Abstract
The development of more efficient and durable multi-targeted therapeutic drug against hepatocellular carcinoma (HCC) has recently been of growing interest to tackle chemoresistance. Several studies indicate that increased expression of methyltransferase-like (METTL) proteins, including METTL1, METTL3, METTL6, METTL16, and METTL18, are associated with the progression of HCC malignancy, making them potential biomarkers. Here, using a series of computer-aided drug design (CADD) approaches, we identified two first-in-class highly potent catalytic multi-target inhibitors (ZINC70666503 and ZINC13000658 with 87% and 82% predicted drug scores, respectively) of these five methyltransferase-like proteins. The molecular dynamics study supported their conformational stability with these METTL proteins and high selectivity at the pocket of proteins’ adenosine moiety of S-Adenosyl Methionine. Further in vitro experiments revealed significant anti-proliferative activity and effects on the cell cycle of ZINC13000658 against two HCC cell lines, HepG2 and SNU-449. This work provides evidence that multitargeted METTL may have stronger inhibition of HCC cell proliferation. Further in vivo validation, toxicity analysis as well as molecular insights will determine the therapeutic utility against HCC.
Similar content being viewed by others
Introduction
Being the fourth leading cause of cancer-related deaths, hepatocellular carcinoma (HCC) is one of the deadliest human malignancies worldwide. Infection of Hepatitis B and C virus, diabetes, dietary toxins, heavy alcohol intake, and nonalcoholic fatty liver disease are the major risk factors for HCC1,2,3. The molecular mechanism of hepatocellular carcinoma initiation and progression are still elusive, limiting proper treatment strategies for patients with advanced HCC4. However, in recent years, researchers have identified several intracellular molecules as biomarkers to detect HCC progression and developed anti-HCC drugs5,6. Until now, the Food and Drug Administration (FDA) has approved several anti-cancer agents for treating HCC, such as Sorafenib and Lenvatinib7. Nevertheless, these drugs or other classical chemotherapeutic agents, tyrosine kinase inhibitors, and novel immune-sensitizing strategies failed to exert an efficient and durable response in HCC8,9.
Recent studies have revealed that several members of the human Methyltransferase-like (METTL) protein family play critical roles in hepatocellular carcinoma (HCC) progression10. These proteins contain a conserved methyltransferase domain with an S-adenosyl methionine (SAM) binding site, enabling them to catalyze methyl group transfer to RNA, DNA, proteins, and other biomolecules. METTL1, for example, promotes HCC cell proliferation and migration by suppressing PTEN signaling11,12, while METTL3 influences glycolytic activity and metastasis via the UBC9/SUMOylated METTL3/Snail axis13,14. METTL6 and METTL18 are upregulated in HCC and contribute to tumor cell proliferation, invasion, and migration15,16. METTL16, although reported as a favorable prognostic marker, has also been implicated in m6A methylation and transcriptomic regulation in HCC17. Similarly, METTL5 and METTL9 overexpression in HCC is associated with poor survival outcomes, influencing tumor progression18,19. These findings highlight the METTL family as both biomarkers and potential therapeutic targets in HCC.
Despite these insights, no existing therapies selectively inhibit the catalytically active (SAM-binding) domains of these METTL proteins, apart from METTL3-specific inhibitors such as STM2457 and STC-1520,21. Due to the success in targeting SAM-dependent methyltransferases in microbial systems and in DNA methyltransferases for cancer therapy22,23,24,25, we hypothesized that SAM-competitive inhibition of multiple overexpressed METTL proteins could offer a novel therapeutic avenue for HCC. To explore this, we employed a computer-aided drug design (CADD) pipeline incorporating pharmacophore modeling, virtual screening, molecular docking, DFT analysis, and molecular dynamics simulation—followed by experimental validation—to identify multi-target METTL inhibitors with favorable pharmacological properties. Computational drug screening plays a vital role in modern drug discovery by enabling the rapid, cost-effective prioritization of lead compounds with high target affinity and favorable pharmacokinetic properties. This is especially valuable in oncology, where the complexity of tumor biology demands highly selective and mechanism-based inhibitors. By narrowing down potential candidates before in vitro and in vivo testing, CADD accelerates early-stage therapeutic development and increases the translational potential of candidate drugs. Hence, we considered studying a set of SAM-dependent microbial RNA methyltransferase inhibitors, human DNMT inhibitors, and all identified chemical inhibitors of METTL3 to identify novel SAM-competitive small molecule inhibitors for developing anti-HCC therapeutics; a novel perspective of drug repurposing26,27,28,29,30,31,32. Furthermore, we evaluated in vitro inhibitory effects of the identified drugs on hepatocellular carcinoma cells proliferation.
Materials and methods
Survival analysis of HCC patient data
Survival analysis was conducted using TCGA and Human Protein Atlas patient data to assess the prognostic significance of METTL gene expression levels. Clinical information, including age, sex, race, cancer stage, survival status, and follow-up time, was extracted and preprocessed. Gene expression was categorized into “High” and “Low” groups based on a TPM (Transcripts per Kilobase Million) cutoff. Kaplan–Meier (KM) survival curves were generated using the survival (Surv()) and survminer (ggsurvplot()) R packages, with statistical significance evaluated by log-rank tests (p < 0.05). Gene expression profiles were visualized through TPM distribution graphs, providing insights into expression group distinctions. All analyses and visualizations were performed in R.
Retrieval of METTL proteins and preparation of drug compounds
The crystal structure of the methyltransferase-like protein METTL1 [PDB ID: 7PL1], METTL3-14 complex [PDB ID: 7O2I], METTL6 [PDB ID: 7F1E], METTL16 [PDB ID: 6B91], and METTL18 [PDB ID: 4RFQ] with the resolution of 1.85 Å, 3.00 Å, 2.59 Å, 1.94 Å, and 2.40 Å, respectively, were retrieved from the RCSB Protein Data Bank (PDB)33. Furthermore, the three-dimensional (3D) PDB structures of 39 microbial tRNA methyltransferase inhibitors, 20 DNA methyltransferase inhibitors, and 14 METTL3 small molecule inhibitors were retrieved from the PubChem database (Supplementary Table 1)34.
Receptor-based screening of drug compounds and active sites identification
Using AutoDock Vina software for molecular docking approaches, selected compounds were screened against all METTL proteins35. At first, we processed the crystal structure of all METTL proteins by separating the ligand molecules, removing the water and other complex molecules, and adding polar hydrogen by AutoDock Vina44. After preparing the PDB structures of the drug candidates, the highest binding affinity and interactive amino acids were assessed by exposing them to all five proteins. The grid boxes with box shape-size (x, y, z) and shape-center (x, y, z) were set for METTL1, METTL3, METTL6, METTL16, and METTL18 (Supplementary Table 2), according to the proteins’ binding sites taken from UniProtKB for molecular docking36. PyMOL was further used to analyze all protein–ligand interactions and visualize the interaction sites (http://www.pymol.org/pymol). Finally, based on the highest binding energy, we selected the top compounds for each protein and analyzed the protein–ligand complexes to identify interacted residues by Protein–Ligand Interaction Profiler (PLIP)37. Additionally, the non-covalent interactions (hydrogen bonds, water bridges, salt bridges, halogen bonds, hydrophobic interactions, π-stacking, π-cation interactions, and metal complexes) in those protein–ligand complexes were also analyzed by PLIP.
Quantum mechanical (QM) calculation
The electronic properties of drug molecules are of significant interest in drug designing, which can be effectively studied through Density Functional Theory (DFT) analysis38. In this investigation, the DFT calculation was implemented by employing B3LYP (Becke exchange functional, which combined Lee, Yang, and Parrs (LYP) correlation functional)39. For single-point DFT analysis by ORCA version 4.2.1, top inhibitors with the highest molecular docking score were treated with B3LYP and RIJCOSX approximation after minimal geometry optimization by Avogadro 1.2.0n40,41,42. In this calculation, we analyzed frontier molecular orbitals, namely highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), their energy gap difference, and chemical potentials. Then, the frontier energies (ε) of HOMO and LUMO were used to measure the hardness and softness of selected compounds. The hardness, η and softness, S of the drugs were measured by the Parr and Pearson interpretation equation and Koopmans theorem equation43,44. Finally, we used IboView to visualize and calculate the HOMO and LUMO of the compounds. From this, we can evaluate an atom’s ability to receive electrons and higher reactivity, indicating a higher softness value. The following equations measure chemical hardness and softness. Hardness (η) = (I − A)/2; Softness (S) = 1/η. In these equations, ‘I’ refer to the ionization potential (− EHOMO) and ‘A’ denotes the electron affinity (− ELUMO).
