanced NSCLC harboring EGFR exon 19 deletion and exon 21 L858R mutations were analyzed. Among these, the interested HLA-A subtypes of exon 19 deletion were composed of HLA-A*03:01 (7.1%), *30:01 (3.7%),*11:01 (82.1%), and *68:01 (7.1%). The interested HLA-A subtypes of exon 21 L858R were HLA-A*30:01 (28.5%), *33:03 (57.1%), and *34:01 (14.4%). Multivariate Cox-regression analysis revealed that the interested HLA-A subtypes were not an independent factor of progression-free or overall survival. No correlation was found between HLA-A subtypes and either inflammatory TIME or the presence of intra-tumoral CD8 TILs. HLA-A subtypes did not correlate with prognostic outcomes in sensitized EGFR mutations. The diverse binding affinity with EGFR peptides was not translated into the TIME patterns.
Similar content being viewed by others
Introduction
The prevalence of epidermal growth factor receptor (EGFR) mutated advanced non-small cell lung cancer, a leading cause of mortality in Thailand, is up to almost 50%1. EGFR tyrosine kinase inhibitors (EGFR TKIs) are currently adopted as standard treatment. Even through the evolution of EGFR TKIs, progressive disease was inevitable. Usual EGFR-mutated advanced non-small cell lung cancer (NSCLC) was correlated with low PD-L1, CD8 tumor-infiltrating lymphocytes (TILs)2,3. The tumor immune microenvironment (TIME) affects the outcome of EGFR TKIs. High expression of PD-L1 and/or CD8 TILs was correlated with poor response to EGFR TKI treatment2,3,4,5.
Human leukocyte antigen (HLA) class I molecules had a significant role in both activation and proliferation of CD8 + T-cells, leading to the destruction of cancerous cells or cancer cell immune evasion that leads to cancer progression6. Recent research has highlighted the significance of HLA diversity in immunotherapy treatment response by recognizing and responding to neoantigens7. Therapeutic cancer vaccines are a novel immunotherapy that utilizes neoantigen epitopes8,9. Neoantigens derived from the EGFR mutation were also immunogenic, with a specific HLA-A subtype, which consequently affected the clinical response in acquired resistance to EGFR TKIs10,11. The action of neoantigens went through peptide-induced antigen-specific T lymphocytes. Tumor-specific mutations were considered immunogenic neo-epitopes, depending on their recognition by the immune system, T-cells, due to HLA restriction. HLA-A, -B, and -C are the primary and highly polymorphic molecules of the classical HLA class I gene12. Functional binding affinity was confined exclusively to HLA-A and is associated with a favorable prognosis in early-stage non-small cell lung cancer with common EGFR alterations13 and prevalence of lung cancer in the Japanese population14. High-affinity HLA-A allotypes differed between EGFR L858R and the exon 19 deletion15. Those might have consequences for activating the CD8 T-cell response. This study aims to investigate the correlation of HLA-A subtypes, focusing on the most common HLA allele in the Thai population, HLA-A*11:01, HLA-A*24:02, HLA-A*02:03, HLA-A*33:03, and HLA-A*02:0716, with the impact of CD8 + TILs, tumor PD-L1 TPS, and the outcome of EGFR TKIs therapy on EGFR-mutated NSCLC participants.
Materials and methods
Study participants
We conducted a prospective cohort at The King Chulalongkorn Memorial Hospital in Bangkok, Thailand. The study included participants aged ≥ 18 years diagnosed with EGFR-mutated recurrence or advanced-stage NSCLC. EGFR mutation testing was conducted using single gene testing Cobas® mutation test v2. or diver alteration gene panel. All participants received EGFR TKIs (1st -3rd generation) as first-line treatment, according to the provided physician. The pretreatment assessment and response evaluation were conducted as a standard practice of the institute. Demographic characteristics were obtained from the hospital’s electronic medical records. I confirm that all experiments were performed in accordance with the Declaration of Helsinki. The Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, approved the study. (IRB No. 894/63 and 580/66). Written informed consent was obtained from all participants. The Bureau of Registration Administration, Ministry of Interior, Bangkok, Thailand, validated the participant’s death date.
