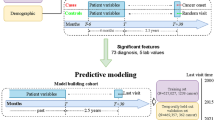
Abstract
This study aimed to develop and validate a practical nomogram for differentiating between benign and malignant pancreatic masses. A total of 494 patients with pancreatic mass lesions, confirmed by histopathology, were enrolled from Wuhan Union Medical College Hospital between January 2020 and May 2022. The participants were randomly divided into development and validation groups in a 7:3 ratio. Using multivariate logistic regression, the nomogram was constructed based on five independent predictors: blood type, CA19-9, IgG4, anorexia, and weight loss. The model’s performance was assessed using receiver operating characteristic (ROC) curve analysis and calibration curves. In the development and validation sets, the areas under the ROC curve were 0.932 and 0.957, respectively. The nomogram demonstrated a high net benefit in the clinical decision curve analysis. Based on the model, pancreatic malignancy risk was classified as low (< 4%), moderate (4%–71%), and high (> 71%). This nomogram provides an easy-to-use, efficient tool for the early differentiation of pancreatic malignancies and could be implemented in primary, secondary, and emergency care settings to facilitate the timely referral of patients to higher-level hospitals.
Similar content being viewed by others
Introduction
Pancreatic cancer (PC) is a highly invasive malignant tumor, and it is increasingly becoming a common cause of cancer mortality1. It has a very poor prognosis, with a median survival of about 10 to 12 months with treatment and 5 to 6 months without treatment2 and despite decades of ongoing efforts, the five-year survival rate remains below 9%3. PC is linked to a poor prognosis caused by multiple factors, including non-specific symptoms, delayed diagnosis, aggressive and resistant tumor biology with early distant metastasis, and the absence of precise and sensitive biomarkers or early disease imaging techniques4,5. The primary treatment options for PC are surgery and chemotherapy. Unfortunately, only 15–20% of PC patients are eligible for surgery at the time of diagnosis6.
Patients with pancreatic diseases, whether malignant or benign pancreatic masses, exhibit similar symptom manifestations (such as jaundice) and biochemical features (such as elevated CA19-9 or abnormalities in liver function tests)5. Patients with PC are often asymptomatic in the early stages, and a delayed diagnosis of PC is a frequent occurrence7. Therefore, early detection of PC remains challenging. Most patients with PC and benign pancreatic masses present as solid or cystic masses on imaging studies, lacking specific imaging features in the early stages. Due to the completely different treatment approaches and prognoses of benign pancreatic diseases and PC, identification of benign and malignant pancreatic masses as early as possible is needed to avoid misdiagnosis, ensure accurate diagnosis, and select appropriate treatment strategies8.
To date, several risk prediction models for PC have been developed in populations with diabetes, family history of PC, and the general population; however, these prediction models focus on identifying individuals at high risk of PC in different populations for further intervention9,10,11,12,13,14. In contrast, only a limited number of studies have focused on differentiating benign from malignant pancreatic masses, most of which rely on imaging-based methodologies such as computed tomography (CT), magnetic resonance imaging (MRI), or endoscopic ultrasound (EUS) combined with serum tumor markers15,16,17,18. These models require advanced imaging modalities or specialized equipment, which limits their applicability in primary or resource-limited settings. Therefore, it is very necessary to establish a nomogram model for distinguishing between benign and malignant pancreatic masses based on routine laboratory test results and clinical symptoms.
In this retrospective study, we analyzed clinical and laboratory data from patients with pancreatic mass lesions treated at our institution between January 2020 and May 2022. The objective was to identify independent predictive factors and to develop and validate an easy-to-use nomogram for differentiating benign from malignant pancreatic masses, providing a practical nomogram for early diagnosis and clinical decision-making across primary, secondary, and emergency care settings, especially where advanced imaging is inaccessible.