Pharmacophore modeling and virtual screening of the ZINC database
Pharmacophore features modeling was done by using PharmaGIST, that are essential for the interaction of inhibitors with METTL proteins45. This study used the topmost inhibitors to generate the pharmacophore model. Then, the generated pharmacophore model was imported to ZINCPharmer, and a library of chemical compounds was created by screening the ZINC database to carry out further virtual screening46,47. Finally, a virtual screening of the library of compounds against all studied HCC-associated METTL proteins was conducted by AutoDock Vina to find the best drug candidates.
Drug likeness properties analysis of the candidate small molecule inhibitors
Absorption, Distribution, Metabolism, and Excretion (ADME) properties were assessed by the SwissADME server to successfully evaluate the physiochemistry, lipophilicity, water solubility, drug-likeness, and pharmacokinetics properties of potential drug candidates47. SwissADME computed the physicochemical descriptors (Formula, Molecular weight, Molar Refractivity, TPSA), lipophilicity (Log Po/w (iLOGP), Log Po/w (XLOGP3), Log Po/w (WLOGP), Log Po/w (MLOGP), Log Po/w (SILICOS-IT), Consensus, Log Po/w), pharmacokinetic parameters (GI absorption, CYP1A2 inhibitor, CYP2C19 inhibitor, CYP2C9 inhibitor, CYP2D6 inhibitor, CYP3A4 inhibitor, Log Kp; skin permeation) and water solubility (Log S: SILICOS-IT, Solubility). Besides, Blood Brain Barrier (BBB) permeability was also computed by adopting the BOILED-Egg yolk method for the screened compounds. Additionally, admetSAR was used to investigate the undesired effects of toxicity, and bioavailability48. Finally, drug likeliness and drug score as well as tumorigenicity, mutagenicity, and reproductive effects were assessed by OSIRIS Property Explorer (https://www.organic-chemistry.org/prog/peo/).
Evaluation of the docking performance
We further evaluated our AutoDock Vina docking results of final drug candidates against studied proteins using two popular online docking servers, SwissDock and CB-Dock, followed by site-specific re-docking by AutoDock Vina49,50. SwissDock can automatically set up the protein and ligand structures and provide convenient visualization and analysis of docking predictions. On the other hand, CB-Dock can automatically predict binding modes and binding sites of a given protein and calculate the centers and sizes with a novel curvature-based cavity detection approach. Thereby, it becomes easier to predict proteins’ binding site cavities where they interact with the inhibitors. For site specific re-docking by AutoDock Vina, the grid box size (x, y, z) and centre (x, y, z) was set as 27 Å × 30 Å × 27 Å and 46 Å × − 20 Å × 6 Å; 27 Å × 27 Å × 27 Å and − 43 Å × 31 Å × 26 Å; 27 Å × 27 Å × 27 Å and 39 Å × − 25 Å × 15 Å; 27 Å × 27 Å × 27 Å and − 7 Å × 8 Å × 20 Å; and 27 Å × 27 Å × 27 Å and 6 Å × 8 Å × 28 Å for METTL1, METTL3, METTL6, METTL16, and METTL18, respectively. After re-docking with three different docking tools, we further selected the best conformations of final drug candidates for interaction analysis by PLIP.
To further investigate the competitive binding potential of selected compounds with SAM at the active site of METTL3 (PDB ID: 7O2I), we conducted an additional series of structure-based molecular docking experiments using the CB-Dock server. Specifically, we examined two lead ZINC compounds to assess their binding affinities and interaction profiles relative to SAM. First, each compound was docked individually into the METTL3 binding pocket to predict the most favorable binding modes and binding energies. Subsequently, competitive binding simulations were performed in two directions: (i) the ZINC compounds were docked into the SAM-bound METTL3 conformation, and (ii) SAM was docked into the METTL3 conformations pre-bound with two ZINC compounds. This approach allowed us to examine the mutual influence of ligand binding on site occupancy and interaction energetics. Binding affinities were recorded, and protein–ligand interaction profiles were visualized.
Finally, to evaluate the selectivity and broader inhibitory potential of the two lead compounds, we conducted molecular docking against additional SAM-dependent methyltransferases, including human DNA (cytosine-5)-methyltransferase 1 (DNMT1) (PDB ID: 7SFG), Catechol O-methyltransferase (COMT) (PDB ID: 3BWY), and the viral methyltransferase (2'-O-methyltransferase) from SARS-CoV-2 (PDB ID: 7JIB). CB DOCK was used for all docking runs. Binding energies were recorded, and key residue interactions were visualized and analyzed.
Molecular dynamics simulation
Molecular Dynamics (MD) is an important step for evaluating the stability of the receptor-ligand complex because trajectories obtained from molecular dynamics simulation could be used to predict the stability of the protein-inhibitors complexes in an aqueous solution by analyzing the best docked pose predicted through the docking analysis51. Hence, 100 ns MD simulation was employed to evaluate the binding stability of the final two compounds to all studied METTL proteins at their active site cavity. GROMACS software suit was used for the simulation with GROMOS96 43a1 forced field for 100 ns52. We used the PRODRG web server for the preparation of the ligand and generation of ligand topology53. The 5000 steps steepest descent method was used for the minimization process and SPC was selected as water model for correct density and dielectric permittivity of water. The approximate number of frames per simulation was set to 1000 and NVT/NPT equilibration system was operated under 300 K temperature and 1 bar pressure. Additionally, during molecular dynamics simulation study, the ligand-receptor root mean square deviation (RMSD), RMS fluctuation, number of hydrogen bonds, and radius of gyration (Rg) were also calculated to determine the stability and compactness of protein inhibitor complexes. We further performed the MM/GBSA (molecular mechanics, the generalized born model, and solvent accessibility) analysis by using Linux operating system to calculate the ligand binding free energies and ligand strain energies for docked lead compounds with METTL proteins54.
In vitro HCC cell proliferation assay
The human hepatocellular carcinoma cell line HepG2, and SNU-449 were purchased from the American Type Culture Collection (Manassas, Virginia). ZINC70666503 (OSSL_860087) and ZINC13000658 (CSSS00027042412) were purchased from Princeton BioMolecular Research, Inc., USA and Chemspace, respectively and prepared in dimethyl sulfoxide (DMSO) at 100 mmol/L and stored in aliquots in − 80 °C. HepG2 and SNU-449 cell lines were cultured in DMEM and RPMI-1640 culture media, respectively, containing 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2. After reaching approximately 80% confluency, the cells were harvested using trypsin–EDTA, counted, and seeded into 96-well plates at a density of 7000 cells per well. Following a 24-h incubation period to allow cell attachment and stabilization, the cells were treated with various concentrations of ZINC70666503 and ZINC13000658. The working solution of the drug was prepared using appropriate culture media instead of DMSO to avoid potential toxicity, and the control group was treated with culture media alone to ensure accurate comparison. The IncuCyte Live-Cell Analysis System (Sartorius) was used to monitor cell proliferation in real-time. Cell proliferation was measured up to 116 h as cell count normalized to day zero to determine the effects of the drugs on cell proliferation and graphs were prepared using GraphPad prism (for all groups, n = 4). To assess cell cycle dynamics in SNU-449, treated cells were harvested after 48 h, stained with propidium iodide (PI), and analyzed via flow cytometry. Apoptotic effects were further investigated by staining the nuclei with Hoechst 33,342 at a concentration of 2 µg/mL after 48 h of treatment, followed by imaging with an Olympus ApexView APX100 fluorescence microscope with 40 × objective. Furthermore, to evaluate the anti-proliferative effects of ZINC70666503 and ZINC13000658 beyond hepatocellular carcinoma models, two prostate cancer cell lines 22Rv1 and VCaP; purchased from the American Type Culture Collection (Manassas, Virginia) were selected based on their METTL gene expression profiles. Cells were treated with each compound at indicated concentrations, and real-time proliferation monitoring was performed using the IncuCyte live-cell imaging system over 92 h. Moreover, gene expression data (nTPM values) for METTL family members across HepG2, SNU-449, VCaP, and 22Rv1 cell lines were obtained from the Human Protein Atlas (HPA) database. The heatmap was generated using these normalized transcript levels to visualize differential METTL expression profiles across cancer types. For statistical analysis, we employed One way Anova to assess differences between groups, considering P values < 0.05 as statistically significant. The statistical analysis was conducted using GraphPad Prism 10.4.