Tumor immune microenvironment assessment
Tissue samples were collected upon the diagnosis of advanced-stage non-small cell lung cancer. CD8 Tumor-infiltrating lymphocytes (TILs) and PD-L1 were evaluated by immunohistochemistry. The interpretation was made by one pathologist (S.S.) who was blinded to clinical outcomes. TILs were assessed using immunohistochemistry staining to evaluate the expression of CD8 + T-cells according to the guidelines established by the International TILs Working Group in 2014 17. The evaluation was based on the spatial location of CD8 + TILs, intra-tumoral or stromal. Intra-tumoral CD8 + TILs were defined as CD8 + TILs with direct cell-cell contact with carcinoma cells. The results will be presented as percentages based on tumor cells17. High intra-tumoral CD8 + TILs were defined as intra-tumoral CD8 + TILs ≥ 10%18. PD-L1 was assessed using the Dako FLEX 22C3 and presented by the tumor proportion score19,20. High PD-L1 was defined as PD-L1 TPS ≥ 15% as previously demonstrated the correlation with prognosis21. The definition of inflammatory tumor microenvironment was defined as either PD-L1 TPS ≥ 15% or intra-tumoral CD8 + TILs ≥ 10% as previously reported4,18,21,22.
Blood specimen correction and HLA typing evaluation
Blood samples were collected before the participants started EGFR TKIs treatment in an EDTA tube, centrifuged, and kept at -80 °C until further process. The evaluation of HLA typing was performed by buffy coat using whole-exome sequencing technology. Briefly, DNA from the buffy coat was extracted using a Qiagen blood mini kit following manufacturer protocol. Library preparation was proceeded using SureSelectXT V6 + UTR library prep kit (Illumina, San Diego, CA, USA). The sequencing was conducted using NovoSeq to generate 150 bp paired-end reads at Macrogen Inc. (Seoul, Korea). We analyzed data through bcbio-nextgen version v1.2.923 with target sequences of approximately 90 Mb. Unmapped BAM was generated from Fastq raw data, aligned with hg38 reference using BWA version 0.7.1724, and processed by using the GATK best practice pipeline through Genome Analysis Toolkit recommendation (GATK version 4.1.0.0 including MarkDuplicates, base quality score recalibration, indel realignment, duplicated removal. We identify high polymorphic HLAs using OptiType algorithm25 which shown high accuracy26. The HLA-A was classified into interested and non-interested subtypes based on the binding affinity of the EGFR mutation subtype, which had been previously reported15. The higher binding affinity of the HLA-A subtype represented potential neoantigen. The interested HLA-A was also correlated with favorable prognostic outcomes in resectable NSCLC15. For EGFR L858R alteration, interested HLA-A subtypes were HLA-A*30:01, HLA-A*31:01, HLA-A*33:01, HLA-A*33:03, HLA-A*34:01, HLA-A*66:02, HLA-A*68:01, HLA-A*68:03, HLA-A*68:04, and HLA-A*68:05. While EGFR exon 19 deletion, interested HLA-A subtypes were HLA-A*03:01, HLA-A*03:02, HLA-A*11:01, HLA-A*30:01, HLA-A*34:02, HLA-A*68:01. The presence of one allele of interested HLA-A was considered positive for interested HLA-A.
Sample size calculation
The author was calculated based on Dimou A., et al., who reported that the interesting HLA-A alleles, i.e., HLA-A*11:01, HLA-A*24:02, HLA-A*02:03, HLA-A*33:03, and HLA-A*02:07, exhibited greater binding efficacy to either EGFR L858R or exon 19 deletion peptides. The prevalence of HLA-A*11:01, HLA-A*24:02, HLA-A*02:03, HLA-A*33:03, and HLA-A*02:07 of the Thai population was 26%, 11%, 11%, 11%, and 8%, respectively as previously reported by Satapornpong et al.16. The prevalence of the inflammatory TIME reported by Matsumoto et al. was 13.5%4. We proposed a hypothesis that an interested HLA-A results in a four-fold higher frequency of inflamed TIME compared to uninterested HLA-A subtypes. Using a proportion sample size calculation27to achieve a Type 1 error rate of 5% and a power of 80%, the sample size was calculated to be 74, without continuity correction.
