Abstract
Immune response during the progression of premalignant lesions and their molecular subtype to colorectal cancer (CRC) remains unclear. Using gene expression data from 135 normal (NLs), 176 conventional adenomas (AD), 42 serrated polyps (SER), and 2760 CRC samples, we estimated overall immune activity (ImmuneScore) and tumor-infiltrating lymphocyte (TIL) abundance using the xCell tool. We evaluated association of the ImmuneScore and TILs with CRC progression using adjusted multivariable regression models. Immunohistochemistry (IHC) staining for five immunological markers was conducted on NL, early- and late-stage AD, and carcinoma tissues from 75 study participants. The consensus molecular subtypes (CMS) of adenomas and carcinomas were classified using random forest methods, and the association of immune activity with CRC progression was assessed. Immune activity consistently decreased from NLs through premalignant lesions to adenocarcinoma, more prominently in AD than SER (AD vs. NL: odds ratio = 0.86, 95% CI = 0.84‒0.88; SER vs. NL: 0.89, 0.85‒0.93). Similar patterns were observed in B cells, CD4 + effector memory T cells, CD8 + naïve T cells, and CD8 + cytotoxic T cells. IHC staining of these immunological markers verified their roles in CRC progression. Our analysis revealed that CMS3 is a major subtype of AD. Consistently, higher immune activity was observed in premalignant lesions than in CRCs of the CMS3 subtype. This study provides additional insights into alterations in immune response and their important role in CRC premalignant lesion progression and subtypes.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer-related deaths worldwide, accounting for over 1.9 million new cases and approximately 935,000 deaths in 2020 alone, according to GLOBOCAN estimates1. Most CRC progress from normal epithelium to precancerous lesion adenoma to carcinoma over the years, accumulating genetic and molecular changes2,3,4,5. Therefore, colorectal adenoma characterization can aid CRC prevention and early detection, one of the most promising strategies6,7,8,9. Conventional adenomas (ADs) and serrated polyps (SERs) are premalignant colorectal lesions with unique molecular heterogeneity and malignant potential. Truncating mutations in APC activate the WNT pathway and cause chromosomal instability in ADs, following previous research3,10,11,12. ADs subsequently accumulate gain-of-function mutations in oncogenes (chiefly KRAS) and loss-of-function mutations in tumor suppressor genes such as TP53. Conversely, SERs harbor BRAF mutations, mirroring the microsatellite instability (MSI-H) of CRCs, and are distinct from ADs in that tumorigenesis is not initiated by APC genetic disruption13,14.
The immune system, particularly tumor-infiltrating lymphocytes (TILs), promotes or suppresses cancer progression by exerting tumor-promoting or -suppressing effects. Bindea et al. demonstrated intratumoral immune cell spatiotemporal dynamics and their association with CRC tumor stage and recurrence15. TILs have been observed in diverse precancerous lesions, including pancreatic cancer16, oral squamous cell carcinoma17, lung cancer18,19,20, myeloma, and breast cancer21. Recently, Chang et al. reported the immune profile of premalignant CRC lesions in patients with Lynch syndrome22. However, knowledge of the immune response in precancerous lesions during normal to CRC progression from tissue-based studies, mainly via distinct adenoma (AD or SER) pathways, is lacking. Two recent studies revealed that colorectal adenomas can be classified as consensus molecular subtypes (CMSs) utilizing a CRC-derived classification system8,9. CMS classification stratifies tumors into four distinct subtypes (CMS1–CMS4), each characterized by unique biological features, including immune activation (CMS1), epithelial differentiation (CMS2), metabolic dysregulation (CMS3), and mesenchymal features with stromal invasion (CMS4). Applying CMS classification to adenomas may improve our understanding of their malignant potential and guide subtype-specific prevention strategies. However, immune response in CMS-based adenoma precancerous lesions is unknown.