Results
Prioritization of prognostic METTL proteins for therapeutic targeting
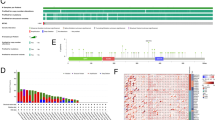
Survival analysis of all protein members of the METTL gene family was performed using patient data obtained from The Cancer Genome Atlas (TCGA) and Human Protein Atlas (HPA) databases. TCGA data indicates that several METTL genes, including METTL1, METTL2A, METTL4, METTL5, METTL6, METTL8, METTL9, METTL18, and METTL23, are associated with hepatocellular carcinoma (HCC) outcomes and are highly expressed in tumor tissues (Fig. 1A,B). Among these, METTL23, METTL3, METTL6, METTL5, and METTL8 were validated as unfavorable prognostic markers in HPA, whereas METTL16, METTL2A, METTL22, METTL14, METTL15, METTL7A, METTL7B, and METTL25B were predicted as favorable markers (data not shown). To shortlist candidate METTL proteins for downstream structural and inhibitor-binding analysis, we considered both biological relevance and structural feasibility. Specifically, five METTL proteins— includes both validated (METTL3, METTL6) and predicted (METTL1, METTL18) unfavorable prognostic markers and METTL16 as a favorable prognostic marker—were prioritized based on the following criteria: (i) their previously reported involvement in HCC progression and oncogenic pathways11,12,13,14,15,16,17,18,19, and (ii) the availability of high-resolution crystal structures (≤ 2.0 Å) with defined SAM-binding pockets in the RCSB Protein Data Bank. High-resolution crystal structures are essential for reliable molecular docking, pharmacophore modeling, and molecular dynamics simulations. Although METTL2A, METTL4, METTL5, and METTL9 showed prognostic significance in the survival analysis, they were excluded from further analysis because of their lack of experimentally solved 3D structures in PDB with complete or well-defined SAM-binding domains, which is crucial for accurate in silico modeling; and limited literature evidence linking these METTLs; particularly METTL2A directly to HCC biology, making their functional relevance less substantiated for this specific cancer context. This strategic selection allowed us to focus on METTL proteins that are not only prognostically relevant but also structurally tractable and biologically implicated in HCC, ensuring both clinical and computational feasibility for identifying potential SAM-competitive inhibitors targeting diverse prognostic profiles.
Survival analysis of METTL Gene Family Members in hepatocellular carcinoma. (A) Kaplan–Meier survival curves represent the overall survival probability for patients with low versus high expression of METTL family genes (METTL1, METTL2A, METTL3, METTL4 METTL5, METTL6, METTL8, METTL9, METTL18, and METTL23) in hepatocellular carcinoma. High expression groups are shown in red, while low expression groups are shown in blue, with the number of samples indicated in parentheses. The p-values denote statistical significance based on log-rank tests. The data were obtained from The Cancer Genome Atlas (TCGA) and Human Protein Atlas databases. (B) The boxplot depicts gene expression profile for each gene in this cancer type.
Structural insights into inhibitor binding hotspots of METTL proteins
Based on the scoring function of AutoDock Vina, we gained insight into the structural and atomistic scale interactions involving the binding modes of five METTL proteins and the 73 retrieved inhibitors. Remarkably, the maximum listed inhibitors showed good binding affinity and lower binding energy than SAM against all studied METTL proteins, the catalytic activator of methyltransferase proteins (Supplementary Table 3). We found CID76318201, a DNMT inhibitor having the highest negative binding energy (− 12.6 kcal/mol) when interacting with the METTL6. Moreover, CID135449332 (− 10.7 kcal/mol), CID5494506 (− 10.7 kcal/mol), CID344265 (− 11.5 kcal/mol) were also found to be the topmost METTL1, METTL3, and METTL18 inhibitors with high binding affinities, respectively.
Molecular docking and studying drug-binding active site have been substantially used in the drug discovery process to predict binding site complementarity between a therapeutic target and a drug ligand and to understand the molecular interaction between the target and drug candidates55. To identify these five METTL proteins’ drug binding active sites, we analyzed the protein–ligand interactions at the docked sites of proteins for all 73 compounds and analyzed by PyMOL, and protein–ligand interaction profiler (PLIP). To evaluate the conserved interaction patterns across METTL family members, we analyzed the top five compounds ranked by binding affinity from our molecular docking studies for each METTL protein (Supplementary Table 4). For each protein, a set of recurrent interacting residues was identified, forming what we define as the drug binding cavity, representing the consistent binding environment targeted by multiple compounds. For example, in METTL1, residues such as 53I, 56G, 57Y, 77E, 79R, 110N, 111A, 112M, 129F, 130L, 131F, 132P, 133D, 135H, 136F, 143W, 144R, 145I, 208T, 209E, and 210E were repeatedly involved in ligand interactions. These align closely with the SAM-binding residues, particularly around the adenosine-binding pocket (Fig. 2). Similarly, METTL3 exhibited a high degree of conservation among interacting residues across its top binders. Importantly, we specifically highlighted in each ligand-binding complex those residues that are also involved in SAM binding, allowing direct comparison between compound binding and the natural cofactor interaction (Supplementary Table 4). This overlap further confirms that the ligands target the same functional pocket as SAM. Furthermore, analysis of the METTL18-inhibitors complexes showed that the binding of the ligands was stabilized through the non-covalent bonds with the active site residues, mostly, 89E, 90P, 172T, 216Q, 218Y, 270W, 295T, and 297Y. These drug binding cavity residues for each METTL protein were then used as targeted regions in subsequent docking analyses, enabling more focused and accurate evaluation of ligand binding within the conserved catalytic pocket. This approach reinforced the hypothesis that the identified compounds interact specifically with the shared SAM-binding cleft, supporting their potential as multi-METTL inhibitors with a conserved binding mode. The binding modes of energetically top five inhibitors inside the active sites of respective METTL proteins are depicted in Fig. 2, and the interacting binding site residues are given in Supplementary Table 4. As seen from data listed in Supplementary Table 3, 12 inhibitors showed average docking scores > -9.0 kcal/mol and interacted at the METTL proteins’ identified active site cavities.
Structural insights into inhibitor binding hotspots of METTL proteins. Structural analysis of active site of Methyltransferase-like 1, 3, 6, 16, 18 (METTL1, METTL3, METTL6, METTL16, and METTL18) protein (left) and active site amino acids residues (right). Superimposed top five compounds that showed highest binding affinity specifically bound at the active site cavity of METTL1 by interacting with the amino acids depicted in right panel. Here, superimposed top 5 drug compounds for each protein are depicted in lemon and interacted amino acids are in sky blue lines.
Generated pharmacophore model paved the way to predict SAM-competitive METTL inhibitors
Multi-target compounds need to have comparatively higher chemical reactivity, chemical potential, and softness to interact with multiple target proteins56. To identify drug compounds with more chemical reactivity profiles, we chose energetically top inhibitors from molecular docking study for doing DFT calculations, particularly frontier molecular orbitals energy and further pharmacophore modeling. In the DFT calculations, we calculated the hardness and softness values of 12 energetically top inhibitors and also their chemical potential (Supplementary Table 5). After calculating the softness and chemical potentiality from EHOMO-ELUMO values, we had five compounds (2 tRNA methyltransferase inhibitors, 2 DNMT inhibitors, and 1 METTL3 inhibitor) with softness values of greater than 0.6 eV (Fig. 3A), suggesting the compounds’ considerably higher chemical instability and high chemical reactivity in compared to others. CID139030487 generated the lowest HOMO–LUMO gap (HLG) with the highest softness value of 0.69, where CID135403798, CID344265, CID24858111, and CID155167581 gave softness value of 0.65eV, 0.63eV, 0.63eV, 0.60eV, and 0.65eV respectively.