Materials and methods
Study subjects
The study population consists of patients with pancreatic mass lesions who were diagnosed and treated at Wuhan Union Hospital between January 2020 and May 2022. In particular, the pancreatic mass is defined as a lesion that may be cystic, solid, or cystic-solid, ranging from approximately a few millimeters to over ten centimeters in diameter, with various shapes that may include sausage-like or irregular forms; all pancreatic masses are confirmed through imaging examinations such as ultrasound, CT, and MRI. The inclusion criteria for this study are as follows: (i) patients aged 18 years or older; (ii) presence of pancreatic mass lesions (including cystic, solid, and cystic-solid masses) on imaging studies; (iii) confirmed pathological diagnosis of pancreatic tissue; (iv) and no history of other malignancies. The exclusion criteria are as follows: (i) severe data missing or incompleteness; (ii) unclear histopathological diagnosis results; (iii) history of other malignancies or secondary pancreatic malignant tumors. This retrospective study was conducted in accordance with relevant guidelines and regulations. The study protocol was approved by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology ([2023] Ethics Review Number 0259). Given the retrospective nature of the study, the need for informed consent was waived by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology.
Data collection
We collected clinical data using the hospital’s electronic medical records system. The demographics, symptoms, coexisting conditions, and laboratory values of the patients were collected and analyzed. Laboratory values include immunoglobulin G4 (IgG4); total bilirubin (TB); direct bilirubin (DB); alanine aminotransferase (ALT); aspartate aminotransferase (AST); alkaline phosphatase (ALP); gamma-glutamyltransferase (γ-GGT); albumin (ALB); serum creatinine; D-dimer; alpha-fetoprotein (AFP); carcinoembryonic antigen (CEA); carbohydrate antigen 19 − 9 (CA19-9); and cancer antigen 125 (CA125). Definitions of clinical variables and parameters are provided in the Supplementary Materials.
Statistical analysis
Based on the histopathology results of the pancreatic mass biopsy, the patients were divided into two groups: benign pancreatic masses and pancreatic malignancy. The entire dataset was randomly divided into training and validation sets in a 7:3 ratio19,20. The randomization was performed using R software by an independent statistician to ensure unbiased and reproducible allocation. The training set was utilized for model development, while the validation set was employed for model validation. Normally distributed continuous variables were presented as means with standard deviations (SD), and parametric t-tests were utilized to assess statistical significance between the two groups. For variables that were not normally distributed, medians with interquartile ranges (IQR) were utilized for description, and nonparametric Mann-Whitney U tests were utilized for comparisons between the two groups. For categorical variables, we presented the counts and percentages of patients in each category. Proportions were compared using either the chi-square test or Fisher’s exact test. Missing data were addressed using multiple imputation methods with the mice package, which are primarily based on the assumption that the missing data are missing at random (MAR).
We initially conducted univariate analysis in the training set to identify potential predictive factors for malignant pancreatic masses, and variables with statistical significance in univariate analysis were included in multivariate logistic regression analysis. The sample size was adequate for the number of predictors considered, adhering to the commonly accepted rule of at least 10 events per variable, which helps reduce overfitting and supports model stability19. Next, the selected factors were further used in a multivariable logistic regression analysis to identify independent predictive factors; only the variables with a P value less than 0.05 were selected as independent risk factors. After the process of univariate and multivariate screening, the final prognostic factors were used to construct a nomogram for predicting the probability of pancreatic malignancy. Based on the regression coefficient, each included variable corresponded to a point on the scale for each value. The total score was calculated as the sum of the scores assigned to each variable for each patient. The association between the total scores and the probability of pancreatic malignancy was depicted on the bottom of the nomogram. Subsequently, calibration curves were constructed to evaluate the consistency between the predicted probability from the nomogram and the actual proportion. Moreover, the concordance index (C-index) was calculated to assess the discriminatory ability of our model. A relatively corrected C-index was computed for internal validation using 1000 bootstrap resampling. The validation set was utilized for internal validation. In addition, to evaluate the clinical utility of our established nomogram, we conducted a decision curve analysis to assess the net benefits at various threshold probabilities. We compared the predictive performance of the nomogram and the CA19-9–only model using the DeLong test to assess differences in their areas under the receiver operating characteristic (ROC) curves (AUCs). Eventually, in order to investigate the impact of age on the outcome, patients were categorized into four age groups: <54, 54–64, 64–74, and ≥ 74 years.
Statistical significance was defined as a two-sided P value < 0.05. All statistical analyses were performed using SPSS (IBM SPSS Statistics, version 27) and R software (version 4.2.2, https://www.r-project.org/).