The present study characterized the immune response using large, comprehensive population-based gene expression data from colorectal 135 normal mucosa (NLs), 176 ADs, 42 SERs, and 2760 CRC samples. We evaluated the associations of overall immune activity (ImmuneScore) and TILs with CRC progression using adjusted multivariable linear regression models. We verified five immunological protein markers associated with CRC progression using immunohistochemistry (IHC) staining of NL, early- and late-stage AD, and carcinoma tissues from 75 study participants. We used the random forest method to classify adenoma and carcinoma CMS and examined how the ImmuneScore affected subtype progression to CRC. Our study provides insights into immune response alterations in the tumor microenvironment and biological mechanisms that advance premalignant lesions and their subtypes in CRC.
Methods
Data collection and processing
Supplementary Fig. 1 shows the overall study design. We focused on four tissue types: NLs (135) from healthy controls, ADs (176) and SERs (42) from patients with adenomas, and adenocarcinomas (2760) from CRC patients. We collected samples from both publics including The Cancer Genome Atlas (TCGA) and proprietary sources (Supplementary Table 1)23,24,25 and using SYNAPSE (http://www.synapse.org)26. Supplementary Digital Content describes raw gene expression data processing (Supplementary Method 1).
Estimating abundance of TILs and evaluating their associations with CRC progression
We evaluated abundance of TILs using a gene signature-based xCell approach on the normalized expression matrix27. The relative abundance of each immune cell type in each sample was determined using enrichment scores. The overall activity, defined as the sum of the enrichment scores for immune cells, stroma, and microenvironment, was estimated as ImmuneScore, StromaScore, and MicroenvironmentScore.
To investigate the associations between alterations in abundance of immune cells (as a continuous variable) and CRC progression group (NL, AD, SER, and CRC), we used multivariable linear regression models adjusted for age, sex, and platform. Odds ratios (ORs) and 95% confidence intervals (CIs) were derived from the comparisons of AD vs. NL, SER vs. NL, and CRC vs. NL. P for trend was independently calculated for the NL-AD-CRC and NL-SER-CRC sequences. Bonferroni corrected P < 0.0007 was applied to report statistical significance (0.05/67 tests).
Similarly, to investigate the association between the abundance of immune cell type-specific genes (as a continuous variable) and CRC progression group, we used multivariable linear regression models adjusted for the covariates mentioned above. ORs and 95% CIs were derived from the same group comparisons as mentioned above. Bonferroni correction established P < 0.001 (0.05/40 tests). Analyses were conducted using R software v4.2.2 (R Project for Statistical Computing, Vienna, Austria).
Experimental verification of TIL levels in NLs, ADs, and CRC using IHC staining
We evaluated the expression of CD3+, CD8+, CD20+, CD56+, and programmed death-ligand 1 (PD-L1) in samples representing different stages of NL early-stage and late-stage AD-CRC sequences by IHC using different datasets from the Tenth People’s Hospital affiliated with Tongji University School of Medicine, Shanghai, China. We retrieved the records of 75 patients who underwent endoscopic resection or colorectal surgery at our hospital between January 2018 and March 2019. This study was approved by the Committee of Tenth People’s Hospital, affiliated with Tongji University. All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from all subjects and/or their legal guardians. Formalin-fixed and paraffin-embedded tissue blocks were collected from the pathological department’s archives at the hospital and categorized as follows: (1) polypoid hyperplasia, (2) low-grade adenoma (early-stage AD), (3) high-grade adenoma (late-stage AD), and (4) adenocarcinoma. All tissue specimens (paraffin-embedded blocks or 5-micron thick slides for IHC analyses) were collected from the archives of the referral pathology departments.