Statistical analysis
Categorical data was analyzed using the Chi-square or Fisher exact test. Continuous data was analyzed using the Mann-Whitney test. The correlation between the HLA-A and either inflammatory TIME or intra-tumoral CD8 TILs was calculated using the Chi-square test. Progression-free survival (PFS) was defined by the time of initiation EGFR-TKIs treatment to the date of objective disease progression or death from any cause. Overall survival (OS) was determined by the time of initiation of EGFR-TKIs treatment to the date of death from any cause. The data was censored on December 31, 2023, for alive or non-progressive disease participants. Multivariate analyses of clinical factors, HLA-A subtype, and tumor immune microenvironment expression level were performed using a Cox proportional hazards model. The Kaplan-Meier method was used to evaluate survival, and the log-rank test was used to evaluate the significance of the difference between groups. The significance level was defined as p-value < 0.05. Statistical analysis was performed using SPSS version 29.0.
Results
Demographic characteristics
Between October 1, 2019, and November 29, 2022, 170 participants with EGFR-mutated advanced-stage NSCLC who received treatment with EGFR TKIs were enrolled. The median follow-up of participants in this cohort was 24.8 months [IQR 14.2–36.4]. At the time of censoring, 85.3% of participants had disease progression from first-line EGFR TKI, while 57% of participants had died. Baseline characteristics are presented in (Table 1). The median age of participants was 65 [range 57–73] years. 81% of the participants were non-smokers. The prevalence of EGFR mutations was 58.8% for exon 19 deletion and 34.7% for L858R mutations. First-generation EGFR TKIs and third-generation EGFR TKIs were used in 78.2% and 16.5% of the participants, respectively. The prevalence of liver metastases at diagnosis was 18.2%. HLA-A assessment through whole-exome sequencing was conducted in ninety-one participants. The demographic characteristics of participants evaluated for HLA-A typing were consistent with those of the overall cohort. TIME was assessed in the pretreatment specimen of 100 participants. The study scheme is illustrated in (Fig. 1).
HLA typing and the clinical outcome
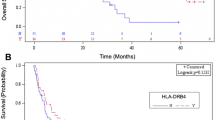
Among an overall cohort of 91 participants, we selected only 86 participants with common sensitizing mutations, including EGFR exon 19 del or exon 21 L858R alteration. The details of the HLA-A subtypes, depending on mutation types, are shown in (Fig. 2). The most common HLA-A subtypes were HLA-A*11:01 (21.5%) and HLA-A*33:03 (12.7%), compatible with the general Thai population16. The prevalence of participants with EGFR exon 19 deletion and L858R mutation was 64% and 36%, respectively. The interested HLA-A subtypes in advanced NSCLC participants with exon 19 deletion (n = 28) consisted of HLA-A*03:01 2 alleles (7.1%), HLA-A*30:01 1 allele (3.7%), HLA-A*11:01 23 alleles (82.1%), and HLA-A*68:01 2 alleles (7.1%). While the interested HLA-A subtypes in advanced NSCLC participants with exon 21 L858R alteration (n = 7) were composed of HLA-A*30:01 2 alleles (28.5%), HLA-A*33:03 4 alleles (57.1%), and HLA-A*34:01 1 allele (14.4%). The median progression-free survival in the interested vs. non-interested HLA-A subtype was 16.4 [95% CI: 12.3–22.4] months and 12.5 [95% CI: 7.9–19.7] months, respectively (Fig. 3A). Multivariate Cox regression analysis revealed that the generation of EGFR TKIs was an independent factor of disease control. Second or third-generation EGFR TKIs were better than first-generation EGFR TKIs with the HR of PFS 0.36 [95% CI: 0.18–0.74, p-value 0.005]. The presence of liver metastasis was also an independent prognostic factor with the HR of PFS 2.98 [95% CI: 1.49–5.98, p-value 0.002] (Table 2). The results were consistent with other reports28,29,30,31. The overall survival in the interested vs. non-interested HLA-A subtype was 31.8 [95% CI: 18.9–61] months and 34.8 [95% CI: 20.6–40.7] months, respectively (Fig. 3B). Smoking status and presence of liver metastasis were the independent factors of OS with the multivariate HR of 2.75 [95% CI: 1.30–5.82, p-value 0.008] and HR of 3.65 [95% CI: 1.72–7.76, p-value < 0.001], respectively (Table S1). The HLA-A subtypes of interest in advanced NSCLC with exon 19 and 21 alterations did not correlate with progression-free or overall survival.
