Abstract
To investigate the biological functions of TOP2A in HCC using bioinformatics and single-cell RNA sequencing (scRNA-seq) methods, and to explore the functional consequences of TOP2A in HepG2 and Huh-7 cells with TOP2A knockdown. This study aims to provide new theoretical insights and potential therapeutic targets for the early diagnosis and targeted treatment of HCC. Through the analysis of single-cell RNA sequencing data, the immunological significance of TOP2A and its relationship with the diagnosis and prognosis of HCC were studied. The expression levels of TOP2A in HCC and normal liver epithelial cells were validated using quantitative PCR (QPCR). siRNA technology was employed to interfere with TOP2A expression in HepG2 and Huh7 cells, and a TOP2A knockdown cell line was established. The transfection efficiency of siRNA was measured using QPCR and Western Blotting. The impact of TOP2A knockdown on the proliferation of HCC cells was assessed using the CCK8 assay. The effect of TOP2A on the migration ability of HCC cells was observed through a wound-healing assay. The influence of TOP2A knockdown on the invasive ability of HCC cells was examined using the Transwell assay. The effect of TOP2A knockdown on the apoptosis of HCC cells was evaluated using flow cytometry. This study integrated single-cell RNA sequencing and functional analyses identified TOP2A as a critical oncogenic driver in HCC. TOP2A expression was significantly upregulated in HCC tissues and strongly correlated with poor patient prognosis (P < 0.05). In vitro experiments showed that the efficiency of TOP2A knockdown mediated by siRNA was over 70% (P < 0.05), and it significantly inhibited the proliferation, invasion and migration of HCC cells (P < 0.05), promoted cell apoptosis (P < 0.05). Mechanistically, scRNA-seq revealed that TOP2A may promotes Tmem-to-Tex differentiation through the SPP1-CD44 axis. These findings position TOP2A as both a prognostic biomarker and a potential therapeutic target for overcoming immune evasion in HCC. TOP2A is significantly upregulated in HCC, and its expression level can serve as an independent prognostic biomarker. Knockdown of TOP2A can significantly inhibit the malignant phenotype of HepG2 and Huh7 cells. Moreover, the analysis shows that TOP2A may promote immune escape by modulating the SPP1-CD44 axis and remodeling the immune microenvironment. These findings not only reveal the crucial role of TOP2A in the progression of HCC, but also indicate that it may become a potential target for immunotherapy in HCC, providing a new theoretical basis for improving the precise treatment strategies for HCC patients.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is a malignant tumor that poses a serious threat to human health1. Its occurrence and development are often associated with risk factors such as viral infections2and non-alcoholic fatty liver disease3. As a common malignant tumor, the incidence and mortality rates of HCC are increasing year by year. However, its treatment and prognosis remain unsatisfactory. Most patients are diagnosed at intermediate or advanced stages due to the absence of early symptoms, delaying the optimal treatment window4. Therefore, studying the pathogenic mechanisms of HCC and discovering new intervention targets are of great significance for improving the therapeutic outcomes of HCC.
Topoisomerase IIα (TOP2A) is a protein located in the nucleus and mitochondria, involved in the formation and binding of DNA double-strand breaks and the regulation of DNA topology, thereby influencing gene transcription, replication, and other functions5. TOP2A is an important tumor suppressor gene, and its high expression is associated with various malignant tumors, such as HCC6, medulloblastoma7, lung cancer8 and pancreatic cancer9. Studies have found that inhibiting TOP2A can promote the aging of HCC cells, thereby suppressing the progression of HCC10. Circular RNA cDCBLD2 increases the mRNA stability of TOP2A, inhibiting cell apoptosis and thereby influencing the development of HCC11. Additionally, TOP2A promotes the epithelial-mesenchymal transition (EMT) of HCC cells by affecting the Snail pathway, further facilitating HCC metastasis12, This mechanism may be related to the miR-139-5p-AC078846.1/AC124798.1/SNHG3 signaling axis13. Furthermore, activating transcription factor 2 promotes the transcription of WHSC1, upregulating TOP2A and inducing HCC through the regulation of the PI3K/AKT signaling pathway14.
This study initially investigated the mechanism of TOP2A in HCC through scRNA-seq, and further evaluated the mechanism of TOP2A in HCC through experiments, especially the influence of knockdown of TOP2A on the biological behavior of HCC, in order to provide a basis for the early diagnosis and treatment of HCC.