Tissue microarray (TMA) blocks were prepared from 75 formalin-fixed paraffin-embedded tissue samples (polypoid hyperplasia, n = 16; low-grade adenoma, n = 15; high-grade adenoma, n = 18; and adenocarcinoma, n = 26), as previously described28. The detailed TMA preparation methods are presented in the Supplementary Digital Content (see Supplementary Method 2). To investigate the association between immune marker levels and CRC progression stage (polypoid hyperplasia, low-grade adenoma, high-grade adenoma, and adenocarcinoma), we performed multivariable linear regression adjusted for age and sex. ORs and 95% CIs were derived from pairwise comparisons between two groups, resulting in a total of six comparisons. Nominal P < 0.05 was used to verify the statistical associations29. Additionally, matched adenoma and carcinoma immunostaining was performed in 13 patients. We compared the immune marker levels for matched adenoma and carcinoma samples from the same patients using the Wilcoxon signed-rank test.
CMS classification for colorectal adenoma and CRC
A subset of the expression matrices for adenomas and carcinomas was extracted from the normalized expression matrix. The genes were re-mapped to Entrez IDs using the “biomaRt” package in R. The random forest method of the CMS classifier23 was implemented in a subset matrix. The sample was classified as a specific CMS subtype with a posterior probability of ≥ 0.5.
Differentially expression analysis and gene set enrichment analysis
Differentially expressed gene (DEG) analyses comparing AD vs. NL, SER vs. NL, CRC vs. NL, CRC vs. AD, CRC vs. SER, and SER vs. AD were conducted using the “limma” package in R, with tissue types and evaluated batches from the “SVA” package for matrix design and linear model fitting. DEGs were significant by a > 1.5-fold change in expression and a false discovery rate (FDR) < 0.05. For pathway enrichment analyses, gene set enrichment analysis (GSEA) was performed on these DEGs using 50 hallmark gene sets, with significant gene sets determined by the Benjamini–Hochberg adjusted P < 0.05.
Results
Alterations of the immune response in CRC progression
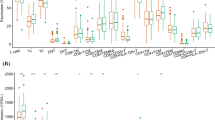
We estimated ImmuneScore, StromaScore, MicroenvironmentScore, and TIL abundances for 64 immune cell types using gene expression data from 3,113 samples (135 NLs, 176 ADs, 42 SERs, and 2760 CRCs; Supplementary Table 2). Among these 3 overall scores and 64 immune cell types in the tumor microenvironment, we observed that ImmuneScore, and MicroenvironmentScore, and 27 TILs demonstrated a significant decrease, whereas StromaScore and 20 TILs related to stroma showed a significant increase in correlation with CRC progression in NL-AD-CRC and/or NL-SER-CRC sequences, at a Bonferroni-corrected level (Fig. 1a and Supplementary Table 3). Notably, the ImmuneScore consistently decreased as carcinogenesis progressed from normal mucosa through premalignant lesions to adenocarcinoma in both NL-AD-CRC and NL-SER-CRC pathways (P < 0.0001). The AD, SER, and CRC groups showed significantly lower immune response expression than the NL group, with a more prominent decrease in AD than in SER (AD vs. NL: OR = 0.86, 95% CI = 0.84‒0.88; SER vs. NL: 0.89, 0.85‒0.93; CRC vs. NL: 0.89, 0.88‒0.9). A similar trend was observed for B cells, CD4 + effector memory T cells, CD8 + naïve T cells, CD8 + cytotoxic T cells, plasma cells, DCs, and NK cells (Fig. 1a and Supplementary Table 3).
Alterations of the immune response during the progression of premalignant lesions to colorectal cancer (a) The alterations of tumor-infiltrating lymphocytes (TILs) in P-values (left panel) and predicted fraction (right panel) (b) The alterations of immune cell-type-specific genes for P-values (left panel) and expression levels (right panel).
Alterations of the immune cell-type-specific genes in CRC progression
Trend analyses revealed that 28 out of 40 immune cell type-specific genes significantly decreased, whereas two significantly increased in correlation with CRC progression in NL-AD-CRC and/or NL-SER-CRC sequences, at a Bonferroni-corrected level (Fig. 1b and Supplementary Table 4).