The correlation between HLA-A subtype, TIME, and clinical outcomes
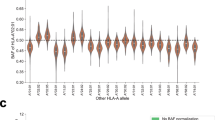
Adequate tissue blocks (n = 100) were requested for tumor PD-L1 and CD8 immunohistochemistry. The correlation analysis was performed in 49 overlapping participants (57%) who had both HLA-A subtypes and at least one evaluable TIME, either PD-L1 or CD8. The correlation of each interested HLA-A subtype and TIME is shown in Table S2. Intra-tumoral and stromal CD8 of each of the HLA-A subtypes was primarily absent, consistent with other reports32,33. Those details, according to the exon 19 deletion and the exon 21 alteration, are shown in Table S3. Inflammatory TIME is an independent factor of EGFR TKIs outcome, with the HR of PFS 3.02 [95% CI; 1.23–7.39, p-value 0.016] (Table S4) and HR of OS 3.99 [95% CI; 1.45–10.96, p-value 0.007] (Table S5). These results were also consistent with previous reports4,22. We found no significant correlation between interested and non-interested HLA-A subtypes with patterns of TIMEs, either inflammatory TIME or the presence of intra-tumoral CD8 TILs (Table 3). Incorporate clinical characteristics, HLA-A subtypes, and TIME in overlapped datasets. We didn’t find an impact of HLA-A subtypes on the treatment outcome (Table S4, S5). The prevalence of HLA-A subtypes according to the duration of EGFR TKIs disease control (< 12 months vs. ≥12 months) was consistent (Figure S1). The advanced generation of EGFR TKIs (2nd to 3rd ) provided more benefits in PFS than the 1st generation EGFR TKIs, but not for OS. The multivariate HR of PFS and OS were 0.28 [95% CI: 0.11–0.71, p-value 0.007] (Table S4) and 0.41 [95% CI: 0.12–1.41, p-value 0.158], respectively (Table S5).
Discussion
Human leukocyte antigen (HLA) class I was mandatory for immune recognition and subsequent immune response to cancer-killing. Impaired HLA-I expression is one of the mechanisms of tumor escape34. In the precision medicine era, HLA class I, especially HLA-A, has been correlated with the prevalence of EGFR-mutated NSCLC14 and is prognostic in early-stage EGFR-mutated NSCLC15. Here, we aim to evaluate the effect of HLA-A subtypes in EGFR-mutated NSCLC who received EGFR TKIs, the outcome of treatment, and correlate with the TIME pattern. The HLA-A allele frequencies observed in our study closely resemble those previously reported16. The most common HLA-A subtypes were HLA-A*11:01, -A*33:03, -A*24:02, -A*02:01, -A*02:07, and -A*02:03. To correlate with the binding activity of EGFR mutation subtypes as previously reported15we could define those HLA-A subtypes as interested and uninterested. However, we did not find a correlation between HLA-A categories and the outcome of EGFR TKIs. HLA-A subtypes were not found to be associated with the outcome. The univariate HR of PFS and OS were 0.86 [95% CI: 0.53–1.40, p-value 0.60] and 1.34 [95% CI: 0.74–2.43, p-value 0.30], respectively. However, EGFR exon 19 deletion (80%) was more prominent in the interested HLA-A group than the exon 21 L858R (20%). The 2nd -3rd generation EGFR TKIs, which had more efficacy than the 1st generation, were more applied in the interested HLA-A group (28.6%) than the uninterested HLA-A group (15.7%). The prognostic effect of HLA-A subtypes in early-stage NSCLC was not translated into advanced NSCLC with EGFR-mutated patients who received EGFR TKIs treatment.
