Abstract
Headaches are a nearly universal human experience traditionally diagnosed based solely on symptoms. Recent advances in imaging techniques and artificial intelligence (AI) have enabled the development of automated headache detection systems, which can enhance clinical diagnosis, especially when symptom-based evaluations are insufficient. Current AI models often require extensive data, limiting their clinical applicability where data availability is low. However, deep learning models, particularly pre-trained ones and fine-tuned with smaller, targeted datasets can potentially overcome this limitation. By leveraging BioMedCLIP, a pre-trained foundational model combining a vision transformer (ViT) image encoder with PubMedBERT text encoder, we fine-tuned the pre-trained ViT model for the specific purpose of classifying headaches and detecting biomarkers from brain MRI data. The dataset consisted of 721 individuals: 424 healthy controls (HC) from the IXI dataset and 297 local participants, including migraine sufferers (n = 96), individuals with acute post-traumatic headache (APTH, n = 48), persistent post-traumatic headache (PPTH, n = 49), and additional HC (n = 104). The model achieved high accuracy across multiple balanced test sets, including 89.96% accuracy for migraine versus HC, 88.13% for APTH versus HC, and 83.13% for PPTH versus HC, all validated through five-fold cross-validation for robustness. Brain regions identified by Gradient-weighted Class Activation Mapping analysis as responsible for migraine classification included the postcentral cortex, supramarginal gyrus, superior temporal cortex, and precuneus cortex; for APTH, rostral middle frontal and precentral cortices; and, for PPTH, cerebellar cortex and precentral cortex. To our knowledge, this is the first study to leverage a multimodal biomedical foundation model in the context of headache classification and biomarker detection using structural MRI, offering complementary insights into the causes and brain changes associated with headache disorders.
Similar content being viewed by others
Introduction
Headaches are a common condition affecting a substantial portion of the global population1. Early detection and accurate diagnosis are essential for effective management and treatment. With advancements in modern imaging techniques and artificial intelligence (AI) technologies, there is an increasing ability to develop automated headache detection systems to support clinicians in making precise diagnoses. In automated headache detection, supervised machine learning algorithms have been increasingly utilized to identify and classify various types of headaches, including migraine2. These methods rely on labeled (annotated) datasets to learn the complex patterns that distinguish headache conditions from normal cases. By integrating diverse patient data including clinical records, machine learning approaches can directly associate patient characteristics with headache diagnoses. Messina and Filippi3 reviewed how machine learning methodologies have been used in headache research. Primarily for migraine, machine learning algorithms have leveraged patient-provided information along with brain imaging data to differentiate those with migraine from HC and subtype those with migraine according to differing symptoms4,5.
Deep learning techniques have further advanced this field by enabling comprehensive analysis of whole brain imaging data, such as MRI scans. Such methods are now widely used for different tasks such as brain tumor detection6,7 anomaly detection8,9 metastases detection10. Convolutional Neural Networks (CNNs)11including architectures, such as ResNets, have been used successfully with multimodal MRI data for migraine classification11,12. Recently, Siddiquee et al.5 showed that a ResNet-18-based 3D classifier demonstrated promising performance in classifying images into categories such as HC, migraine (Mig), acute post-traumatic headache (APTH), and persistent post-traumatic headache (PPTH). Nevertheless, a significant challenge in developing such systems is the limited availability of imaging data and their annotations, particularly for less common headache subtypes.
Conventional supervised deep learning methods require large quantities of labeled data to guide the model training. Because gathering extensive annotated medical data is both time-intensive and costly, alternative approaches have been developed. Alzubaidi et al.13 presented a comprehensive list of tools to address the issue of data scarcity in deep learning, including transfer learning, self-supervised learning, deep synthetic minority oversampling, and other techniques. Transfer learning leverages pre-trained models and fine-tunes them on available medical data, substantially reducing the need for large-scale annotations. In addition, self-supervised learning trains models on unlabeled data by solving auxiliary tasks, which helps the model learn useful representations without needing large, labeled datasets. Both approaches can potentially alleviate the challenges of limited annotated data by reducing the reliance on large, labeled datasets. However, in medical imaging, traditional pre-trained models such as those developed on ImageNet14 are often suboptimal due to domain differences (e.g., ImageNet is trained on natural image dataset). A medical image pre-trained model may provide advantages for medical tasks with significantly fewer samples than those pre-trained on ImageNet15. However, significant challenge exists in developing medical imaging pre-trained models due to the lack of large volume of annotated medical images. Training Vision Transformers (ViTs) from scratch on limited data is known to result in poor generalization, largely due to their lack of inherent inductive biases such as spatial locality and hierarchical feature representation16. An innovative emerging idea is to leverage text and images in a multimodal fashion and construct a contrastive learning problem which is annotation-free.