Results
Demographic characteristics
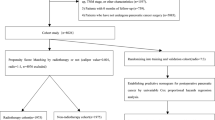
According to the inclusion criteria, 552 patients with pancreatic mass lesions were initially included. Among them, 46 patients had significant missing data, 10 patients had secondary pancreatic malignancy or a history of previous malignant tumors, and 2 patients had an unknown histopathological diagnosis. The remaining 494 patients meeting the inclusion criteria were further analyzed, as shown in Fig. 1. Among them, there were 112 patients with benign pancreatic masses and 382 patients with pancreatic malignancy. Additionally, the specific histopathological types of benign pancreatic masses in 112 patients and malignant pancreatic masses in 382 patients are presented in Supplementary Fig. 1. The clinical characteristics of the population were displayed in Supplementary Table 1. The median age of the study population was 59 years (IQR: 53–67), and 61.13% of patients were male (302). Moreover, the most common symptom at the onset of illness was abdominal discomfort (445 patients, 90.08%), followed by weight loss (353 patients, 71.46%), anorexia (330 patients, 66.80%), and jaundice (100 patients, 20.24%). In addition, diabetes (173 patients, 35.02%), hyperlipidemia (173 patients, 35.02%), hypertension (127 patients, 25.71%), and smoking (122 patients, 24.70%) were the most common coexisting medical conditions, and (158 patients, 31.98%) patients suffered from other comorbidities, such as alcohol, hepatitis, and hyperuricemia. The blood type A was the most prevalent, representing 162 patients (32.79%), while the remaining blood types O, B, and AB accounted for 155 patients (31.38%), 127 patients (25.71%), and 50 patients (10.12%), respectively. There were many differences in laboratory values between patients with benign pancreatic tumors and patients with pancreatic malignancy (Supplementary Table 1). In routine blood tests, patients with malignant pancreatic masses had higher levels of TB, DB, ALP, γ-GGT, D-dimer, CEA, CA125, and CA19-9 compared to patients with benign pancreatic masses, while IgG4 levels were the opposite.
Our study population was randomly divided into the development set (n = 346) and the validation set (n = 148). There was no statistical difference in the proportion of malignant pancreatic masses between the two groups (development set: 76.88%, validation set: 78.38%, P>0.05). The characteristics, symptoms, coexisting conditions, and laboratory values of the patients were similar in the development and validation sets (Table 1).
Development of the nomogram model
Firstly, all the characteristics, symptoms, coexisting conditions, and laboratory values in the development set were included in the univariate analysis to screen potential predictors for predicting pancreatic malignancy in patients with pancreatic mass (Table 2). Next, the potential predictors—including age, weight loss, anorexia, abdominal discomfort, blood type, hypertension, diabetes, IgG4, TB, DB, ALP, D-dimer, CEA, CA125, and CA19-9—with P-values < 0.05 in the univariate analysis were further included in the multivariate logistic regression to screen for independent predictive indicators. Eventually, five independent predictive factors were identified, including blood type, CA19-9, IgG4, anorexia, and weight loss (Table 3). The easy-to-use nomogram for predicting pancreatic malignancy in patients with pancreatic mass was developed based on these five prognostic factors, which is illustrated in Fig. 2 and available online (https://xinyangcodecraft.shinyapps.io/dynnomapp/). The quantitative method was made accessible for clinicians to predict the probability of pancreatic malignancy in patients with pancreatic mass. For each prognostic parameter, each patient was assigned a point, and the coefficients of the variables were scaled to a score between 0 and 100 to reflect their relative importance. By adding the corresponding scores, the probability of a pancreatic malignant mass in the pancreas mass population can be predicted directly and conveniently. The cutoff values were determined as 84 and 130 points, corresponding to estimated probabilities of 0.04 and 0.71 (Fig. 2). The higher the total number of scores, the more likely the patient is to have malignancy.
Nomogram for predicting pancreatic malignancy in patients with pancreatic mass. The linear predictor is an axis of linear predictions that are converted to probability values by a certain transformation function. To use this nomogram, the first step is to identify the option of each variable on its axis and then draw a vertical line to the point axis with each option as the origin. By adding the points of each variable, the total points can be calculated. Finally, with this total point, we can obtain the predicted risk value by drawing a vertical line from the total point axis to the risk of pancreatic malignancy.