Generated pharmacophore model paved the way to predict SAM-competitive inhibitors. (A) Frontier molecular orbital analysis of 5 compounds with the highest softness and chemical potential. HOMO (Highest occupied molecular orbitals) are in green while LUMO (Lowest unoccupied molecular orbitals) are in blue. (B) Generated ligand structure-based pharmacophore model which was used for virtual screening of ZINC database with 230 million compounds. The generated model has a four aromatic ring structure (violet). (C) Molecular docking results of top scored ZINC compounds with five METTL family proteins. Chemical structure of (D) ZINC70666503, and (E) ZINC13000658.
Next, to identify novel multi-METTL protein targeted drug candidates, at first, we selected these top five inhibitors with the highest frontier molecular orbital properties for generating a ligand structure-based pharmacophore model and then screened the ZINC database. The generated pharmacophore model has four aromatic groups. (Fig. 3B). Aromatic groups enhance drug binding by facilitating π–π and hydrophobic interactions with target proteins, improving affinity and specificity. Then, the model was imported to ZINCPharmer to virtually screen the ZINC database. A maximum of 0.2 Å RMSD, 5 rotation bonds, and molecular weight between 350 and 550 from sphere centers were set as input parameters for ZINCPharmer, and a total of 483 hits was retrieved to make a ligand library for further molecular docking-based screening. This ligand library was further screened against all five METTL proteins by AutoDock Vina tools to select the candidate compounds. After shortlisting the compounds according to the highest binding affinity score, we checked the binding pattern and interaction with individual METTL proteins by PyMOL to finalize the list of compounds that interact at the protein’s drug-binding catalytic active sites. Molecular docking and subsequent protein–ligand interaction site analysis demonstrated 15 drug candidates with energetically top-scored (average binding energy > − 10.0 kcal/mol) for targeting METTL1, METTL3, METTL6, METTL16, and METTL18. Interestingly, further protein–ligand interaction analysis by PyMOL indicated that 13 out of above-mentioned 15 potent compounds have considerably occupied the catalytic SAM binding pockets of all METTL proteins. Among these 13 compounds, ZINC70666503 demonstrated the highest binding energy (13.1 kcal/mol) when interacting with METTL18 (Supplementary Table 6). Intriguingly, the binding affinity of this compound to other METTL proteins was also notable. Surprisingly, most of the 13 compounds exhibited the highest binding affinity while interacting with METTL18 (Fig. 3C). The docking results of all 483 compounds from the ZINC database are shown in Supplementary Table 6.
Two compounds are predicted as safe METTL targeted drug candidates with SAM-competitive binding patterns
To identify promising drug candidates with favorable pharmacokinetic profiles and minimize the risk of late-stage failures, in silico physicochemical properties, lipophilicity, water-solubility, and drug-likeness of 18 prospective drug candidates (CID139030487, CID135403798, CID344265, CID24858111, CID155167581 and 13 top-scored ZINC compounds) were evaluated by SwissADME (Supplementary Table 7). Among them, 3 compounds violated the Lipinski rule of five and showed undesired physiochemical properties in the case of molecular weight (MW), molecular refractivity (MR), and topological polar surface area (TPSA), which are considered as very important properties of a drug compound. Moreover, OSIRIS Property Explorer and admetSAR predicted the toxic and immunotoxic effects as well as cytochromes P450 (CYPs) isoforms inhibition of the several compounds. After initial ADMET properties screening, nine potential drug compounds with no predicted adverse effects were shortlisted (details are included in Supplementary Table 7). Finally, drug score prediction by OSIRIS Property Explorer predicted ZINC70666503 (Fig. 3D) and ZINC13000658 (Fig. 3E) as the top drug candidates, with drug scores of 87% and 82%, respectively. Additionally, ZINC70666503 has the blood barrier permeate ability with no AMES toxic effects (Table 1). Conversely, ZINC13000658 might be AMES toxic but not a blood–brain barrier permeable one which is important for drugs with a peripheral target as minimize BBB penetration ability might be required to reduce the possibility of undesired pharmacological events and avoid CNS side effects57.
Subsequently, site-specific protein–ligand re-docking of these final two drug compounds and CID155167581 (STM2457) against all METTL proteins with different docking server indicated mostly similar results to the previous results with AutoDock Vina. In both AutoDock Vina and CB-Dock tools, the binding affinity score was found to be almost the same for all three compounds with previous docking results (Supplementary Table 8). In most cases, ZINC70666503 and ZINC13000658 had higher binding affinity than CID155167581, SAM competitive inhibitor of METTL3. Among our top 2 drug candidates, ZINC70666503 showed the lowest binding energy and the highest binding affinity to all METTL proteins (Supplementary Table 8). After docking, we identified the residues involved in the interaction and surprisingly found that both ZINC70666503 and ZINC13000658 interacted at the SAM binding catalytic domain of every METTL protein. As can be seen in Fig. 4A,B, the interacting binding sites are at the interface of the studied METTL protein’s active site residues demonstrated earlier (Fig. 2). Most surprisingly, binding site residues of the interacting sites of proteins-inhibitors complexes revealed that both ZINC70666503 and ZINC13000658 interacted with the amino acid residues that are the primary residues responsible for SAM binding at respective proteins’ active sites (Supplementary Table 9). Additionally, competitive docking analysis demonstrated that both ZINC13000658 and ZINC70666503 exhibit stronger binding affinities to METTL3 (− 10.3 and − 9.5 kcal/mol, respectively) compared to SAM (− 8.5 kcal/mol) (Supplementary Fig. 1A). When docked into SAM-bound METTL3, the compounds showed moderately reduced affinities (− 7.7 and − 7.9 kcal/mol), indicating partial steric interference. In contrast, SAM’s binding affinity dropped substantially when docked into METTL3 pre-bound with either ZINC compound (− 5.0 and − 5.3 kcal/mol), suggesting that these ligands impair SAM access to its canonical binding site (Supplementary Fig. 1B). These observations support a competitive binding mechanism, whereby ZINC13000658 and ZINC70666503 effectively displace or block SAM from engaging its functional binding pocket on METTL3.
Two compounds are predicted with SAM-competitive binding patterns. Structural representations of the binding sites and interactions of METTL proteins with (A) ZINC70666503 and (B) ZINC13000658. The ribbon diagrams highlight secondary structural elements, with helices in pink, beta-sheets in yellow, and loops in white. Key amino acid residues involved in binding are labeled and shown in stick representation, with hydrogen bonds and other interactions indicated by dashed lines. The ligands are represented in ball-and-stick models, and the specific residue-ligand interactions are annotated. Both compounds are predicted to bind within the SAM catalytic pocket, forming critical interactions that could competitively block SAM and inhibit METTL methyltransferase activity.
Moreover, docking analysis also revealed that both ZINC13000658 and ZINC70666503 bound favorably to the SAM-binding sites of DNMT1, COMT, and SARS-CoV-2 nsp10/nsp16 2’-O-methyltransferase, with binding energies comparable to those observed for METTL family proteins (Supplementary Fig. 2). Both compounds occupied the conserved SAM-adjacent pocket and engaged with catalytically relevant residues, suggesting a shared binding mode across different SAM-utilizing enzymes (Supplementary Table 10). However, their binding affinities were generally lower than those observed for SAM itself, in contrast to the METTL family proteins where these inhibitors showed comparable or stronger binding energies relative to SAM. This suggests a preferential inhibitory effect toward METTLs, though some degree of cross-reactivity remains as well as a potential for broader methyltransferase inhibition, highlighting the chemical scaffolds’ utility beyond METTL targeting.