Tumor immune microenvironment (TIME) impacts cancer development and progression. Several publications also reported TIME, especially PD-L1 TPS and CD8 + TILs, as prognostic factors in EGFR-mutated advanced NSCLC who received EGFR TKIs treatment4,22. EGFR-mutated NSCLCs were generally associated with low-PD-L1 and CD8 TILs32,33. These were the consequences of EGFR signaling that prevented the recruitment of effector CD8 TILs by down-regulation of cytokines35. The presence of either PD-L1 or CD8 TILs might imply crosstalk between EGFR signaling and other pathways that limit the efficacy of EGFR TKIs treatment. Using our inflammatory TIME definition, PD-L1 ≥ 15% or intra-tumoral CD8 TILs ≥ 10%, the prevalence of inflammatory TIME in our study population was 16.3%, comparable with 13.5% in the previous study using a different definition4. HLA-A subtypes were not correlated with either inflammatory TIME or the presence of intra-tumoral CD8 TILs.
Lastly, we would like to express the study’s limitations. First, most participants received first-generation EGFR TKIs, which are not currently standard treatment. Thailand’s universal coverage, social security coverage, and CSMBS policy were mandatory to provide only first-generation EGFR TKIs treatment in the first-line setting for advanced EGFR-mutated NSCLC. Second, we explored only the HLA-A subtypes, which are part of MHC class I and are correlated with clinical outcome in early-stage disease. Third, the overlapped cohort (n = 49) was lower than our initial expectation, which might represent inadequate power of interpretation. In our study, the authors didn’t find any correlation between specific HLA-A subtypes and pretreatment specimens of CD8 + TILs, tumor PD-L1 TPS, or the clinical outcome of EGFR TKIs. The author cautions against the potential clinical applications of specific HLA-A subtypes and immunotherapy in treatment-naïve advanced EGFR-mutant NSCLC.
Data availability
The HLA-A typing and demographic data supporting this study’s findings are in a supplementary file (Table S6).
Abbreviations
- EGFR :
-
Epidermal growth factor receptor
- EGFR TKIs:
-
EGFR tyrosine kinase inhibitors
- HLA:
-
Human leukocyte antigen
- NSCLC:
-
Non-small cell lung cancer
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- TILs:
-
Tumor-infiltrating lymphocytes
- TIME:
-
Tumor immune microenvironment
References
Sriuranpong, V. et al. High frequency of mutation of epidermal growth factor receptor in lung adenocarcinoma in Thailand. Cancer Lett. 239, 292–297. https://doi.org/10.1016/j.canlet.2005.08.029 (2006).
Dong, Z. Y. et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 6, e1356145 https://doi.org/10.1080/2162402X.2017.1356145 (2017).
Toki, M. I. et al. Immune marker profiling and programmed death ligand 1 expression across NSCLC mutations. J. Thorac. Oncol. 13, 1884–1896. https://doi.org/10.1016/j.jtho.2018.09.012 (2018).
Matsumoto, Y. et al. Impact of tumor microenvironment on the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with EGFR-mutant non-small cell lung cancer. Cancer Sci. 110, 3244–3254. https://doi.org/10.1111/cas.14156 (2019).