CLIP17 (Contrastive Language-Image Pretraining) is a multimodal AI model developed by OpenAI that can understand images and text together. Pre-trained models using CLIP thus can serve as foundation models tailored to tasks with smaller datasets without the need for annotations. In this study, we explored using the pre-trained CLIP based model for headache classification when having access to a relatively limited number of brain MRIs. The CLIP model comprises an image encoder, implemented as a Vision Transformer (ViT)18 and a text transformer19 encoder, which is trained jointly on image-text pairs using a contrastive learning approach. We utilize BioMedCLIP20a foundational model based on the CLIP framework that incorporates ViT-B (base ViT model) as the image encoder with ~ 86 million parameters, and PubMedBERT21 as the text encoder. As its name suggests, this foundation model is specifically trained for biomedical applications. It is pre-trained on the PMC-15 M dataset, which contains 15 million biomedical image-text pairs, using the CLIP framework with contrastive learning. While this foundation model was first released through archival in March 2023, it was published in the New England Journal of Medicine AI in Dec. 2024. During the period, researchers already explored the advantages of contrastive language-image pretraining in medical applications including hematological image classification22 differentiating between normal and malignant cells in digital pathology23. Notably, the applications make use of both images and text.
Although BioMedCLIP is a multimodal model combining ViT and PubMedBERT, in this study we exclusively use the ViT image encoder component only for MRI-based headache classification, due to absence of textual annotations in MRI datasets. To address this, our study uniquely utilizes BiomedCLIP’s image encoder independently for fine-tuning on our institutional brain MRI dataset. The image encoder of this foundation model captures intricate biomedical patterns from extensive pre-training on millions of image-text pairs. These learned representations inherently carry relevant anatomical insights beneficial for diagnostic tasks without explicit textual data. Thus, exclusively leveraging the pre-trained image encoder strategically maximizes transferable biomedical knowledge. By leveraging pre-trained image encoder, we can effectively fine-tune the model on our relatively small dataset of T1-weighted brain MRIs for headache classification. This strategy not only circumvents the limitations of scarce annotated data but also harnesses the deep, contextual understanding embedded in the model, thereby enhancing performance in specialized diagnostic applications.
Our dataset consists of individuals with three headache types, Migraine, APTH, PPTH, and HC. Migraine, a primary headache type, affects approximately 14% of the general population24,25 and is a leading cause of disability. Migraine is one of the most common headache types among patients seeking care in outpatient clinics. PTH (APTH and PPTH), a secondary headache type, is a headache attributed to a traumatic injury to the head. In this study, we included participants who had PTH due to mild traumatic brain injuries (mTBI) and who had headache persistence for 3 months or less (APTH) or for longer than 3 months (PPTH). We fine-tuned the BioMedCLIP model on an MRI dataset of 721 subjects including 424 from IXI public dataset, and 297 participants imaged at Mayo Clinic, including Migraine, APTH, PPTH, and HC. Specifically, we only used the pre-trained image encoder, a ViT model, from the foundation model and fine-tuned it further using cross-entropy loss to adapt it for headache classification using 3D MRI data. We propose a novel slice-based multiple-instance learning evaluation strategy to assess our model’s performance. We employed Grad-CAM26 (Gradient-weighted Class Activation Mapping), a model explanation technique to identify the brain regions associated with different headache types. This method enabled us to extract critical biomarkers that contribute to the model’s classification decisions for Migraine, APTH, and PPTH. One reason we chose to include 424 IXI subjects in this study is to support comparison study to the published work26 to demonstrate the advantages of foundation model. We demonstrate that the pre-trained model with fine-tuning considerably outperforms models trained from scratch by comparing the classification metrics and biomarkers derived from the two approaches. In summary, we make the following contributions in this study:
-
We propose a method to fine-tune the pre-trained image encoder (ViT) from the multimodal foundation model for headache classification using structural brain MRI. By leveraging its prior training on millions of biomedical image–text pairs, we transfer rich domain-specific representations to a single-modality MRI task, despite the absence of paired text annotations.
-
Our proposed model demonstrates superior and consistent accuracy across multiple cross-validation folds compared to training a model from scratch given same imaging data.
-
We identify potential biomarkers by extracting and ranking brain regions significantly influencing the model’s predictions, thereby providing insights into neuroanatomical correlations of headache phenotypes.
Materials and methods
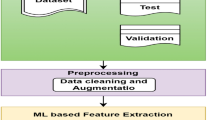
Dataset
We collected T1-weighted structural MRI data from 96 individuals with Migraine, 48 with APTH, and 49 with PPTH, diagnosed in accordance with the diagnostic criteria of the International Classification of Headache Disorders (ICHD)27,28. Participants with APTH and PPTH were enrolled at the Mayo Clinic Arizona or the Phoenix Veterans Administration, but all participants had MRI at the Mayo Clinic Arizona. Additionally, we included 104 HC from the Mayo Clinic and extended our dataset incorporating MRI scans of 424 HC from the publicly available IXI dataset18. For each data set, the information of participants is summarized in (Table 1).