Consistent with the predicted reduction in B cells, a significant decrease in B cell-specific genes (BLK, MS4A1, CD19, and HLA-DOB) was observed during the progression of carcinogenesis. The expression of more genes was significantly lower in ADs than in SERs. More importantly, the expression of CD8 + T cell-specific genes (CD8A and PRF1), T cell-specific genes (CD2, CD247, CD28, CD3G, CD6, IL2RB, and ZAP70), and cytotoxic T cell-specific genes (GZMK, GZMH, and KLRF1) significantly decreased from NLs to adenoma to cancer (Fig. 1b and Supplementary Table 4). Notably, we observed significant alterations in the expression of genes associated with T-cell exhaustion. The expression of one immune checkpoint gene (CTLA4) was significantly upregulated from NLs to adenomas (both ADs and SERs) and cancers, whereas the opposite trend was observed for the remaining four genes (BTLA, CD244, CD160, and PDCD1) (Supplementary Table 4). These results suggested that the immune response to premalignant lesions alters during colorectal carcinogenesis.
Verification of immunological markers in CRC progression using IHC staining
A panel of immunoregulatory cell subpopulations, including T (CD3 and CD8) and B (CD20) lymphocytes, NK cells (CD56), and the immune checkpoint molecule PD-L1 was analyzed using IHC staining of tissue samples from 75 study participants (Supplementary Table 5). The percentage of CD3 + cells in the epithelium was significantly lower in high-grade than in low-grade adenomas (Fig. 2 and Supplementary Table 6). Similarly, the percentage of CD8 + cells in the epithelium is significantly lower in high-grade adenomas than in low-grade adenomas, with a more pronounced decrease in adenocarcinomas than in high-grade adenomas. The percentages of other immune markers, including CD20+, CD56+, and PD-L1, were significantly higher in the polypoid hyperplasia, low-grade adenoma, and high-grade adenoma groups than in the adenocarcinoma group. Among these, only the comparison between high-grade adenoma and adenocarcinoma is shown in Fig. 2, with all P values < 0.0001 (Supplementary Table 6). Additionally, immunostaining of matched adenoma and carcinoma samples from 13 individuals showed patterns similar to those of the above immune markers (Supplementary Fig. 2 and Supplementary Table 7). Analysis of this matched dataset revealed that the percentages of CD3+ (P = 0.048), CD8+ (P = 0.0007), CD20+ (P = 0.017), and CD56+ (P = 0.002) cells were significantly higher in adenomas than in adenocarcinomas. In contrast, PD-L1 and pan-CK (epithelial marker) levels were not significantly different between adenomas and adenocarcinomas. IHC staining for these immunological markers supported the trends observed in CRC progression.
Heterogeneity in immune response in different colorectal adenoma subtypes
Next, we investigated whether colorectal adenomas elicited a heterogeneous immune response. Based on the aggregated expression data from colorectal adenomas and carcinomas, we first classified colorectal adenomas and carcinomas using random forest from the CMS classifier23. We observed 144 adenomas (130 ADs and 14 SERs); 74 adenomas (46 ADs and 28 SERs) were not classified (Fig. 3a and Supplementary Table 8). Among the ADs, 71.6% (126/176) were classified as CMS3, 1.7% (3/176) were classified as CMS2, and 0.6% (1/176) were classified as CMS1. Among the SERs, 33.3% (14/42) were classified as CMS3 subtype. None of the adenomas showed CMS4. Furthermore, we evaluated the ImmuneScores of the adenoma and carcinoma subtypes. For carcinomas, original CMS labels23 were used to evaluate the immune response. Among carcinomas, the highest ImmuneScore was observed in the CMS1 and CMS4 subtypes, and the lowest ImmuneScore was observed in the CMS2 subtype (Fig. 3b), which corresponded to the high immunogenicity of CMS1 and poor immunogenicity of CMS230. Notably, a similar ImmuneScore pattern was observed among the CMS adenoma subtypes, with the highest ImmuneScore observed in CMS1, although it was higher in CMS2 than in CMS3 (Fig. 3b). Corresponding to the aforementioned observation of ImmuneScores in adenomas and carcinomas, our results showed a higher immune score for each subtype in adenomas than in carcinomas (Fig. 3b).