Validation and assessment of the nomogram model
Based on the probabilities obtained from logistic regression analysis, the ROC analysis demonstrated that the area under the ROC curve of the nomogram model was 0.932 in the development set (Fig. 3A) and 0.957 in the validation set (Fig. 3B), indicating good discrimination. In the development set, the Youden index was maximized at 0.722, resulting in a cutoff of 0.710 (equivalent to a 71% risk according to the nomogram), with sensitivity and specificity values of 0.835 and 0.888, respectively. The model’s effectiveness in identifying individuals with the condition is demonstrated by its sensitivity (0.835), which indicates an 83.5% likelihood of accurate prediction when the condition is present. This high sensitivity is essential in clinical settings to avoid missing cases and facilitate early intervention. Specificity (0.888) reflects the model’s ability to accurately identify individuals without the condition, indicating an 88.8% likelihood of correct prediction when the condition is absent. High specificity is important for reducing unnecessary anxiety and avoiding additional testing for those who do not have the condition. Furthermore, the calibration curves showed that the nomogram had a similar performance compared with the ideal model in the development and validation sets, suggesting the good predictive power of our nomogram (Figs. 3C, D). Moreover, the C-index of our nomogram was 0.932 and was corrected to 0.918 through bootstrapping validation, which showed that the nomogram model had good predictive power.
ROC curve of the predictive model for the development set (A) and the validation set (B), and the calibration curves for predicting pancreatic malignancy in patients with pancreatic mass in the development set (C) and the validation set (D). The x-axis represents the predicted pancreatic malignancy probability, and the y-axis denotes the actual proportion of pancreatic malignancy. The diagonal dotted line indicates the best prediction by an ideal model, the apparent line represents the uncorrected performance of the nomogram, while the solid line shows the bias-corrected performance. 1000 bootstrap repetitions in the development set; mean absolute error = 0.014; n = 346, and 1000 bootstrap repetitions in the validation set; mean absolute error = 0.024; n = 148. AUC, area under the ROC curve.
Clinical practicality of the nomogram
The nomogram demonstrated superior diagnostic performance compared to CA19-9 alone. The AUC of the nomogram was 0.932 (95% CI: 0.905–0.959), significantly higher than that of CA19-9 (AUC: 0.869; 95% CI: 0.829–0.910; Delong P < 0.001) (Table 4). Decision curve analysis further confirmed the clinical utility of the nomogram. Compared to CA19-9, the nomogram provided a greater net benefit across a wide range of threshold probabilities (4–71%) in the development set (Fig. 4). In other words, when the prognostic risk value falls within the range of 4–71%, further imaging and an EUS-guided fine needle aspiration (EUS-FNA) biopsy are strongly recommended. In addition, if the predicted risk value is less than 4%, the likelihood of pancreatic malignant mass is very low, and continuous monitoring and follow-up are sufficient. Furthermore, if the predicted risk value is higher than 71% and the probability of malignant pancreatic mass is extremely high, immediate referral to higher-level hospitals is crucial for timely treatment decisions, such as surgery or chemotherapy. On the basis of these results, pancreatic malignancy risk can be classified as low (< 4%), moderate (4–71%), and high (> 71%) with the nomogram model.
Decision curve for the proposed predictive nomogram. Black line: assume no patients are intervened and the net benefit is zero; gray line: assume all patients are intervened; red and blue lines: patients are treated if the proposed model exceeds a threshold. For example, as shown in the decision curves, if the probability of pancreatic malignancy for patients with pancreatic mass is between 4% and 71%, then the proposed nomogram model can be beneficial for making the decision to undergo further diagnostics.
Association of age with predicted outcomes
In addition, the OR was 3.000 (95% CI, 1.342–6.706) for the 75-years-old age group compared with the youngest group, suggesting that the elderly have a greater likelihood of pancreatic malignancies. The trend test revealed an association between increasing age and an increased likelihood of pancreatic malignancy, although the second and third age groups did not show a significant difference in disease remission compared with the first age group (Supplementary Table 2).
Discussion
The treatment and prognosis of pancreatic malignancies and benign pancreatic diseases differ; it is necessary to quickly identify benign and malignant pancreatic masses to avoid misdiagnosis, ensure accurate diagnosis, and select appropriate treatment options. Therefore, we constructed a predictive nomogram model using individual symptoms (anorexia and weight loss) and routine laboratory values (blood type, CA19-9, and IgG4) to make an accurate estimation of the nature of benign and malignant pancreatic masses. The nomogram prediction model is simple, practical, and highly maneuverable, providing a visual means for quantitatively predicting clinical outcomes and letting frontline clinicians make timely preliminary assessments of the nature of pancreatic masses.