Materials and methods
Experimental reagents and instruments
Human HCC cells (HepG2 and Huh-7 from Yimo Biotech), TOP2A inhibitor (Fukesi), Lipofectamine 3000 transfection reagent (Thermo Fisher), DMEM medium (Gibco), penicillin-streptomycin (Meilun), fetal bovine serum (Gibco), sterile PBS buffer (biosharp), 0.25% EDTA trypsin (Gibco), CCK8 kit (Beyotime), primers (Sangon Biotech), RNA extraction kit (Accurate Biology), RNA reverse transcription kit (Accurate Biology), SYBR Green kit (Accurate Biology), 4% PFA fixative (biosharp), T25 culture flask (NEST), 6-well plate, 24-well plate, 96-well plate (NEST), PCR instrument (Bio-Rad), multifunctional microplate reader (Bio-Rad).
Data collection and scRNA-seq analysis
We analyzed all the scRNA-seq data downloaded from the GEO database and conducted preliminary quality control on the scRNA-seq data. The raw scRNA-seq data were preprocessed using CellRanger and Seurat in R software. The scRNA-seq cohort was used for subsequent analysis based on the following criteria: (1) Genes must be expressed in at least 3 cells; (2) Cells must express at least 50 genes; (3) Mitochondrial gene expression must not exceed 5%. Batch effects were removed using the “harmony” R package, and the scRNA-seq data were standardized using the “Seurat” R package. After identifying highly variable genes, principal component analysis (PCA) was performed using the “RunPCA” function. Unsupervised cell clusters were determined using the “FindClusters” function (selecting the top 20 principal components, resolution = 1.0). Automated cell type annotation was performed using SingleR (v1.8.0), which provides rapid and standardized classification with significantly improved objectivity, efficiency, and cross-dataset consistency compared to manual annotation, while maintaining compatibility with multi-omics integration approaches. Notably, novel cell populations without established reference signatures still require manual validation. The clusters were visualized through dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP). Markers for each cluster were identified using the “FindAllMarkers” function under the following criteria: |logFC| > 1.0, adjusted P value < 0.05.
InferCNV analysis
prepares the original count matrix, annotation file and gene/chromosome location file according to the instructions on the InferCNV GitHub page (https://github.com/broadinstitute/inferCNV). We use the R software package inferCNV (v1.6.0) to calculate the somatic large-scale chromosomal CNV score for each hepatocyte, with normal liver cells serving as the reference cells. We apply the default parameters (cut-off value = 0; denoising = 0.1).
Cell-to-cell interaction analysis
The R software package “CellChat” is used to conduct analysis of intercellular interactions based on the expression of specific ligands and receptors. This tool quantitatively infers and analyzes the intercellular communication networks from scRNA-seq data. By leveraging multi-faceted learning and quantitative comparison, CellChat classifies signaling pathways and depicts conserved and context-specific pathways in different datasets.
Cellular development trajectory analysis
Using Monocle 2 (v2.18.0), the cell trajectories of hepatocytes were inferred, assuming one-dimensional “pseudo-time” to describe the high-dimensional expression values of single cells. The cell trajectories and positions were presented in a two-dimensional graph with a tree structure, which was after log-normalization and DDR tree dimensionality reduction.
CIBERSORTx analysis and functional analysis
Utilizing CIBERSORTx, an analytical tool designed for interpolating gene expression profiles and estimating the abundance of cell types within mixed cell populations (https://cibersortx.stanford.edu/). Gene set variation analysis (GSVA) was conducted to estimate biological functions and signaling pathways in batch RNA-seq and scRNA-seq data.
Cell culture
The cryopreserved cells were retrieved from liquid nitrogen. After thawing, the cells were quickly inoculated into DMEM medium and cultured in a cell incubator at 37 °C, with saturated humidity and 5% CO2. The old medium was aspirated and discarded, and the adherent cells were gently washed with 1× PBS buffer by shaking the culture flask. The PBS was removed using a negative pressure aspirator with a new sterile pipette tip, and this process was repeated twice. Trypsin digestion solution was then added to digest the cells, followed by an equal volume of medium to stop the digestion. The cell suspension was transferred to a centrifuge tube and centrifuged at 1000 rpm for 5 min. The supernatant was discarded, and the cell pellet was resuspended in 2 mL of complete medium. The cell density was adjusted under an inverted microscope, and the cells were inoculated into culture flasks and cultured in a 37 °C, 5% CO2 incubator with saturated humidity. The above steps were repeated every 48 h. After three generations of culture, cells in the logarithmic growth phase were subcultured for subsequent experiments.