Biological characterization of the progression of premalignant lesions to CRC
A total of 3814 DEGs, including 2051 between NL and AD (3363 between NL and SER) and 462 between AD and CRC (448 between SER and CRC), were identified using a threshold fold change > 1.5 and an FDR < 0.05 (Fig. 4a, left panel). We observed that the majority of alterations occurred in > 1000 DEGs in the premalignant stage. Approximately 200 additional DEGs were identified between adenomas and cancer. Notably, a greater abundance of changes in the upregulated and downregulated DEGs occurred in SER than in AD (Fig. 4a, right panel).
We identified 26 unique significantly enriched gene sets: five between NL and AD, one between AD and CRC, 11 between NL and SER, one between SER and CRC, and eight between NL and CRC (Fig. 4b and Supplementary Table 9). Upregulated pathways in AD versus NL included E2F targets, hypoxia, TNF-α signaling via NFKB, and MYC targets, while downregulated pathways included apical junctions. The upregulated pathway in CRC versus AD was mainly involved in the cell cycle (epithelial-mesenchymal transition). Upregulated pathways in SER versus NL included all aforementioned pathways in AD versus NL, oxidative phosphorylation, MTORC1 signaling, adipogenesis, G2M checkpoint, and bile acid metabolism, while downregulated pathways included apical junctions and myogenesis. The upregulated pathway in CRC versus SER was myogenesis.
Discussion
The present study characterized the immune response during the progression of premalignant lesions to CRC by evaluating TIL abundance and comparing immune cell-specific gene expression. To the best of our knowledge, this is the first study to comprehensively compare the immune responses between premalignant lesions and cancers, specifically characterizing adenomas as ADs and SERs, and our study made two major findings. We observed a decreased immune response during the progression of premalignant lesions to CRC; however, this decrease was more prominent in the AD pathway than in the SER pathway. We validated these results for the AD pathway by pathological verification. Second, most premalignant lesions were classified as CMS3 subtypes, with a higher ImmuneScore observed in premalignant lesions than in CRCs within CMS3. Additional differential gene expression analysis suggested that several cancer-related signaling pathways may play a role in the immune response during CRC progression.
The transition from immunosurveillance to immunosuppression is crucial for the progression from premalignant to cancerous ones31,32,33. Consistent with previous studies, our results showed a progressive decrease in the immune score from normal mucosa to precancerous adenoma and ultimately to adenocarcinoma, indicating diminished immune activity during CRC development. We also observed a decrease in the number of immune cells during CRC progression, which was consistent with previous findings. For example, Liu et al. have reported that the number of tumor-infiltrating CD8 + T cells is inversely correlated with tumor progression34. The activation of DCs, which is central to the initiation of immunity, contributes to tumor escape from immune surveillance35. NK cells, which play a role in innate immunity, have also been implicated in tumor surveillance36. Moreover, the concurrent decrease in ImmuneScore and TILs, alongside an increase in StromaScore and stroma-associated immune cells, suggests a shift from active immune surveillance to a more immunosuppressive, stroma-dominant tumor microenvironment. These stromal components may act as physical and biochemical barriers that limit immune cell infiltration and function, thereby contributing to immune evasion and tumor progression. This finding highlights the potential importance of stromal-immune interactions in early colorectal carcinogenesis.