Hsu, K. H. et al. PD-L1 strong expressions affect the clinical outcomes of osimertinib in treatment Naive advanced EGFR-mutant non-small cell lung cancer patients. Sci. Rep. 12, 9753. https://doi.org/10.1038/s41598-022-13102-7 (2022).
Rodriguez, J. A. HLA-mediated tumor escape mechanisms that May impair immunotherapy clinical outcomes via T-cell activation. Oncol. Lett. 14, 4415–4427. https://doi.org/10.3892/ol.2017.6784 (2017).
Naranbhai, V. et al. HLA-A*03 and response to immune checkpoint Blockade in cancer: an epidemiological biomarker study. Lancet Oncol. 23, 172–184. https://doi.org/10.1016/S1470-2045(21)00582-9 (2022).
Gupta, R. G., Li, F., Roszik, J. & Lizee, G. Exploiting tumor neoantigens to target cancer evolution: current challenges and promising therapeutic approaches. Cancer Discov. 11, 1024–1039. https://doi.org/10.1158/2159-8290.CD-20-1575 (2021).
Blass, E. & Ott, P. A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 18, 215–229. https://doi.org/10.1038/s41571-020-00460-2 (2021).
Li, F. et al. Rapid tumor regression in an Asian lung cancer patient following personalized neo-epitope peptide vaccination. Oncoimmunology 5, e1238539 https://doi.org/10.1080/2162402X.2016.1238539 (2016).
Li, F. et al. Neoantigen vaccination induces clinical and Immunologic responses in non-small cell lung cancer patients harboring EGFR mutations. J. Immunother Cancer 9 https://doi.org/10.1136/jitc-2021-002531 (2021).
Robinson, J. et al. Distinguishing functional polymorphism from random variation in the sequences of > 10,000 HLA-A, -B and -C alleles. PLoS Genet. 13, e1006862. https://doi.org/10.1371/journal.pgen.1006862 (2017).
So, T. et al. Unfavorable prognosis of patients with non-small cell lung carcinoma associated with HLA-A2. Lung Cancer. 32, 39–46. https://doi.org/10.1016/s0169-5002(00)00204-x (2001).
Uramoto, H. et al. Correlation between HLA alleles and EGFR mutation in Japanese patients with adenocarcinoma of the lung. J. Thorac. Oncol. 5, 1136–1142. https://doi.org/10.1097/JTO.0b013e3181e0b993 (2010).
Dimou, A. et al. HLA class I binding of mutant EGFR peptides in NSCLC is associated with improved survival. J. Thorac. Oncol. 16, 104–112. https://doi.org/10.1016/j.jtho.2020.08.023 (2021).
Satapornpong, P. et al. Genetic diversity of HLA class I and class II alleles in Thai populations: contribution to Genotype-Guided therapeutics. Front. Pharmacol. 11, 78. https://doi.org/10.3389/fphar.2020.00078 (2020).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs working group 2014. Ann. Oncol. 26, 259–271. https://doi.org/10.1093/annonc/mdu450 (2015).
Ngamchokwathana, C. Correlation of tumor microenvironment signature in advanced stage non-small cell lung cancer with EGFR mutation who received EGFR-TKIs. Ann. Oncol. 34 https://doi.org/10.1016/S0923-7534(23)01931-2 (2023).
Schwartz, L. H. et al. RECIST 1.1-Update and clarification: from the RECIST committee. Eur. J. Cancer. 62, 132–137. https://doi.org/10.1016/j.ejca.2016.03.081 (2016).
Marletta, S. et al. Atlas of PD-L1 for pathologists: indications, scores, diagnostic platforms and reporting systems. J. Pers. Med. 12, 1073. https://doi.org/10.3390/jpm12071073 (2022).
Cruz-Rico, G. et al. Association of lung adenocarcinoma subtypes according to the IASLC/ATS/ERS classification and programmed cell death ligand 1 (PD-L1) expression in tumor cells. Pathol. Oncol. Res. 27, 597499. https://doi.org/10.3389/pore.2021.597499 (2021).