So far, there is a range of risk prediction models for PC; however, these prediction models primarily aim to pinpoint high-risk individuals for potential intervention within various population groups9,10,11,12,13,14,21. Secondly, most of these models were developed by applying complex statistical methods, such as artificial neural networks and machine learning algorithms, to data encompassing PC risk factors, clinical symptoms, and circulating markers. The complexity of these models often leads to difficulties in interpretation and hinders clinicians from using them in practice. Contrary to the above models, a nomogram offers an easy way for clinicians to integrate complex information, allowing doctors to specifically interpret the impact of each predictor variable on the outcome and make evidence-based decisions directly at the point of care22. On top of that, this model improves the precision of clinical predictions and contributes to the risk stratification of diseases, leading to more effective patient management23. Thus, we established and validated a nomogram prediction model in this study.
To our knowledge, there are a few studies utilizing nomograms to predict the nature of pancreatic masses. Nomograms based on multiparametric MRI radiomics characteristics and clinical independent risk factors (CEA, CA19-9)15, as well as those based on CT, have been reported to distinguish pancreatic carcinoma from mass-forming chronic pancreatitis16. Furthermore, Fan et al.17 developed a nomogram integrating EUS, age, and CA19-9 to differentiate benign from malignant pancreatic lesions, while Agarwal et al.18 constructed a nomogram based on EUS and CA19-9 to predict malignancy risk in chronic pancreatitis-associated masses. Similar to our study, the importance of CA19-9 in predicting benign and malignant pancreatic masses was also highlighted. Although there is currently no standardized imaging screening program for pancreatic masses, medical imaging undoubtedly plays a crucial role in screening and early detection, preoperative assessment and staging, differential diagnosis, follow-up, and treatment evaluation of PC24. However, the use of MRI, CT, or EUS is limited in primary care settings, such as community hospitals, and the cost of these examinations is relatively expensive; therefore, the feasibility of distinguishing benign and malignant pancreatic masses in such settings is poor. In contrast, our study focuses on readily available and inexpensive clinical parameters—individual clinical symptoms (anorexia, weight loss) and routine laboratory values (blood type, CA19-9, and IgG4). This approach fills an important clinical gap by providing a practical tool for early differentiation of pancreatic masses in primary, secondary, and emergency care settings where advanced imaging modalities may not be accessible. Furthermore, the sample sizes of the above studies were small, including only 138, 102, 151, and 107 patients, respectively. Whereas the number of patients in our study was 494, indicating our research has a relatively large sample size, the selection bias was relatively small. Additionally, the AUC of the CT nomogram was 0.87, and the AUC of the EUS-based nomogram was 0.865. In comparison, the AUC of our nomogram (0.932) was higher than both, indicating that our model has a stronger ability to differentiate between benign and malignant pancreatic masses. In conclusion, our nomogram first distinguished the nature of pancreatic disease based on routine laboratory test results (blood type, CA19-9, and IgG4 levels) and clinical symptoms (anorexia and weight loss), and the predictive indicators are more readily available and less expensive. Therefore, our nomogram demonstrates strong predictive accuracy and meets the need for a simple, cost-effective, and widely accessible tool to diagnose malignant pancreatic masses, facilitating early clinical decision-making and timely referral.
Notably, this is the first prediction model that includes blood type to predict benign and malignant pancreatic masses. Our study found that patients with non-O blood types had a significantly higher risk of malignancy compared to those with blood type O, consistent with previous reports25,26. Several biological mechanisms may underlie this association. First, ABO locus variants (such as rs505922, rs495828, rs657152, and rs630014) affect glycosyltransferase activity, altering glycosylation of cell surface molecules and influencing tumor behavior25,26. Second, ABO antigens, as glycoconjugates, mediate cell adhesion and signaling, potentially impacting tumor progression and immune recognition27. Third, ABO blood groups are linked to systemic inflammation markers like TNF-α and sICAM-1, which relate to pancreatic carcinogenesis27,28.