Cell transfection
Cells with good growth status and 90% confluence were digested with trypsin and resuspended in antibiotic-free medium to prepare a single-cell suspension at a density of 5 × 104/mL. The suspension was seeded into a 24-well plate at 500 µL per well and cultured in a 37 °C, 5% CO2 incubator for 24 h. When the cell confluence reached 70%, the designed siRNA and negative control (si-NC) were mixed with transfection reagent and added to the 24-well plate containing HCC cells at 50 µL per well. The plate was gently shaken to ensure even distribution. The plate was then placed in a 37 °C, high-humidity, 5% CO2 incubator and cultured for 24 and 48 h. The expression level of TOP2A in HCC cells was detected using QPCR and Western Blotting.
Western blot analysis
The cell proteins were extracted using RIPA buffer, quantified by BCA method, then subjected to 10% SDS-PAGE electrophoresis and transferred onto a PVDF membrane. The membrane was incubated with 5% milk for blocking, followed by incubation with TOP2A primary antibody (1:1000) and HRP secondary antibody (1:5000). The protein bands were placed in the imaging instrument, and an appropriate amount of developed solution was added, adjusted and exposed. The experimental results were recorded.
Cell CCK8 proliferation assay
After transfection, cells from each group were seeded into a 96-well plate at a certain density. After 24 h of adhesion in a 37 °C, 5% CO2 incubator, 100 µL of prepared CCK-8 solution was added to each well, and the plate was incubated further. After 1 h, the a bsorbance (OD value) at 450 nm was measured using a microplate reader. The cell proliferation rate was calculated as follows: Cell proliferation rate (%) = (OD value of experimental group - OD value of blank group) / (OD value of control group - OD value of blank group) × 100%.
Cell scratch assay
Cells in the logarithmic growth phase were digested and resuspended, and the cell suspension was counted under a microscope. Cells were seeded into a 6-well plate at 1000 cells per well and cultured in a 37 °C, 5% CO2 incubator for 24 h. When cell confluence reached 80%, a cross-shaped scratch was made using a 200 µL pipette tip perpendicular to a ruler. The medium was aspirated, and the cells were washed with sterile PBS to remove cell debris. The plate was then placed back into the incubator. The scratch area was observed and photographed under an inverted microscope at 0, 24, and 48 h. Three parallel experiments were set up, and the results were analyzed using statistical software.
Cell invasion assay
Complete medium was added to a 24-well plate. Cells that had been serum-starved were digested and resuspended, then added to the upper chamber of a Transwell insert. The insert was placed into the 24-well plate and cultured for 24–48 h. The cells were fixed, stained, and photographed under a 20× microscope. Five random fields were selected for counting.
Cell apoptosis detection
Cells were digested with 0.25% trypsin, collected, and resuspended in a 5 mL flow tube. After adding 5 µL of Annexin V and mixing thoroughly, the cells were incubated in the dark at room temperature for 5 min. Then, 10 µL of propidium iodide (PI) solution (20 µg/mL) and 400 µL of PBS were added, and the cells were analyzed using flow cytometry.
Statistical analysis
GraphPad 9.5 software was used for statistical analysis and graphing. All experiments were repeated three times, and data were expressed as mean ± standard deviation. Independent sample t-tests were used for comparisons between groups, with P < 0.05 considered statistically significant.
Results
Identification and validation of TOP2Aby multi-omics
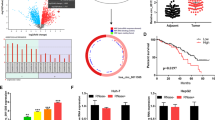
Firstly, in all datasets, it was found that the expression of TOP2A was upregulated in tumor tissues compared with normal tissues (all P < 0.05, Figs. 1A-D). Additionally, we analyzed the expression of TOP2A at the single-cell level. Initially, all cells underwent quality control and dimensionality reduction clustering. Based on the expression analysis of marker genes specific to each subgroup, the cells were classified as NK/T cells, B cells, bone marrow cells, endothelial cells, liver cells, and hepatic stellate cells (HSCs), etc. (Supplementary Figures S1A-G). To further validate the upregulation of TOP2A in HCC cells, we isolated hepatocytes from primary tumors and normal tissues and divided them into several different cell clusters. Among them, Normal Cluster_1 was mainly composed of normal tissue cells and was used as a reference, while other clusters showed higher CNV scores (Fig. 1E). Therefore, those identified as normal liver cells, and the other clusters were identified as HCC cells (Fig. 1F, G). Overall, the upregulation of TOP2A in HCC was verified through transcriptomics and scRNA-seq.