Molecular Dynamics Simulation and MM/PB(GB)SA analysis from post molecular dynamics trajectory suggest strong stability of ZINC70666503 and ZINC13000658
Although molecular docking is a widely used technique, reliable predictions for binding affinities, drug-receptor intermolecular interactions, solvent effects, dynamics must be considered to strengthen in silico study. Therefore, molecular dynamics simulation for the METTL1, METTL3, METTL6, METTL16, and METTL18 in complex with ZINC70666503 and ZINC13000658 was performed to determine the stability and rigidity of protein–ligand complexes in a specific artificial cellular environment at nanosecond scaled. Root mean square deviation (RMSD) of METTL proteins and ZINC70666503 showed a constant binding pattern and no significant fluctuation during the 100ns time frame. However, a slight fluctuation was observed for RMSD of METTL6 near 70 ns when complexed with ZINC13000658. However, primarily stable conditions with no dramatic inconstancy had been observed in the RMSD of all METTL proteins in complex with these two inhibitors (Fig. 5A).
Molecular Dynamics Simulation and MM/PB(GB)SA analysis from post molecular dynamics trajectory. (A) RMSD plots show stable binding conformations of ZINC70666503 and ZINC13000658 across METTL proteins over 100 ns, with minimal fluctuation, indicating persistent binding—except for moderate deviation in the METTL6 complex. (B) RMSF analysis indicates limited residue-level fluctuations, supporting conformational stability. (C) Radius of gyration analysis reveals high compactness of the protein–ligand complexes. (D) Binding free energy analysis by MM/PB(GB)SA demonstrates strong and favorable interaction energies (> − 300 kcal/mol) for most complexes for over the 100 ns simulation time, suggesting that both compounds, particularly ZINC13000658, maintain energetically stable interactions and could function as effective METTL inhibitors under physiological conditions. ESE: Electrostatic energy; VDW: Van-der Waals contribution.
Besides, flexibility estimation of protein and ligand complex by RMSF indicated that the fluctuations were confined in the 1 nm range, which indicates higher stability (Fig. 5B). On the other hand, the lower Rg value indicates high compactness, and the larger value evidences the dissociation of the inhibitors from the protein. For both ZINC70666503 and ZINC13000658, lower Rg values were also measured (Fig. 5C). Moreover, H-bond estimation during this simulation time showed ZINC13000658 had a maximum number of 7 hydrogen bonds when interacting with METTL18 (Supplementary Fig. 3). This ligand also exhibited a more significant number of interacting H-bond numbers than ZINC70666503 with other METTL proteins. An average of 4 hydrogen bonds formed during METTL protein’s interactions with ZINC13000658 and two hydrogen bonds between METTL proteins and ZINC70666503 interactions (Supplementary Fig. 3). Finally, we adopted MM/PB(GB)SA analysis to calculate the ligand-binding free energy to the desired protein by calculating electrostatic energy (ELE), Van der Waals contribution (VDW), and total binding energy. This energy calculation showed that the value of total binding energy in the METTL18 in complex with ZINC70666503 is − 355.124 kcal/mol, which is higher than the other complexes for this compound (Fig. 5D). On the other hand, ZINC13000658 had a value of − 388.081 kcal/mol when complexed with METTL6. Most importantly, except for that of the METTL1-ZINC70666503 complex, most of the protein–ligand complex arguably showed significant total binding energy (> − 300 kcal/mol) for over the 100 ns simulation time.
ZINC13000658 inhibits HCC cells proliferation in vitro
To validate in silico findings and evaluate the in vitro effects of ZINC70666503 and ZINC13000658, a cell proliferation assay was performed on two hepatocellular carcinoma cell lines, HepG2, and SNU-449 using IncuCyte (Supplementary Fig. 4A & 4B). ZINC13000658 demonstrated a dose-dependent inhibition of cell proliferation in both cell lines (Fig. 6A,B and Supplementary Fig. 4A). Specifically, the IC50 values for ZINC13000658 were 5.632 μM for HepG2 cells and 6.184 μM for SNU-449 cells, and significant inhibition was observed at 4 μM for HepG2 and 6 μM for SNU-449 cells (Supplementary Fig. 4C) indicating high antiproliferative activity. Additionally, phase-contrast imaging revealed a marked reduction in cell confluency upon treatment with ZINC13000658, demonstrating significant growth inhibition in HepG2 and SNU-449 cells (Fig. 6C & Supplementary Fig. 4D & 4E). Flow cytometric analysis of the cell cycle showed that control cells predominantly maintained a typical G1 phase population, whereas increasing concentrations of the compounds induced G1 cell cycle arrest, as evidenced by a pronounced shift in peak frequencies (Fig. 6D). Furthermore, Hoechst staining highlighted distinct nuclear morphological changes in treated cells, including nuclear fragmentation, which is a characteristic of apoptosis (Fig. 6E). These analyses were performed at 48 h, as this time point demonstrated the onset of statistically significant inhibition, making it ideal for detecting early changes in the cell cycle and apoptosis. In contrast, ZINC70666503 did not show any observable inhibition of cell proliferation in either HepG2 or SNU-449 cells while treating cells up to 100 μM, suggesting a lack of efficacy of this compound in these models (Supplementary Fig. 4B). However, these findings collectively indicate the therapeutic potential of ZINC13000658 by inhibiting proliferation and inducing cell cycle arrest and apoptosis in HCC cells. Additionally, to assess the anti-proliferative potential of ZINC70666503 and ZINC13000658 beyond hepatocellular carcinoma, we treated two aggressive prostate cancer cell lines (22Rv1 and VCaP) with a similar METTL profile observed in HCC and monitored real-time cell proliferation using the IncuCyte imaging system. As shown in Supplementary Fig. 5A and B (left panels), 5 μM conc ZINC13000658 markedly inhibited proliferation in both cell lines compared to vehicle controls (Supplementary Fig. 5A and B, middle panels). Representative IncuCyte images (right panels) further illustrate the reduced cell density under treated conditions.
ZINC13000658 inhibits proliferation of liver cancer cells in in vitro. (A,B) Cell proliferation assays in SNU-449 and HepG2 cells show a clear dose-dependent inhibitory effect of ZINC13000658, with IC50 values determined from fitted curves (n = 4), indicating strong anti-proliferative activity. (C) Phase-contrast microscopy displays reduced confluency upon compound treatment, consistent with impaired growth. (D) Flow cytometry analysis reveals G1-phase cell cycle arrest after 48h treatment, suggesting cell cycle disruption as a key mechanism. (E) Hoechst 33,342 staining highlights nuclear condensation and fragmentation, confirming apoptotic cell death. (F) A schematic illustrates the proposed mode of action: ZINC13000658 interferes with SAM binding to METTL proteins, thereby inhibiting methylation-dependent pathways critical for hepatocellular carcinoma progression.
To contextualize this effect, we examined the expression of METTL family genes in HepG2, SNU-449, 22Rv1, and VCaP cells (Supplementary Fig. 5C). Notably, prostate cancer cells displayed high expression of METTL3, METTL5, and METTL9, mirroring the expression profile seen in the HCC lines suggesting that elevated METTL expression may underlie the sensitivity to ZINC13000658 and support its broader therapeutic potential.
Discussion
Hepatocellular carcinoma (HCC) progression is driven by inactivation of tumor suppressors, activation of oncogenes (e.g., K-ras, BRAF), dysregulated tyrosine kinases, and aberrant signaling pathways58. While targeted and immunotherapy exists, their limited efficacy and emerging resistance highlight the need for more potent therapeutic strategies59. Recent studies underscore the oncogenic roles of METTL family proteins in HCC, influencing cell survival, proliferation, and metastasis, with high expression correlating with poor prognosis 11,12,13,14,15,16,17,18,19. In addition, no multi-targeted inhibitors have been developed against METTL proteins in HCC60. Therefore, this study aimed to identify potent, selective METTL inhibitors by modeling SAM-competitive ligands targeting conserved features of METTL and related methyltransferases.