Although CA19-9 remains the most widely used serum biomarker for PC, its diagnostic performance is hindered by several limitations. Elevated CA19-9 levels are not specific to malignancy and may also occur in benign conditions such as cholangitis, pancreatitis, and obstructive jaundice, thereby reducing its specificity29,30. Furthermore, it lacks sufficient sensitivity for early-stage PC detection31. In contrast, our study demonstrated that a model combining clinical symptoms and laboratory values performs better than CA19-9 alone in predicting pancreatic malignancy.
Based on the predictive outcomes of this nomogram, graded health management of malignant pancreatic masses becomes feasible by discerning the categorization of low risk (< 4%), moderate risk (4–71%), and high risk (> 71%) for malignant pancreatic masses. The threshold values were set at 84 and 130 points, aligning with estimated probabilities of 0.04 and 0.71, respectively. For the patients with a low risk of pancreatic malignancy (< 4%), continuous monitoring and rigorous follow-up are needed; for the patients at moderate risk of pancreatic malignancy (4–71%), further imaging and EUS-FNA biopsy are strongly recommended, and referral to experienced pancreatic centers is warranted even if EUS-FNA results are negative; for the patients with a high risk of pancreatic malignancy (> 71%), instant referral to higher-level hospitals is a priority, and treatment decisions such as surgery or chemotherapy would be given as soon as possible. This risk classification helps clinicians promptly identify patients in need of early intervention while minimizing the burden of invasive testing, such as EUS-FNA, and saving medical resources. Furthermore, patients have the right to be informed about the probability of their current pancreatic malignancy, and the risk category assists them in making decisions about further diagnostic and treatment plans in consultation with their clinicians. Additionally, utilizing our nomogram may facilitate better clinician-patient communication in the medical setting, which is too important to ignore. Therefore, our prediction model has great potential to be applied in primary, secondary, and emergency care settings for early differentiation of suspicious pancreatic malignancy patients and instant referral of these patients to tertiary hospitals or experienced pancreatic centers. This model, in short, has good clinical practicability for providing a basis for clinical decision-making. However, it’s emphasized that this risk prediction model isn’t intended to replace current screening tools and diagnostic methods, such as imaging and EUS-FNA, but rather to complement them. Besides, we recommend rigorous follow-up for low-risk patients.
Previous studies have shown that age is an independent risk factor for PC32. Our age-stratified results revealed a positive correlation: the older the patient, the greater the likelihood of a malignant pancreatic mass. Specifically, our findings indicate that 53.33% of patients over 75 years old had malignant pancreatic masses, emphasizing the significance of age stratification in the pancreatic mass prediction model. Consequently, early detection programs may hold greater potential for reducing PC mortality in older populations, as 50% of PC patients are diagnosed after the age of 70 years32.
This study has several limitations. First, the model was developed and validated in a relatively small cohort from a single center, which may limit its generalizability to broader populations and clinical settings. Second, although the benign group included diverse conditions such as pancreatic cysts, chronic pancreatitis, and autoimmune pancreatitis, the small sample sizes within these subgroups limited our ability to assess prediction scores or misclassification rates, restricting evaluation of model specificity. Additionally, the absence of external validation using independent datasets—combined with the high proportion of malignant cases in our cohort—raises the possibility of overestimated model performance. Despite these limitations, the nomogram demonstrates strong predictive capability and offers a practical, accessible tool to support early diagnostic decision-making and improve clinician–patient communication. Therefore, further rigorous external validation in different centers is necessary before clinical implementation to ensure the generalizability and clinical benefits of this model in the future.
Conclusion
In conclusion, we developed and validated an easy-to-use nomogram for predicting pancreatic malignancy masses based on blood type, CA19-9, IgG4, anorexia, and weight loss. This nomogram gives a quantitative assessment of the likelihood of pancreatic malignancy in patients with pancreatic masses, helps clinicians in primary, secondary, and emergency care settings to decide on appropriate imaging examinations, EUS-FNA, or instant referrals of patients with suspicious pancreatic malignancy to tertiary hospitals or experienced pancreatic centers, facilitates better clinician-patient communication in the medical setting, and provides a basis for clinical decision-making.
Data availability
The fully de-identified datasets analyzed in this study are provided in the supplementary materials. Additional materials are available from the corresponding author upon reasonable request.