Knockdown of TOP2A inhibits the proliferation, migration, invasion and apoptosis of HCC cells
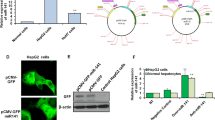
To investigate the expression level and biological function of TOP2A in HCC, we first measured the expression of TOP2A in HCC using QPCR. We found that, compared to the normal liver epithelial cell line THLE-2, TOP2A expression was significantly upregulated in the HepG2 HCC cell line and Huh-7 HCC cell line (P < 0.05, Fig. 2A,B). Next, we constructed a HCC cell line with knocked-down TOP2A. Specifically, we used siRNA technology to interfere with TOP2A expression in the HCC cell line to achieve knockdown, and then verified the transfection efficiency of siTOP2A using QPCR (Fig. 2C,D) and Western blotting (Figures 2E, F). In order to investigate the effects of knockdown of TOP2A on the proliferation, migration and invasion abilities of HCC cells, we conducted CCK8, cell plate scratch, Transwell and apoptosis experiments in two HCC cell lines. The experimental results demonstrated that the knockdown of TOP2A inhibited the proliferation of HepG2 cells and Huh-7 cells (P < 0.05, Fig. 2G,H). Furthermore, after knockdown of TOP2A, the migration ability and invasion ability of HepG2 cells and Huh-7 cells were inhibited (Fig. 2I,J) and (Fig. 2K, L). Using the Annexin-FITC and PI double staining method and observing the apoptosis of cells through flow cytometry, The experimental results demonstrated that knockdown of TOP2A promoted apoptosis in HepG2 cells and Huh-7 cells (Fig. 2M, N).
(A) QPCR assay detecting the expression of TOP2A in normal liver epithelial cells THLE-2 and HepG2 HCC cell line; (B) QPCR assay detecting the expression of TOP2A in normal liver epithelial cells THLE-2 and Huh-7 HCC cell line; (C) QPCR assay detecting the interference level of siTOP2A in HepG2 HCC cell line; (D) QPCR assay detecting the interference level of siTOP2A in Huh-7 HCC cell line; (E) Western blotting detecting the interference level of siTOP2A in HepG2 HCC cell line; (F) Western blotting detecting the interference level of siTOP2A in Huh-7 HCC cell line; (G) Knocking down TOP2A inhibited the proliferation ability of HepG2 cells; (H) Knocking down TOP2A inhibited the proliferation ability of Huh-7 cells; (I) Knocking down TOP2A weakened the migration ability of HepG2 cells and statistical significance; (J) Knocking down TOP2A weakened the migration ability of Huh-7 cells and statistical significance; (K) Knocking down TOP2A weakened the invasive ability of HepG2 cells and statistical significance; (L) Knocking down TOP2A weakened the invasive ability of Huh-7 cells and statistical significance; (M) Flow cytometry analysis of apoptosis in HepG2 cells after TOP2A gene knockout (the x-axis represents FITC, the y-axis represents PI) and statistical analysis of the number of early and late apoptotic cells in HepG2 cells after TOP2A gene knockout; (N) Flow cytometry analysis of apoptosis in Huh-7 cells after TOP2A gene knockout (the x-axis represents FITC, the y-axis represents PI) and statistical analysis of the number of early and late apoptotic cells in HepG2 cells after TOP2A gene knockout (*P < 0.05,**P < 0.01,***P<0.001).
Overexpression of TOP2A induces T cell exhaustion
TiME plays a crucial role in the occurrence, development and prognosis of HCC. We evaluated 21 infiltrating immune cells between the high and low TOP2A subgroups using CIBERSORTx. The results showed that based on TOP2A expression, HCC tissues were classified into the TOP2A high subgroup and the TOP2A low subgroup. Compared with the TOP2A low subgroup, the proportion of CD8 T cells and Macrophages M1 in the TOP2A high subgroup was significantly higher (both P < 0.05, Fig. 3A); conversely, in the TOP2A high subgroup, CD4 memory resting T cells (Tmem) and Macrophages M2 were lower (both P < 0.05, Fig. 3A). Taking into account the observed differences in Treg and T cells between the low and high TOP2A levels in the batch RNA analysis, we isolated NK/T cells for further study. After dimensionality reduction, clustering and annotation (Fig. 3B), we stratified NK/T cells into NK, effector T cells (Teff), memory T cells (Teff), Teff and exhausted T cells (Tex) (Fig. 3C). Differential analysis revealed that the low TOP2A subgroup showed increased infiltration of Tmem and Treg and decreased infiltration of Tex (all P < 0.05, Figs. 3D-E). Cellular development trajectory analysis indicated that Tmem differentiated into Tex over pseudotime (Supplementary Figures S2A, B). Overall, these findings suggest that overexpression of TOP2A may lead to T cell exhaustion and subsequent immune escape of HCC.
TOP2A affects immune cell infiltration in TiME. (A) The differential infiltration of immune cells by CIBERSORTx analysis. (B) The UMAP plot shows the extraction of cell subpopulations from hepatocytes. (C) UMAP plot exhibited the cell subpopulations of NK/T cells. (D,E) The different infiltrating of Tmem cells, Tex cells and Treg cells between TOP2A low and TOP2A high groups.