To identify potent inhibitors targeting HCC-associated METTL proteins, we screened 74 small molecules using molecular docking. The top five compounds exhibited strong binding affinities and consistent interaction patterns within the SAM-binding pockets of METTL1, METTL3, METTL6, METTL16, and METTL18 (Fig. 2), suggesting conserved druggable regions across these enzymes. For METTL3, the key interacting residues closely matched those reported by Yankova et al. including 378I, 395D, 406Y, 431W, 457W, 511 K, 513 K, 532E, 534F, 536R, and 549N; supporting the reliability of our docking approach20. Notably, this study is the first to report potential catalytic site residues involved in ligand binding for METTL1, METTL6, METTL16, and METTL18 (Fig. 2 & Supplementary Table 4).
To further refine our search for multitarget inhibitors, we prioritized compounds with high chemical softness (softness > 0.6 eV), as determined by frontier molecular orbital (FMO) analysis, indicating their potential for broad target engagement. A ligand-based pharmacophore model, built from these top candidates, was used to screen the ZINC database, yielding 483 hits. Subsequent docking and interaction analysis identified ZINC70666503 and ZINC13000658 as lead compounds. Both displayed favorable ADMET properties, including minimal predicted inhibition of major cytochrome P450 enzymes (CYP3A4, CYP2D6, CYP2C9, CYP2C19), with ZINC13000658 showing no CYP interaction. Consistent docking results across three independent tools further validated their strong and selective binding to the SAM-binding pockets of METTL proteins, highlighting their potential as effective multi-target methyltransferase inhibitors.
Moreover, we propose the selectivity of these two compounds towards all five SAM-dependent HCC-associated METTL proteins in a SAM-competitive mechanism (Supplementary Table 9). The competitive docking results also support a SAM-competitive inhibition mechanism, as both ZINC13000658 and ZINC70666503 effectively reduce SAM binding affinity to METTL3 by occupying or obstructing its canonical binding site (Supplementary Fig. 2). Although SAM is a shared cofactor among various methyltransferases, extended docking showed these compounds also bind DNMTs, COMTs, and viral methyltransferases, indicating a conserved interaction mode (Supplementary Fig. 2). However, their affinity was generally lower than SAM in non-METTL targets, while remaining stronger or comparable in METTLs, suggesting preferential selectivity. This selectivity likely stems from the pharmacophore-based screening approach, which was designed using shared features of known METTL inhibitors. This ligand-based approach inherently biases the resulting candidates toward molecules that are structurally and functionally optimized for METTL protein interaction, providing a rationale for their observed selectivity. Importantly, DNMT1 and DNMT3A are often overexpressed or mutated in cancers and contribute to aberrant methylation of tumor suppressor genes61. COMT regulates catecholamine and estrogen metabolism, with roles in neuroendocrine and hormone-related cancers62. Viral RNA methyltransferases like SARS-CoV-2 nsp16 are essential for viral mRNA capping and immune evasion63. Although the cross-reactivity observed with these enzymes suggests potential off-target effects, it also opens avenues for repurposing these compounds as broad-spectrum epigenetic or antiviral agents, especially in contexts where SAM-dependent methylation plays a pathological role.
Our study is the first to identify ZINC70666503 and ZINC13000658 as novel inhibitors of METTL1, METTL6, METTL16, and METTL18. Unlike existing METTL3 inhibitors such as STM2457, STC-15, Quercetin, and Eltrombopag, which have uncertain safety profiles, both compounds demonstrated higher binding affinities, better pharmacokinetics, and no predicted toxicity. Molecular dynamics simulations confirmed their stable interactions with METTL proteins, with ZINC13000658 forming more hydrogen bonds, indicating stronger potential to disrupt SAM binding (Supplementary Fig. 3). While most METTL–ligand complexes exhibited stable RMSD trajectories over 100 ns, the METTL6–ZINC13000658 complex showed a transient fluctuation around 70 ns, likely due to local conformational adjustments within the SAM-binding pocket. The trajectory quickly stabilized, and both Rg and RMSF values remained within acceptable ranges, indicating structural integrity. Such mid-simulation shifts are common in methyltransferase MD studies and may reflect productive reorientations that enhance binding64. This flexibility could contribute to the favorable binding energetics observed, though further simulations may clarify its functional relevance. However, ZINC13000658 demonstrated potent, dose-dependent inhibition of cell proliferation in HepG2 and SNU-449 HCC cell lines over ZINC 70,666,503. This compound also induces G1 phase arrest, reduced cell confluency, and caused nuclear fragmentation, indicating apoptosis (Figs. 6).
While ZINC70666503 demonstrated strong in silico performance, including high binding affinity across five METTL proteins, and stable molecular dynamics, it failed to produce significant anti-proliferative effects in HepG2 and SNU-449 HCC cell lines even at 100 μM (Supplementary Fig. 4). This disconnect highlights a critical challenge in early-stage drug discovery, where computational predictions may not fully capture biological efficacy. One possible explanation is limited cellular uptake; despite predicted BBB permeability, ZINC70666503 may have poor membrane permeability, be subject to active efflux, or become sequestered intracellularly, reducing its effective cytoplasmic concentration. Additionally, real-world issues such as low solubility, aggregation, or degradation in culture medium could diminish its availability. It may also require metabolic activation or target molecular contexts absent in the tested cell lines. Moreover, METTL expressions in HepG2 and SNU-449 may not correlate with functional dependency; even with elevated levels, these cells might not rely on the targeted METTLs for proliferation. While docking data suggests SAM-binding pocket occupancy, ZINC70666503 may not engage key catalytic residues necessary for inhibition. In contrast, ZINC13000658, though slightly lower in computational ranking, showed significant anti-proliferative effects in both HCC and prostate cancer models, likely due to enhanced hydrogen bonding, solubility, and intracellular pharmacokinetics. These findings emphasize the need for integrated validation pipelines, combining computational predictions with biochemical assays and cellular models, and underscore the importance of evaluating cellular uptake, target engagement (e.g., cellular thermal shift assay and drug affinity responsive target stability), and enzymatic inhibition to effectively prioritize candidate therapeutics targeting epigenetic regulators such as the METTL family.
Overall, we have shown that targeting RNA methyltransferases in the METTL family by small molecule inhibitor, such as METTL1, METTL3, METTL6, METTL16, and METTL18, offers a promising strategy for inhibiting hepatocellular carcinoma (HCC) progression. The success of small-molecule inhibitors in targeting various oncogenic proteins, such as Aurora B kinase, Microtubule-affinity regulating kinase 4 (MARK-4), BCR-ABL, and EGFR, highlights their potential in modulating critical cancer pathways, including those regulated by RNA methyltransferases like METTLs65,66,67. These METTL enzymes regulate critical processes like cell proliferation, migration, invasion, and immune response through m6A and m7G modifications, influencing RNA stability, protein translation, and immune modulation, thereby contributing to tumorigenesis and metastasis11,12,13,14,15,16,17,18,19. Moreover, METTL1, METTL3 and METTL16 have been specifically implicated in rewiring metabolic pathways, including glycolysis and lipogenesis in HCC and colorectal cancer, and drug resistance in oral squamous cell carcinoma, establishing a direct link between METTL activity and cancer cell metabolic adaptation68. These METTL proteins, along with METTL6, play essential roles in RNA epigenetic regulation by catalyzing m6A or m7G modifications on coding and non-coding RNAs, thereby influencing transcript stability, splicing, translation, and degradation. METTL3 enhances the stability and translation of oncogenic transcripts like Snail, MYC, SOCS2, and EGFR, while METTL1 promotes translation of pro-growth genes such as RAC1 and CCND1 via tRNA m7G methylation69,70,71. METTL6 has been linked to methylation of tRNAs and stabilization of mRNAs involved in ribosome biogenesis and protein synthesis. The inhibition of METTL1 has been reported to reduce protein synthesis and suppress tumor progression in lung and bladder cancers. Though less characterized, METTL6, METTL16, and METTL18 are overexpressed in HCC and are associated with aggressive phenotypes and cell cycle progression15,16,17. Inhibition of these enzymes, evident from our observed anti-proliferative effect of ZINC13000658 in HCC cell lines, may suppress tumor growth by destabilizing oncogenic transcripts or impairing translation. However, future studies will be necessary to map the exact transcriptomic alterations and methylation signatures modulated by METTL inhibition in HCC.