References
Mizrahi, J. D., Surana, R., Valle, J. W. & Shroff, R. T. Pancreatic cancer. Lancet (London England). 395, 2008–2020. https://doi.org/10.1016/s0140-6736(20)30974-0 (2020).
Wood, L. D., Canto, M. I., Jaffee, E. M. & Simeone, D. M. Pancreatic cancer: pathogenesis, screening, diagnosis, and treatment. Gastroenterology 163, 386–402e381. https://doi.org/10.1053/j.gastro.2022.03.056 (2022).
Qin, C. et al. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Mol. Cancer. 19, 856. https://doi.org/10.1186/s12943-020-01169-7 (2020).
Singhi, A. D., Koay, E. J., Chari, S. T. & Maitra, A. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology 156, 2024–2040. https://doi.org/10.1053/j.gastro.2019.01.259 (2019).
Pereira, S. P. et al. Early detection of pancreatic cancer. Lancet Gastroenterol. Hepatol. 5, 698–710. https://doi.org/10.1016/s2468-1253(19)30416-9 (2020).
Halbrook, C. J., Lyssiotis, C. A., Pasca di Magliano, M. & Maitra, A. Pancreatic cancer: advances and challenges. Cell 186, 1729–1754. https://doi.org/10.1016/j.cell.2023.02.014 (2023).
Walter, F. M. et al. Symptoms and patient factors associated with diagnostic intervals for pancreatic cancer (SYMPTOM pancreatic study): a prospective cohort study. Lancet Gastroenterol. Hepatol. 1, 298–306. https://doi.org/10.1016/s2468-1253(16)30079-6 (2016).
Umans, D. S. et al. Pancreatitis and pancreatic cancer: a case of the chicken or the egg. World J. Gastroenterol. 27, 3148–3157. https://doi.org/10.3748/wjg.v27.i23.3148 (2021).
Baecker, A. et al. Do changes in health reveal the possibility of undiagnosed pancreatic cancer? Development of a risk-prediction model based on healthcare claims data. PloS One. 14, e0218580. https://doi.org/10.1371/journal.pone.0218580 (2019).
Sharma, A. et al. Model to determine risk of pancreatic cancer in patients with New-Onset diabetes. Gastroenterology 155, 730–739e733. https://doi.org/10.1053/j.gastro.2018.05.023 (2018).
Dong, X., Lou, Y. B., Mu, Y. C., Kang, M. X. & Wu, Y. L. Predictive factors for differentiating pancreatic cancer-Associated diabetes mellitus from common type 2 diabetes mellitus for the early detection of pancreatic cancer. Digestion 98, 209–216. https://doi.org/10.1159/000489169 (2018).
Boursi, B. et al. A clinical prediction model to assess risk for pancreatic cancer among patients with New-Onset diabetes. Gastroenterology 152, 840–850e843. https://doi.org/10.1053/j.gastro.2016.11.046 (2017).
Klein, A. P. et al. An absolute risk model to identify individuals at elevated risk for pancreatic cancer in the general population. PloS One. 8, e72311. https://doi.org/10.1371/journal.pone.0072311 (2013).
Wang, W. et al. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J. Clin. Oncology: Official J. Am. Soc. Clin. Oncol. 25, 1417–1422. https://doi.org/10.1200/jco.2006.09.2452 (2007).
Liu, J., Hu, L., Zhou, B., Wu, C. & Cheng, Y. Development and validation of a novel model incorporating MRI-based radiomics signature with clinical biomarkers for distinguishing pancreatic carcinoma from mass-forming chronic pancreatitis. Translational Oncol. 18, 101357. https://doi.org/10.1016/j.tranon.2022.101357 (2022).
Zhang, H. et al. Two nomograms for differentiating mass-forming chronic pancreatitis from pancreatic ductal adenocarcinoma in patients with chronic pancreatitis. Eur. Radiol. 32, 6336–6347. https://doi.org/10.1007/s00330-022-08698-3 (2022).
Fan, X. et al. Clinical value of the nomogram model based on endoscopic ultrasonography radiomics and clinical indicators in identifying benign and malignant lesions of the pancreas. Front. Oncol. 15,856. https://doi.org/10.3389/fonc.2025.1504593 (2025).