Mechanism of TOP2A in regulating T-cell exhaustion
In order to explore the mechanism by which TOP2A regulates T cell exhaustion, we conducted an analysis of the intercellular communication network using the R software package “CellChat”. The expression of TOP2A in HCC was divided into the TOP2A high subgroup and the TOP2A low subgroup. We analyzed the quantity and intensity of each cell interaction (Fig. 4A, B). The secreted phosphoprotein-1 (SPP1) is expressed in various tumors. In this study, the role of cells in the SPP1 pathway was analyzed, and it was found that TOP2A high HCC is the “sender” and NK/T cells are the “receivers” (Supplementary Table 1). Furthermore, we found that CD44 is also closely related to MIF, LGALS9, etc. These factors are all involved in the T cell exhaustion process (all P < 0.05, Supplementary Table 1). HCC may induce NK/T cell exhaustion through the TOP2A-SPP1-CD44 axis, thereby leading to tumor progression. However, this mechanistic link remains speculative, as direct evidence for its immune-modulating function in HCC is currently lacking. Future studies employing co-culture assays or conditional knockout models are required for experimental validation.
Discussion
HCC is a major disease that poses a serious threat to human life and health. Although surgical resection, radiotherapy, and chemotherapy can achieve relatively good outcomes, the efficacy remains suboptimal15. Therefore, in-depth research into its pathogenesis and the identification of molecular markers are of significant importance for screening high-risk populations and achieving early diagnosis16. Type II topoisomerase (TOP2) plays a crucial regulatory role in transcription, replication, and cell division17,18 and is also critical for maintaining chromosome structure19. TOP2A is an important tumor suppressor gene closely associated with the development of various cancers, such as gastric cancer20, colorectal cancer21, lung cancer22, breast cancer23, and cervical cancer24. Moreover, its high expression is closely related to tumor recurrence and invasion25. Studies have found that TOP2A expression is elevated in HCC, and its high expression is closely associated with poor prognosis in HCC patients26. The occurrence and development of HCC are closely related to the immune mechanism. The immune system plays a crucial role in the proliferative and metastatic behaviors of HCC, but the specific mechanism remains unclear. Single-cell RNA sequencing (scRNA-seq) can conduct high-resolution analysis of individual cell types in HCC, thereby more accurately mapping the interaction patterns of immune cells and identifying new targets. This provides new ideas for the diagnosis and treatment of HCC.
In this article, we first investigated the immunological role of TOP2A in HCC through single-cell sequencing technology. Subsequently, using QPCR technology, we first detected the expression of TOP2A in HCC cells (HepG2, Huh-7) and normal liver epithelial cells THLE-2. The results showed that TOP2A was highly expressed in HCC cell lines, and knockdown of TOP2A expression in HCC cell lines inhibited the proliferation, migration and invasion abilities of HCC cells, emphasizing the crucial role of TOP2A in the development of HCC. This indicates that TOP2A is a potential biomarker for HCC and is involved in the occurrence and development of HCC.
Research has found that TOP2A can promote the progression of various tumors, such as HCC27, prostate cancer28, bladder cancer29, cervical cancer24, osteosarcoma30. We used siRNA technology to interfere with the expression of TOP2A in HCC and verified it through QPCR technology. Through CCK8 experiments and plate clone experiments, it was found that the proliferation ability of HCC cells was inhibited after TOP2A was knocked down; the plate scratch experiment showed that the migration ability of cells decreased, and the Transwell experiment found that the expression of TOP2A was knocked down, which inhibited the invasion ability of HCC cells; the apoptosis ability of cells was detected by flow cytometry, and it was found that the knockdown of TOP2A promoted the apoptosis of HCC cells. The above experimental results indicate that TOP2A is related to the proliferation, migration, invasion and apoptosis ability of HCC cells. Therefore, we speculate that TOP2A has a promoting effect on the proliferation of HCC cells, but the specific molecular mechanism needs further exploration.
T cell exhaustion is currently a hot topic in tumor research. In this study, based on TOP2A expression, we classified HCC tissues into TOP2A-high and TOP2A-low subgroups. The TOP2A-high subgroup exhibited higher levels of CD8 + T cells and M1 macrophages. Research has shown that in the early stages of tumor development, the efficient initiation of CD8 + T cell responses requires sufficient cancer cell death or the presentation of tumor antigens. Even if tumor antigens are captured and presented by antigen-presenting cells (APCs), the absence of pathogen-associated molecular pattern (PAMP)-mediated innate immune activation and the lack of CD4 + T cell help can suppress the processes of T cell priming, activation, and differentiation, ultimately leading to a hyporesponsive state resembling anergy and tolerance31. This may explain why even cancers with highly immunogenic neoantigens can progress. As the cancer advances, CD8 + T cells are continuously stimulated by tumor antigens and enter a state of late-stage dysfunction, leading to immune evasion and promoting tumor progression32. M1 macrophages typically exert pro-inflammatory effects in the tumor microenvironment33, though some studies suggest that M1-like macrophages may also promote cancer progression. For instance, M1 macrophages enhance the motility of HCC cells via the NF-κB/FAK pathway34 and activate the JAK/STAT3 pathway to facilitate metastasis in oral cancer35.