Yoneshima, Y. et al. PD-L1 expression in lung adenocarcinoma harboring EGFR mutations or ALK rearrangements. Lung Cancer. 118, 36–40. https://doi.org/10.1016/j.lungcan.2018.01.024 (2018).
bcbio/bcbio-nextgen v1.2.9. (2021).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/bioinformatics/btp324 (2009).
Szolek, A. et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316. https://doi.org/10.1093/bioinformatics/btu548 (2014).
Thuesen, N. H., Klausen, M. S., Gopalakrishnan, S., Trolle, T. & Renaud, G. Benchmarking freely available HLA typing algorithms across varying genes, coverages and typing resolutions. Front. Immunol. 13, 987655. https://doi.org/10.3389/fimmu.2022.987655 (2022).
Fleiss, J. L., Tytun, A. & Ury, H. K. A simple approximation for calculating sample sizes for comparing independent proportions. Biometrics 36, 343–346 (1980).
Wu, Y. L. et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol. 18, 1454–1466. https://doi.org/10.1016/S1470-2045(17)30608-3 (2017).
Yao, Z. H. et al. Real-World data on prognostic factors for overall survival in EGFR Mutation-Positive advanced Non-Small cell lung cancer patients treated with First-Line gefitinib. Oncologist 22, 1075–1083. https://doi.org/10.1634/theoncologist.2016-0331 (2017).
Soria, J. C. et al. Osimertinib in untreated EGFR-Mutated advanced Non-Small-Cell lung cancer. N Engl. J. Med. 378, 113–125. https://doi.org/10.1056/NEJMoa1713137 (2018).
Choi, M. G. et al. Different prognostic implications of hepatic metastasis according to front-line treatment in non-small cell lung cancer: a real-world retrospective study. Transl Lung Cancer Res. 10, 2551–2561. https://doi.org/10.21037/tlcr-21-206 (2021).
Azuma, K. et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann. Oncol. 25, 1935–1940. https://doi.org/10.1093/annonc/mdu242 (2014).
Madeddu, C. et al. EGFR-Mutated Non-Small cell lung cancer and resistance to immunotherapy: role of the tumor microenvironment. Int. J. Mol. Sci. 23 https://doi.org/10.3390/ijms23126489 (2022).
Hazini, A., Fisher, K. & Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother Cancer. 9 https://doi.org/10.1136/jitc-2021-002899 (2021).
Sugiyama, E. et al. Blockade of EGFR improves responsiveness to PD-1 Blockade in EGFR-mutated non-small cell lung cancer. Sci. Immunol. 5 https://doi.org/10.1126/sciimmunol.aav3937 (2020).
Acknowledgements
This project is funded by the Health Systems Research Institute (Thailand) (Grant number 66-153) to NZ, PS, CV. The biospecimen collection was supported by Biobank, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand.
Funding
The Health Systems Research Institute (Thailand) Grant Number 66–153.
Author information
Authors and Affiliations
Contributions
Author Contributions: NZ contributed to the data acquisition, investigation, analysis, and drafted the manuscript. KS contributed to the data acquisition and analysis. SS contributed to investigation and data acquisition. PS contributed to specimen collection and data acquisition. CA contributed to investigation and analysis. CV contributed to the design, conception, analysis of the data, funding acquisition, and drafted the manuscript. PC contributed to the consultation. All authors critically revised the manuscript, agreed to be fully accountable for ensuring the integrity and accuracy of the work, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
This study was approved by the Institutional Review Board of the Faculty of Medicine at Chulalongkorn University (IRB No. 894/63 and 580/66). All participants provided written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zungsontiporn, N., Srianuwattipong, K., Santisukwongchote, S. et al. The correlation of HLA-A in Thai EGFR-mutated advanced non-small cell lung cancer, outcome, and tumor microenvironment. Sci Rep 15, 30989 (2025). https://doi.org/10.1038/s41598-025-16365-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-16365-y