A recent study by Chen et al. using single-cell resolution demonstrated different immune microenvironmental features in conventional and serrated polyps37. Most immune cell types have been reported to be increased in polyps compared to normal tissues, including CD4 + T cells, although no difference existed between polyp subtypes. Notably, the numbers of CD8 + T, NK, and gdT cells (labeled cytotoxic cells) were significantly higher in SERs than in ADs. Our results are consistent with these findings. CD + Tcm and Tgd cells were increased in both ADs and SERs compared to NLs. However, CD8 + T cells decreased in ADs and SERs compared with NL. Despite these discrepancies, our results revealed a declining pattern in overall immune activity and an increasing pattern in stromal cells during CRC progression. We improved our findings by adjusting for age, sex, and platform and increasing statistical power.
Additionally, the upregulation of CTLA-4 expression from NL to adenoma stages suggests an early establishment of immune checkpoint activation. This finding may have clinical implications, as it indicates that immune evasion mechanisms are already initiated during the premalignant phase. CTLA-4 could serve as a potential early biomarker or therapeutic target for immune modulation in high-risk adenomas, warranting further investigation.
Similar immune patterns were observed among CMS subtypes of adenoma and carcinoma CMS subtypes. This study classified adenomas (ADs and SERs) following CMS, with most adenoma subtypes classified as CMS3. These proportions of adenoma CMS subtypes were consistent with a previous study by Komor et al.8, which reported that the CMS3 ‘metabolic subtype,’ least common among CRCs, was most prevalent among adenomas (73%), followed by CMS1 ‘microsatellite instability immune subtype’ (2%) and CMS2 ‘canonical subtype’ (13%). Furthermore, we investigated the CMS subtypes following ADs and SERs, revealing disparities in CMS3 proportions between ADs (71.6%) and SERs (33.3%), indicating different mechanisms. Importantly, our data revealed a shift from CMS3 in premalignant lesions to CMS4 in CRCs, which was the most dominant subtype among cancer samples in our cohort. CMS4 is defined by mesenchymal characteristics, high stromal content, and poor prognosis9,38. Previous studies have proposed that this transition is driven by activation of epithelial–mesenchymal transition (EMT), largely regulated by TGF-β and WNT signaling pathways39. Our own pathway enrichment results support this notion, particularly in the AD-to-CRC transition, which showed pronounced enrichment of EMT-related genes. These findings align with recent literature suggesting that CMS subtypes are not static but evolve under the influence of tumor-intrinsic changes and the surrounding microenvironment, including stromal remodeling and immune suppression40,41. Together, our results provide molecular and immunological support for the concept that the CMS3-to-CMS4 shift represents a critical juncture in the adenoma–carcinoma sequence.
In addition to CMS classification, some well-known oncogenic and tumor suppressor pathways exhibit significant alterations during the colorectal oncogenic transition. Similarly, a previous study by Elsayed et al. showed that perturbations in the cell cycle and p53 pathways appeared to be notable characteristics of conventional adenoma development in the preadenomatous colon42. The extracellular matrix modeling, cell migration, and metabolic pathways are perturbed during the subsequent transformation of sporadic conventional adenomas into adenocarcinomas. The transitions from NL to AD and NL to SER caused the most significant alterations in our study (Fig. 4a). More prominently, an epithelial-mesenchymal transition from AD to CRC was observed (Fig. 4b). These pathway enrichment studies provided further insights into the potential biological mechanisms underlying the observed immune alterations.
This study has several limitations. The retrospective nature of the public datasets used may introduce selection bias, and the lack of matched normal–adenoma–tumor pairs from the same individual restricts the interpretation of inter-individual variability. Furthermore, the absence of functional validation limits causal inference. Future studies integrating spatial transcriptomics and prospective clinical validation are warranted to build on these findings. Lastly, although the statistical significance remained robust after applying Bonferroni correction with the current group sample sizes, the marked imbalance in group sizes (e.g., 2,760 CRCs vs. 135 NLs, 176 ADs, and 42 SERs) may have inflated associations related to CRC. Future studies using balanced cohorts and well-annotated mutation profiles and clinical data—including tumor location and stage—will be essential for more nuanced interpretation.