Additionally, our proliferation study in prostate cancer cell lines (Supplementary Fig. 5) further supports the broader therapeutic potential of ZINC13000658 beyond hepatocellular carcinoma. The observed anti-proliferative activity of ZINC13000658 in 22Rv1 and VCaP cells, both of which exhibit elevated expression of METTL family genes, including METTL3, METTL5, and METTL9 highlights a possible link between METTL expression and drug sensitivity. This aligns with our earlier observation that high METTL expression correlates with poor prognosis in HCC and suggests that METTLs may serve as both therapeutic targets and biomarkers across multiple malignancies. Indeed, our selected METTL targets are known to be dysregulated in various human cancers beyond HCC. For example, METTL1 is frequently overexpressed in lung, bladder, head and neck, and gastric cancers72,73,74,75,76, while METTL3 is implicated in lung adenocarcinoma, prostate cancer, triple-negative breast cancer, and other aggressive tumors77. These associations reinforce the rationale for investigating METTL-targeting compounds, such as ZINC70666503 and ZINC13000658, in a broader oncologic context where these could be combined with existing first-line and second-line therapies for HCC to combat drug resistance and enhance treatment efficacy. While ZINC70666503 lacked efficacy in HCC models despite strong computational predictions, its potential remains viable in other tumor types where METTL expression or dependency may differ. Collectively, these underscore the translational relevance of METTL-targeted inhibition and support future mechanistic and therapeutic exploration of these compounds across METTL-driven cancers where ZINC70666503 might exhibit more significant therapeutic potential.
However, our study highlights the need for further research to identify which specific METTL proteins are targeted by our compound and to uncover the underlying molecular mechanisms. Direct biochemical assays are needed to confirm the functional inhibition of METTL enzymatic activity by ZINC13000658 and ZINC70666503. Although computational insights remain indirect evidence, biochemical assays such as surface plasmon resonance (SPR) or microscale thermophoresis (MST) are necessary to definitively establish target engagement, specificity, and the competitive nature of inhibition with respect to SAM. Additionally, this study is limited by the lack of mechanistic insight into how METTL inhibition affects downstream gene regulation in HCC. While METTL proteins act via RNA methylation, the specific targets of METTL6, METTL16, and METTL18 in HCC remain poorly defined in the context of HCC and normal physiological condition. This restricts our ability to fully understand the biological consequences of inhibiting these enzymes. To address this, future work will integrate transcriptomic and proteomic approaches, such as RNA-seq, m6A MeRIP-seq, ribosome profiling, and global proteome analysis to define METTL-dependent regulatory networks and assess the impact of inhibitor treatment. These efforts will clarify mechanisms of action and guide therapeutic application of METTL inhibitors like ZINC70666503.
Conclusion
Hepatocellular carcinoma is one the most prominent health issues worldwide with no effective treatment strategies. In this study, two compounds ZINC70666503 and ZINC13000658 showed promising results that could expedite multi-targeting drug development against five METTL proteins involved in HCC. Here we identified that ZINC13000658 could be a more promising option for treating human malignancies than any previously identified METTL3 inhibitors. Remarkably, this compound is the first identified catalytic inhibitors that can act against METTL1, METTL6, METTL16, and METTL18. However, further in vivo efficacy testing and exploration of their anti-tumor activity are still needed.
Data availability
All data supporting the findings of this study are available within the article and its supplementary materials.
References
Chidambaranathan-Reghupaty, S., Fisher, P. B. & Sarkar, D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv. Cancer Res. 149, 1–61 (2021).
Gallage, S. et al. The therapeutic landscape of hepatocellular carcinoma. Med. 2(5), 505–552 (2021).
Lee, S. K. et al. Immunological markers, prognostic factors and challenges following curative treatments for hepatocellular carcinoma. Int. J. Mol. Sci. 22(19), 10271 (2021).
Raja, A. & Haq, F. Molecular classification of hepatocellular carcinoma: Prognostic importance and clinical applications. J. Cancer Res. Clin. Oncol. 148(1), 15–29 (2022).
Garcia-Lezana, T., Lopez-Canovas, J. L. & Villanueva, A. Signaling pathways in hepatocellular carcinoma. Adv. Cancer Res. 149, 63–101 (2021).
Chow, A. K., Yau, S. W. & Ng, L. Novel molecular targets in hepatocellular carcinoma. World J. Clin. Oncol. 11(8), 589–605 (2020).
Luo, X. Y., Wu, K. M. & He, X. X. Advances in drug development for hepatocellular carcinoma: Clinical trials and potential therapeutic targets. J. Exp. Clin. Cancer Res. 40(1), 172 (2021).
Marin, J. J. G. et al. Molecular bases of the poor response of liver cancer to chemotherapy. Clin. Res. Hepatol. Gastroenterol. 42(3), 182–192 (2018).
Marin, J. J. G. et al. Molecular bases of drug resistance in hepatocellular carcinoma. Cancers (Basel) 12(6), 1663 (2020).
Campeanu, I. J. et al. Multi-omics integration of methyltransferase-like protein family reveals clinical outcomes and functional signatures in human cancer. Sci. Rep. 11(1), 14784 (2021).
Tian, Q. H. et al. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J. Mol. Med. (Berl) 97(11), 1535–1545 (2019).
Chen, Z. et al. METTL1 promotes hepatocarcinogenesis via m. Clin. Transl. Med. 11(12), e661 (2021).
Lin, Y. et al. METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med. 9(8), 2859–2867 (2020).
Xu, H. et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics 10(13), 5671–5686 (2020).
Bolatkan, A. et al. Downregulation of METTL6 mitigates cell progression, migration, invasion and adhesion in hepatocellular carcinoma by inhibiting cell adhesion molecules. Int. J. Oncol. 60(1), 4 (2022).
Li, T. H. et al. Identification METTL18 as a potential prognosis biomarker and associated with immune infiltrates in hepatocellular carcinoma. Front. Oncol. 11, 665192 (2021).
Dai, Y. Z. et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m. Cell Mol. Biol. Lett. 27(1), 41 (2022).
Wang, L. & Peng, J.-L. METTL5 serves as a diagnostic and prognostic biomarker in hepatocellular carcinoma by influencing the immune microenvironment. Sci. Rep. 13(1), 10755 (2023).
Bi, F. et al. METTL9-SLC7A11 axis promotes hepatocellular carcinoma progression through ferroptosis inhibition. Cell Death Discovery 9(1), 428 (2023).
Yankova, E. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593(7860), 597–601 (2021).
Ofir-Rosenfeld, Y. et al. STC-15, an oral small molecule inhibitor of the RNA methyltransferase METTL3, inhibits tumour growth through activation of anti-cancer immune responses associated with increased interferon signalling, and synergises with T cell checkpoint blockade. Eur. J. Cancer 174, S123 (2022).
Creusot, F., Acs, G. & Christman, J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2’-deoxycytidine. J. Biol. Chem. 257(4), 2041–2048 (1982).
Pappalardi, M. B. et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2(10), 1002–1017 (2021).
Huang, S. et al. A novel class of selective non-nucleoside inhibitors of human DNA methyltransferase 3A. Bioorg. Med. Chem. Lett. 40, 127908 (2021).
Hu, C. et al. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: Mechanism and clinical application. Clin. Epigenetics 13(1), 166 (2021).
Moroz-Omori, E. V. et al. METTL3 inhibitors for epitranscriptomic modulation of cellular processes. Chem. Med. Chem. 16(19), 3035–3043 (2021).
Li, J. & Gregory, R. I. Mining for METTL3 inhibitors to suppress cancer. Nat. Struct. Mol. Biol. 28(6), 460–462 (2021).
Lee, J. H. et al. Discovery of substituted indole derivatives as allosteric inhibitors of m. Drug Dev. Res. 83(3), 783–799 (2022).