We observed that, compared with the TOP2A-low group analyzed by bulk RNA-seq, the TOP2A-high group analyzed by scRNA-seq showed a reduction in γδ T cells but an increase in exhausted T (Tex) cells. These findings suggest that TOP2A may induce exhaustion in γδ T cells. Furthermore, scRNA-seq revealed that the TOP2A-high group had fewer memory T (Tmem) cells than the TOP2A-low group. Pseudotime trajectory analysis indicated that Tmem cells differentiated into Tex cells over time. CellChat analysis demonstrated that the most relevant signaling pathway between TOP2A-high HCC and NK/T cells was SPP1-CD44. SPP1 is a multifunctional extracellular matrix protein that promotes colon cancer metastasis by facilitating epithelial-mesenchymal transition36,37, enhances drug resistance in HCC through bypass activation of oncogenic signaling38, and triggers macrophage polarization toward the M2 phenotype to accelerate HCC progression39. CD44 is a non-kinase transmembrane glycoprotein overexpressed in various cancers, including HCC40, gastric cancer41, and colon cancer42. STAT5A regulates CD44 to promote gastric cancer proliferation43, while the RNA-binding protein LSM7 binds to CD44, increasing its expression and driving breast cancer metastasis and invasion44. In HCC, CD44 upregulates integrin subunit β2 and VCAM-1 expression, contributing to tumor progression45. Studies have shown that the SPP1-CD44 pathway is involved in immunosuppression and tumor progression. The SPP1-CD44 signaling axis promotes T cell exhaustion, inhibits sustained T cell proliferation, and thereby increases tumor burden in cancers such as HCC46and cholangiocarcinoma47. Additionally, the SPP1/CD44 axis is associated with chemotherapy resistance in solid tumors, as SPP1 can activate CD44 receptor signaling to confer drug resistance in cancer cells48,49. Therefore, we hypothesize that SPP1 may bridge intrinsic tumorigenesis and immune escape in tumor cells via CD44 receptor signaling.
In summary, TOP2A is highly expressed in HCC and plays a critical role in promoting tumor progression, correlating closely with poor prognosis. In this study, we investigated the epigenetic mechanisms of TOP2A by modulating its expression in two HCC cell lines. Our findings demonstrate that TOP2A knockdown significantly suppresses HCC cell proliferation, migration, and invasion while inducing apoptosis. These results highlight TOP2A as a potential therapeutic target and provide novel insights for HCC prevention and treatment strategies. However, this study is still not perfect. Firstly, it lacks clinical samples. This study only used databases and in vitro experiments, and further verification is needed by combining clinical samples. Secondly, it lacks in vivo animal experiments, ignoring the influence of the in vivo environment. Therefore, further in-depth research is needed at the level of animal living bodies.
Data availability
The datasets used in this study are available online in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE14520, and The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) with the accession code TCGA-LIHC.
References
Rich, N. E. Changing epidemiology of hepatocellular carcinoma within the united States and worldwide [J]. Surg. Oncol. Clin. N Am. 33 (1), 1–12 (2024).
Kalantari, L. et al. A state-of-the-art review on the NRF2 in hepatitis virus-associated liver cancer [J]. Cell. Commun. Signal. 21 (1), 318 (2023).
Pugliese, N. et al. Clinical features and management issues of NAFLD-related HCC: what we know so Far [J]. Expert Rev. Gastroenterol. Hepatol. 17 (1), 31–43 (2023).
Buttell, A. & Qiu, W. The action and resistance mechanisms of lenvatinib in liver cancer [J]. Mol. Carcinog. 62 (12), 1918–1934 (2023).
Zhang, K. et al. TOP2A modulates signaling via the akt/mtor pathway to promote ovarian cancer cell proliferation [J]. Cancer Biol. Ther. 25 (1), 2325126 (2024).
Liu, G. et al. Elucidating the prognostic and therapeutic significance of TOP2A in various malignancies [J]. Cancer Genet., 288–289, 68–81. (2024).
Zhang, Y. et al. TOP2A correlates with poor prognosis and affects radioresistance of Medulloblastoma [J]. Front. Oncol. 12, 918959 (2022).