In summary, our findings provide novel insights into the alterations in immune responses and related biological mechanisms during the progression of premalignant lesions and their subtypes in CRC.
Data availability
Aggregate and anonymized data supporting the findings of this study are available in the article and Supplementary Material. The data supporting the findings of this study are available from the corresponding author (Xingyi Guo).
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Fearon, E. R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 6, 479–507 (2011).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Markowitz, S. D. & Bertagnolli, M. M. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl. J. Med. 361, 2449–2460 (2009).
Punt, C. J. A., Koopman, M. & Vermeulen, L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 14, 235–246 (2017).
Borras, E. et al. Genomic landscape of colorectal mucosa and adenomas. Cancer Prev. Res. (Phila). 9, 417–427 (2016).
Nikolaev, S. I. et al. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Res. 72, 6279–6289 (2012).
Komor, M. A. et al. Consensus molecular subtype classification of colorectal adenomas. J. Pathol. 246, 266–276 (2018).
Chang, K. et al. Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2. Ann. Oncol. 29, 2061–2067 (2018).
Cottrell, S., Bicknell, D., Kaklamanis, L. & Bodmer, W. F. Molecular analysis of APC mutations in Familial adenomatous polyposis and sporadic colon carcinomas. Lancet 340, 626–630 (1992).
Fearnhead, N. S., Britton, M. P. & Bodmer, W. F. The ABC of APC. Hum. Mol. Genet. 10, 721–733 (2001).
Rowan, A. J. et al. APC mutations in sporadic colorectal tumors: A mutational ‘hotspot’ and interdependence of the ‘two hits’. Proc. Natl. Acad. Sci. U S A. 97, 3352–3357 (2000).
Thorstensen, L. et al. Genetic and epigenetic changes of components affecting the WNT pathway in colorectal carcinomas stratified by microsatellite instability. Neoplasia 7, 99–108 (2005).
Crockett, S. D., Nagtegaal, I. D. & Terminology Molecular features, epidemiology, and management of serrated colorectal neoplasia. Gastroenterology 157, 949–966e4 (2019).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Beatty, P. L. et al. Immunobiology and immunosurveillance in patients with intraductal papillary mucinous neoplasms (IPMNs), premalignant precursors of pancreatic adenocarcinomas. Cancer Immunol. Immunother. 65, 771–778 (2016).
Foy, J. P. et al. Immunological and classical subtypes of oral premalignant lesions. Oncoimmunology 7, e1496880 (2018).
Beane, J. E. et al. Molecular subtyping reveals immune alterations associated with progression of bronchial premalignant lesions. Nat. Commun. 10, 1856 (2019).
Teixeira, V. H. et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019).
Krysan, K. et al. The immune contexture associates with the genomic landscape in lung adenomatous premalignancy. Cancer Res. 79, 5022–5033 (2019).
Degnim, A. C. et al. Alterations in the immune cell composition in premalignant breast tissue that precede breast cancer development. Clin. Cancer Res. 23, 3945–3952 (2017).
Chang, K. et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. 4, 1085–1092 (2018).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Roepman, P. et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int. J. Cancer. 134, 552–562 (2014).
Van Cutsem, E. et al. Randomized phase III trial comparing biweekly infusional fluorouracil/leucovorin alone or with Irinotecan in the adjuvant treatment of stage III colon cancer: PETACC-3. J. Clin. Oncol. 27, 3117–3125 (2009).
info@sagebase.org, S. B. Synapse | Sage Bionetworks. https://www.synapse.org
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Zhang, S. et al. Loss of Yes-associated protein represents an aggressive subtype of colorectal cancer. J. Cancer. 10, 689–696 (2019).
Varghese, F., Bukhari, A. B., Malhotra, R. & De, A. IHC profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 9, e96801 (2014).