Lee, J. H. et al. Eltrombopag as an allosteric inhibitor of the METTL3-14 complex affecting the m. Pharmaceuticals (Basel) 15(4), 440 (2022).
Du, Y. et al. Discovery of METTL3 small molecule inhibitors by virtual screening of natural products. Front. Pharmacol. 13, 878135 (2022).
Dolbois, A. et al. 1,4,9-Triazaspiro[5.5]undecan-2-one derivatives as potent and selective METTL3 inhibitors. J. Med. Chem. 64(17), 12738–12760 (2021).
Bedi, R. K. et al. Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem 15(9), 744–748 (2020).
Rose, P. W. et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 45(D1), D271–D281 (2017).
Kim, S. et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 49(D1), D1388–D1395 (2021).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31(2), 455–461 (2010).
Consortium, U. UniProt: The Universal protein knowledgebase in 2023. Nucleic Acids Res. 51(D1), D523–D531 (2023).
Salentin, S. et al. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43(W1), W443–W447 (2015).
Ye, N., Yang, Z. & Liu, Y. Applications of density functional theory in COVID-19 drug modeling. Drug Discov. Today 27(5), 1411–1419 (2022).
Lehtola, S. et al. Recent developments in libxc—A comprehensive library of functionals for density functional theory. SoftwareX 7, 1–5 (2018).
Neese, F. Software update: The ORCA program system—Version 50. Wiley Interdiscip. Rev. Comput. Mol. Sci. 12, e1606 (2022).
Hanwell, M. D. et al. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformat. 4(1), 17 (2012).
Bagheri Novir, S. & Aram, M. R. Quantum mechanical simulation of chloroquine drug interaction with C60 fullerene for treatment of COVID-19. Chem. Phys. Lett. 757, 137869 (2020).
Pearson, R. G. Absolute electronegativity and hardness correlated with molecular orbital theory. Proc. Natl. Acad. Sci. U. S. A. 83(22), 8440–8441 (1986).
Parr, R. G. Density functional theory of atoms and molecules. in Horizons of Quantum Chemistry (Springer, 1980).
Schneidman-Duhovny, D. et al. PharmaGist: A webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 36, W223–W228 (2008).
Koes, D. R. & Camacho, C. J. ZINCPharmer: Pharmacophore search of the ZINC database. Nucleic Acids Res. 40, W409–W414 (2012).
Daina, A., Michielin, O. & Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7(1), 42717 (2017).
Cheng, F. et al. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 52(11), 3099–3105 (2012).
Grosdidier, A., Zoete, V. & Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 (2011).
Liu, Y. et al. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 50(W1), W159–W164 (2022).
Hasan, M. et al. Main protease inhibitors and drug surface hotspots for the treatment of COVID-19: A drug repurposing and molecular docking approach. Biomed. Pharmacother. 140, 111742 (2021).
Abraham, M., et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 2015. 1.
Schüttelkopf, A. W. & van Aalten, D. M. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60(Pt 8), 1355–1363 (2004).
Genheden, S. & Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug Discov. 10(5), 449–461 (2015).
Cui, W. et al. Discovering anti-cancer drugs via computational methods. Front. Pharmacol. 11, 733 (2020).
Mumit, M. A. et al. DFT studies on vibrational and electronic spectra, HOMO-LUMO, MEP, HOMA, NBO and molecular docking analysis of benzyl-3-N-(2,4,5-trimethoxyphenylmethylene)hydrazinecarbodithioate. J. Mol. Struct. 1220, 128715 (2020).
Daina, A. & Zoete, V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 11(11), 1117–1121 (2016).
Cucarull, B. et al. Hepatocellular carcinoma: Molecular pathogenesis and therapeutic advances. Cancers (Basel) 14(3), 621 (2022).
Juaid, N. et al. Anti-hepatocellular carcinoma biomolecules: Molecular targets insights. Int. J. Mol. Sci. 22(19), 10774 (2021).
Wilhelm, S. et al. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 5(10), 835–844 (2006).
Zhang, W. & Xu, J. DNA methyltransferases and their roles in tumorigenesis. Biomarker Res. 5(1), 1 (2017).
Janacova, L. et al. Catechol-O-methyl transferase suppresses cell invasion and interplays with MET signaling in estrogen dependent breast cancer. Sci. Rep. 13(1), 1285 (2023).
Kremling, V., et al. SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery. 2024, eLife Sciences Publications, Ltd.
Ye, F. et al. Biochemical studies and molecular dynamic simulations reveal the molecular basis of conformational changes in DNA Methyltransferase-1. ACS Chem. Biol. 13(3), 772–781 (2018).
Ahmed, A. et al. Aurora B kinase: A potential drug target for cancer therapy. J. Cancer Res. Clin. Oncol. 147(8), 2187–2198 (2021).
Anwar, S. et al. Microtubule-affinity regulating kinase 4: A potential drug target for cancer therapy. Cell Signal 99, 110434 (2022).
Zubair, T. & Bandyopadhyay, D. Small molecule EGFR inhibitors as anti-cancer agents: Discovery, mechanisms of action, and opportunities. Int. J. Mol. Sci. 24(3), 2651 (2023).
Park, S. & Hall, M. N. Metabolic reprogramming in hepatocellular carcinoma: Mechanisms and therapeutic implications. Exp. Mol. Med. 57(3), 515–523 (2025).
Chen, M. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67(6), 2254–2270 (2018).
Lin, S. et al. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62(3), 335–345 (2016).
Cheng, W. et al. Novel roles of METTL1/WDR4 in tumor via m7G methylation. Mol. Ther. Oncol. 26, 27–34 (2022).
Ma, C. et al. The molecular mechanism of METTL3 promoting the malignant progression of lung cancer. Cancer Cell Int. 22(1), 133 (2022).
Ying, X. et al. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin. Transl. Med. 11(12), e675 (2021).
Wang, C. et al. Methyltransferase-like 1 regulates lung adenocarcinoma A549 cell proliferation and autophagy via the AKT/mTORC1 signaling pathway. Oncol. Lett. 21(4), 330 (2021).
Chen, J. et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun. (Lond) 42(3), 223–244 (2022).
Ma, X. et al. TSPAN31 regulates the proliferation, migration, and apoptosis of gastric cancer cells through the METTL1/CCT2 pathway. Transl. Oncol. 20, 101423 (2022).
Cai, Y. et al. Novel insights into the m(6)A-RNA methyltransferase METTL3 in cancer. Biomark Res. 9(1), 27 (2021).
Acknowledgements
We wholeheartedly thanked Brian Latimer and the INLET for IncuCyte support
Funding
No specific grant was received for this study. MH is supported by the SUST Research Center (LS/2023/2/17), Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh. Additional support was obtained by the Feist-Weiller Cancer Center at Louisiana State University Health Shreveport.
Author information
Authors and Affiliations
Contributions
M.N.M. Conceptualization, Methodology, Software & Formal analysis, Investigation, and Data interpretation, Writing—original draft, and review & editing. M.S.A. Conceptualization, Methodology, Writing—review & editing. R.I.A. Software & Formal analysis, Writing—review & editing. M.K.S. Software & Formal analysis, Writing—review & editing. M.M.A. In vitro Experiments, Writing—review & editing J.F. Formal analysis, Writing—review & editing. Y.Z Data interpretation, Writing—review & editing O. E. F. Supervision, Project administration. Writing—review & editing. M.J.H. Project administration, Supervision, Writing—review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Ethical approval was not required for this study as it did not involve human participants or animal subjects. All cell lines used in this study (HepG2 and SNU-449) were purchased from the American Type Culture Collection (ATCC), a certified biological resource center. The cell lines were authenticated by ATCC using short tandem repeat (STR) profiling and were routinely tested for mycoplasma contamination, with negative results prior to experimentation.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Morshed, M.N., Parvez, S.A., Akanda, R.I. et al. Identification of novel small molecule inhibitors targeting multiple methyltransferase like proteins against hepatocellular carcinoma. Sci Rep 15, 33087 (2025). https://doi.org/10.1038/s41598-025-16614-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-16614-0