Wu, J. et al. The role of TOP2A in immunotherapy and vasculogenic mimicry in non-small cell lung cancer and its potential mechanism [J]. Sci. Rep. 13 (1), 10906 (2023).
Pei, Y. F., Yin, X. M. & Liu, X. Q. TOP2A induces malignant character of pancreatic cancer through activating β-catenin signaling pathway [J]. Biochim. Biophys. Acta Mol. Basis Dis. 1864 (1), 197–207 (2018).
Wang, K. et al. EZH2-H3K27me3-mediated Silencing of mir-139-5p inhibits cellular senescence in hepatocellular carcinoma by activating TOP2A [J]. J. Exp. Clin. Cancer Res. 42 (1), 320 (2023).
Ruan, Y. et al. cDCBLD2 mediates Sorafenib resistance in hepatocellular carcinoma by sponging miR-345-5p binding to the TOP2A coding sequence [J]. Int. J. Biol. Sci. 19 (14), 4608–4626 (2023).
Dong, Y. et al. Type IIA topoisomerase (TOP2A) triggers epithelial-mesenchymal transition and facilitates HCC progression by regulating snail expression [J]. Bioengineered 12 (2), 12967–12979 (2021).
Wang, X. Q. et al. Noval CeRNA axis-mediated high expression of TOP2A correlates with poor prognosis and tumor immune infiltration of hepatocellular carcinoma [J]. Transl Cancer Res. 12 (12), 3486–3502 (2023).
Bao, Z. M. et al. Activating transcription factor 2 promotes the progression of hepatocellular carcinoma by inducing the activation of the WHSC1-mediated TOP2A/PI3K/AKT axis [J]. Kaohsiung J. Med. Sci. 38 (7), 662–674 (2022).
Chen, L. et al. Human liver cancer organoids: biological applications, current challenges, and prospects in hepatoma therapy [J]. Cancer Lett. 555, 216048 (2023).
Foda, Z. H. et al. Detecting liver cancer using Cell-Free DNA fragmentomes [J]. Cancer Discov. 13 (3), 616–631 (2023).
Xu, T. et al. A signature of circadian rhythm genes in driving anaplastic thyroid carcinoma malignant progression [J]. Cell. Signal. 95, 110332 (2022).
Lee, S. B. et al. Striking efficacy of a vaccine targeting TOP2A for triple-negative breast cancer Immunoprevention [J]. NPJ Precis Oncol. 7 (1), 108 (2023).
Uusküla-reimand, L. & Wilson, M. D. Untangling the roles of TOP2A and TOP2B in transcription and cancer [J]. Sci. Adv. 8 (44), eadd4920 (2022).
Chen, Y. U. et al. E2F1-mediated up-regulation of TOP2A promotes viability, migration, and invasion, and inhibits apoptosis of gastric cancer cells [J]. J. Biosci. 47 (2022).
Priyamvada, P. & Ramaiah, S. Potential signature therapeutic biomarkers TOP2A, MAD2L1, and CDK1 in colorectal cancer: A systems Biomedicine-Based approach [J]. Biochem. Genet. 62 (3), 2166–2194 (2024).
Wu, J. et al. Expression and potential molecular mechanism of TOP2A in metastasis of non-small cell lung cancer [J]. Sci. Rep. 14 (1), 12228 (2024).
Mehraj, U. et al. Cryptolepine targets TOP2A and inhibits tumor cell proliferation in breast cancer cells - An in vitro and in Silico study [J]. Anticancer Agents Med. Chem. 22 (17), 3025–3037 (2022).
Yu, B. et al. TOP2A and CENPF are synergistic master regulators activated in cervical cancer [J]. BMC Med. Genomics. 13 (1), 145 (2020).
Solhusløkk Höse, K. et al. TOP2A expression in pheochromocytoma and abdominal paraganglioma: a marker of poor clinical outcome?? [J]. Endocr. Pathol. 34 (1), 129–141 (2023).
Gao, S. et al. Computational analysis for identification of early diagnostic biomarkers and prognostic biomarkers of liver cancer based on GEO and TCGA databases and studies on pathways and biological functions affecting the survival time of liver cancer [J]. BMC Cancer. 21 (1), 791 (2021).
Cai, H. et al. High expression of TOP2A gene predicted poor prognosis of hepatocellular carcinoma after radical hepatectomy [J]. Transl Cancer Res. 9 (2), 983–992 (2020).
Tian, L. et al. Low level exposure to BDE-47 facilitates the development of prostate cancer through TOP2A/LDHA/lactylation positive feedback circuit [J]. Environ. Res. 263 (Pt 2), 120094 (2024).