Dienstmann, R. et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 17, 79–92 (2017).
Cui, G. Immune battle at the premalignant stage of colorectal cancer: focus on immune cell compositions, functions and cytokine products. Am. J. Cancer Res. 10, 1308–1320 (2020).
Dunn, G. P., Old, L. J. & Schreiber, R. D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 22, 329–360 (2004).
Karlsson, M. et al. Evidence for immunosurveillance in intestinal premalignant lesions. Scand. J. Immunol. 71, 362–368 (2010).
Liu, F. et al. TNFα cooperates with IFN-γ to repress Bcl-xL expression to sensitize metastatic colon carcinoma cells to TRAIL-mediated apoptosis. PLOS ONE. 6, e16241 (2011).
Vicari, A. P., Caux, C. & Trinchieri, G. Tumour escape from immune surveillance through dendritic cell inactivation. Semin Cancer Biol. 12, 33–42 (2002).
Levy, E. M., Roberti, M. P. & Mordoh, J. Natural killer cells in human cancer: from biological functions to clinical applications. J Biomed Biotechnol 676198 (2011). (2011).
Chen, B. et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 184, 6262–6280e26 (2021).
Fessler, E. et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med. 8, 745–760 (2016).
Waldner, M. J. & Neurath, M. F. TGFβ and the tumor microenvironment in colorectal cancer. Cells 12, 1139 (2023).
Fessler, E. & Medema, J. P. Colorectal cancer subtypes: developmental origin and microenvironmental regulation. Trends Cancer. 2, 505–518 (2016).
Rejali, L. et al. Principles of molecular utility for CMS classification in colorectal cancer management. Cancers (Basel). 15, 2746 (2023).
Elsayed, I. et al. Adenoma to carcinoma: A portrait of molecular and immunological profiles of colorectal sporadic tumors. Int. Immunopharmacol. 100, 108168 (2021).
Acknowledgements
Data processing and statistical analysis were conducted at the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University.
Funding
X. G. received support from the National Cancer Institute (R37CA227130). J. C. received support from the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (RS-2023-00210034).
Author information
Authors and Affiliations
Contributions
Authors’ contributionsZhishan Chen: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Investigation (lead); Writing – original draft (lead); Writing – review and editing (equal).Dongwoo Kim: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Investigation (lead); Writing – review and editing (equal).Jifeng Wang: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Investigation (lead); Writing – original draft (lead); Writing – review and editing (equal).Qiongyi Huang: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Yaohui Gao: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Jiayi Zheng: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Linshuoshuo Lyu: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Yinying Li: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Ling Lu: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Ruting Xie: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Jung Sun Kim: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Ah-Reum Lim: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Se Ryeon Lee: Investigation (equal); Writing – review and editing (equal).Jinny Park: Investigation (equal); Writing – review and editing (equal).Hwa Jung Sung: Data curation (equal); Investigation (equal); Writing – review and editing (equal).Dongyan Han: Conceptualization (equal); Data curation (lead); Investigation (equal); Writing – review and editing (equal).Xingyi Guo: Conceptualization (lead); Data curation (lead); Supervision (lead); Formal analysis (lead); Investigation (lead); Writing – original draft (lead); Writing – review and editing (lead).Jungyoon Choi: Conceptualization (equal); Data curation (equal); Formal analysis (lead); Investigation (lead); Writing: original draft (lead); Writing: review and editing (lead).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest. The funders had no role in the design of the study; collection, analysis, and interpretation of the data; writing of the manuscript; or the decision to submit the manuscript for publication. All authors had access to the study data. The corresponding author is responsible for the decision to submit this manuscript for publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Z., Kim, D., Wang, J. et al. Adaptive immune responses associated with the progression of premalignant lesions to colorectal cancer. Sci Rep 15, 33369 (2025). https://doi.org/10.1038/s41598-025-17653-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-17653-3