Agarwal, S., Gunjan, D., Gopi, S. & Saraya, A. Combination of serum CA 19 – 9 and endoscopic ultrasound findings can predict malignancy risk in patients with chronic pancreatitis presenting with pancreatic head mass: A Proof-of-Concept study. Pancreas 53, e168–e175. https://doi.org/10.1097/mpa.0000000000002279 (2024).
Riley, R. D. et al. Calculating the sample size required for developing a clinical prediction model. BMJ (Clinical Res. ed.). 368, m441. https://doi.org/10.1136/bmj.m441 (2020).
He, Y. et al. Development and validation of a nomogram for predicting postoperative early relapse and survival in hepatocellular carcinoma. J. Natl. Compr. Cancer Network: JNCCN. 22, e237069. https://doi.org/10.6004/jnccn.2023.7069 (2023).
Santos, R., Coleman, H. G., Cairnduff, V. & Kunzmann, A. T. Clinical prediction models for pancreatic cancer in general and At-Risk populations: a systematic review. Am. J. Gastroenterol. 118, 26–40. https://doi.org/10.14309/ajg.0000000000002022 (2023).
Park, S. Y. & Nomogram An analogue tool to deliver digital knowledge. J. Thorac. Cardiovasc. Surg. 155, 526. https://doi.org/10.1016/j.jtcvs.2017.12.107 (2018).
Balachandran, V. P., Gonen, M., Smith, J. J. & DeMatteo, R. P. Nomograms in oncology: more than Meets the eye. Lancet Oncol. 16, e173–180. https://doi.org/10.1016/s1470-2045(14)71116-7 (2015).
Yang, J. et al. Early screening and diagnosis strategies of pancreatic cancer: a comprehensive review. Cancer Commun. (London England). 41, 1257–1274. https://doi.org/10.1002/cac2.12204 (2021).
Wolpin, B. M. et al. Pancreatic cancer risk and ABO blood group alleles: results from the pancreatic cancer cohort consortium. Cancer Res. 70, 1015–1023. https://doi.org/10.1158/0008-5472.Can-09-2993 (2010).
Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 41, 986–990. https://doi.org/10.1038/ng.429 (2009).
Paré, G. et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 4, e1000118. https://doi.org/10.1371/journal.pgen.1000118 (2008).
Melzer, D. et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 4, e1000072. https://doi.org/10.1371/journal.pgen.1000072 (2008).
Duffy, M. J. et al. Tumor markers in pancreatic cancer: a European group on tumor markers (EGTM) status report. Annals Oncology: Official J. Eur. Soc. Med. Oncol. 21, 441–447. https://doi.org/10.1093/annonc/mdp332 (2010).
Goonetilleke, K. S. & Siriwardena, A. K. Systematic review of carbohydrate antigen (CA 19 – 9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncology: J. Eur. Soc. Surg. Oncol. Br. Association Surg. Oncol. 33, 266–270. https://doi.org/10.1016/j.ejso.2006.10.004 (2007).
Mayerle, J. et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 67, 128–137. https://doi.org/10.1136/gutjnl-2016-312432 (2018).
Yuan, C. et al. The age-dependent association of risk factors with pancreatic cancer. Annals Oncology: Official J. Eur. Soc. Med. Oncol. 33, 693–701. https://doi.org/10.1016/j.annonc.2022.03.276 (2022).
Funding
This research did not receive any specific funding from any agencies in the public, commercial, or not-for-profit areas.
Author information
Authors and Affiliations
Contributions
All authors were involved in the preparation of this manuscript. Xin Yang designed the study and collected the data. Xin Yang and Shengjie Ding analyzed the data and wrote the manuscript. Xin Yang, Shengjie Ding, Jinlu Guo, Jingze Yang and Fan Du summarized the data. Fan Du provides a quick way to obtain data. Shi Liu and Fan Du substantial contribution to the study design, performed the operation, and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Ethics Committee of Union Hospital Affiliated to Tongji Medical College, Huazhong University of Science and Technology ([2023] Ethics Review Number (0259)).
Informed consent
The requirement for informed consent was waived because of the retrospective study design.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, X., Ding, S., Guo, J. et al. Development and validation of an easy-to-use nomogram for predicting pancreatic malignancy in patients with pancreatic mass. Sci Rep 15, 31509 (2025). https://doi.org/10.1038/s41598-025-17421-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-17421-3