Simon, R. et al. HER-2 and TOP2A coamplification in urinary bladder cancer [J]. Int. J. Cancer. 107 (5), 764–772 (2003).
Xu, Y. et al. Exploration of s new biomarker in osteosarcoma and association with clinical outcomes: (TOP2A+) cancer associated fibroblasts [J]. J. Gene Med. 25 (11), e3528 (2023).
Philip, M. & Schietinger, A. CD8(+) T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 22 (4), 209–223 (2022).
Reina-Campos, M., Scharping, N. E. & Goldrath, A. W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 21 (11), 718–738 (2021).
Gao, J., Liang, Y. & Wang, L. Shaping polarization of Tumor-Associated macrophages in cancer immunotherapy. Front. Immunol. 13, 888713 (2022).
Wang, H. et al. CD68(+)HLA-DR(+) M1-like macrophages promote motility of HCC cells via NF-κB/FAK pathway. Cancer Lett. 345 (1), 91–99 (2014).
You, Y. et al. M1-like tumor-associated macrophages cascade a mesenchymal/stem-like phenotype of oral squamous cell carcinoma via the IL6/Stat3/THBS1 feedback loop. J. Exp. Clin. Cancer Res. 41 (1), 10 (2022).
Xu, C. et al. SPP1, analyzed by bioinformatics methods, promotes the metastasis in colorectal cancer by activating EMT pathway. Biomed. Pharmacother. 91, 1167–1177 (2017).
Liu, X. et al. ANGPTL2 + cancer-associated fibroblasts and SPP1 + macrophages are metastasis accelerators of colorectal cancer. Front. Immunol. 14, 1185208 (2023).
Eun, J. W. et al. Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to Sorafenib and lenvatinib. Cancer Commun. (Lond). 43 (4), 455–479 (2023).
Liu, L. et al. Construction of TME and identification of crosstalk between malignant cells and macrophages by SPP1 in hepatocellular carcinoma. Cancer Immunol. Immunother. 71 (1), 121–136 (2022).
Song, X. et al. Knockdown of CD44 inhibits proliferation, migration, and invasiveness in hepatocellular carcinoma cells by modulating CXCR4/Wnt/β-Catenin axis. Acta Biochim. Pol. 70 (1), 117–122 (2023).
Hou, W. et al. CD44 is a prognostic biomarker and correlated with immune infiltrates in gastric cancer. BMC Med. Genomics. 15 (1), 225 (2022).
Tang, F. et al. CD44(+) cells enhance pro-tumor stroma in the Spatial landscape of colorectal cancer leading edge. Br. J. Cancer. 132 (8), 703–715 (2025).
Wang, S. et al. STAT5A modulates gastric cancer progression via upregulation of CD44. Genomics 115 (3), 110638 (2023).
Li, C. et al. RNA-binding protein LSM7 facilitates breast cancer metastasis through mediating alternative splicing of CD44. Life Sci. 356, 123013 (2024).
Gerardo-Ramírez, M. et al. Deletion of Cd44 inhibits metastasis formation of liver cancer in Nf2-Mutant mice. Cells, 12(9). (2023).
He, H. et al. Multi-dimensional single-cell characterization revealed suppressive immune microenvironment in AFP-positive hepatocellular carcinoma. Cell. Discov. 9 (1), 60 (2023).
Cheng, M. et al. Immunosuppressive role of SPP1-CD44 in the tumor microenvironment of intrahepatic cholangiocarcinoma assessed by single-cell RNA sequencing. J. Cancer Res. Clin. Oncol. 149 (9), 5497–5512 (2023).
Qian, J. et al. Cancer-associated mesothelial cells promote ovarian cancer chemoresistance through paracrine osteopontin signaling. J. Clin. Invest., 131(16). (2021).
Xie, W. et al. Multi-Transcriptomic analysis reveals the heterogeneity and Tumor-Promoting role of SPP1/CD44-Mediated intratumoral crosstalk in gastric cancer. Cancers (Basel), 15(1). (2022).
Author information
Authors and Affiliations
Contributions
Lian Xie: designed the project and wrote the main manuscript text. Fangyin Xu: designed the project and wrote the main manuscript text. Ruixin Shi: collected data and contributed to the discussion and interpretation of the results. Zhendong Zhang: collected data and contributed to the discussion and interpretation of the results. Chenwei Pan: provided guidance on study design.Ping Chen: provided guidance on study design and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, L., Xu, F., Shi, R. et al. Single cell-RNA sequencing reveal TOP2A as a key driver of hepatocellular carcinoma progression. Sci Rep 15, 32009 (2025). https://doi.org/10.1038/s41598-025-16785-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-16785-w