Institutional data set: participant enrollment and characteristics
This study was approved by the Mayo Clinic Institutional Review Board (IRB), the Phoenix Veterans Administration IRB, and the United States Department of Defense Human Research Protection Office, and all participants provided written informed consent for their participation. The authors declare that all the methods in this article were performed in accordance with the relevant guidelines and regulations in the editorial and publishing policies of Scientific Reports (https://www.nature.com/srep/journal-policies/editorial-policies#experimental-subjects). At the time of enrollment, migraine participants were diagnosed with episodic or chronic migraine, with or without aura, based on the most recent edition of the International Classification of Headache Disorders (ICHD-3 beta or ICHD-3). Participants with APTH or PPTH had PTH attributed to mTBI according to the latest ICHD criteria (ICHD-3 beta or ICHD-3). Individuals with a history of moderate or severe traumatic brain injury were excluded from the study. Participants with APTH were enrolled between 0 and 59 days following mTBI, while those with PPTH were enrolled at any point after they had PTH for longer than three months. HC were excluded if they had a history of TBI or any headache type other than infrequent tension-type headache.
Image acquisition was performed using 3 Tesla Siemens scanners (Siemens Magnetom Skyra, Erlangen, Germany) equipped with a 20-channel head and neck coil. Anatomical T1-weighted images were captured using magnetization-prepared rapid gradient echo (MPRAGE) sequences. The imaging parameters for acquiring T1-weighted images were as follows: repetition time (TR) = 2400 ms, echo time (TE) = 3.03 ms, flip angle (FA) = 8°, and voxel size = 1 × 1 × 1.25 mm³.
The participants with Migraine had a mean age of 39.9 years (\(\:\pm\:\)11.6) and 74.7% were female. They had a mean headache frequency of 15.3 days per month; 37 had episodic migraine and 59 had chronic migraine. Additionally, 49 patients reported experiencing aura with at least some of their migraine attacks. The PTH group included 48 participants with APTH, with a mean age of 41.6 years ( ± 12.7) and 60.4% were female. Individuals with APTH experienced their first PTH symptoms an average of 24.5 days (± 14.5) prior to imaging. They reported headaches on 76.3% (±29.6%) of days following the onset of APTH. Their mTBIs were due to motor vehicle accidents (n = 20), falls (n = 21), and direct hits to the head (n = 7). The dataset also included 49 patients with PPTH. The mean age of the PPTH patients was 38.1 years (±10.5 years), and 34.7% were female. PPTH participants experienced headaches on an average of 15.3 days (±7.4 days) per month and they experienced PPTH for an average of 10.8 ± 8 years. The mTBIs leading to PPTH were attributed to various causes, including blast injuries (n = 22), falls (n = 12), sports-related injuries (n = 8), and motor vehicle accidents (n = 7).
Public IXI data set: participant enrollment and characteristics
The IXI dataset, which is an integral part of the “Information eXtraction from Images” project (EPSRC GR/S21533/02)29, is a comprehensive compilation of MRIs from healthy individuals. It was acquired between June 2005 and December 2006 to facilitate medical imaging and neuroimaging research. In addition to Magnetic Resonance Angiography (MRA) images and diffusion-weighted images (DWI) with 15 gradient orientations, the dataset comprises T1, T2, and Proton Density (PD) weighted images. The data were collected at three hospitals in London, each of which utilized a distinct MRI system to capture a variety of imaging parameters. Hammersmith Hospital utilized a Philips 3T scanner (TR = 9.6, TE = 4.6, acquisition matrix = 208 × 208, FA = 8°), Guy’s Hospital employed a Philips 1.5T scanner (TR = 9.813, TE = 4.603, FA = 8°), and the Institute of Psychiatry utilized a GE 1.5T scanner. The images are supplied in the Neuroimaging Informatics Technology Initiative (NIFTI) format, which simplifies their utilization in a variety of imaging software. In total, 277 male and 342 female participants were enrolled. Our final cohort contains 194 male and 230 female subjects with an average age of 42.4 years (± 13.0 years), matching the average ages of the Mayo Clinic dataset. In total, the dataset includes 721 subjects: 424 HC from IXI, and 297 local subjects (96 Migraine, 48 APTH, 49 PPTH, 104 HC).
Image preprocessing
The preprocessing pipeline for the Brain MRI dataset involved several standardized image processing steps to enhance data uniformity and facilitate accurate analysis. Images obtained from the Mayo Clinic were collected in DICOM format and later converted to NIFTI format. Initially, brain extraction was carried out using the Brain Extraction Tool (BET)30 from the FMRIB Software Library (FSL) (Wellcome Centre, University of Oxford, UK) BET efficiently isolates cerebral structures by removing extraneous non-brain tissues, resulting in skull-stripped MR images. This critical step normalizes image contents across the combined datasets, effectively mitigating biases from varying scanners, acquisition protocols or batch effects in both public IXI and institutional dataset. Following these steps, linear image registration was performed using the FMRIB’s Linear Image Registration Tool (FLIRT)31 also from FSL. FLIRT was applied to align individual MR images to the Montreal Neurological Institute (MNI) standard brain atlas (MNI152 1 mm). This normalization step involves linear affine transformations to match the anatomical features and spatial orientation of each subject’s brain to the standardized template. The registration effectively corrects for variations in brain size, shape, and orientation, thus further normalizing the images and enabling robust inter-subject comparisons. Freesurfer was subsequently utilized to execute White Matter Parcellation (wmparc), generating parcellation masks while excluding 14 irrelevant regions, including the left vessel, right vessel, right lateral ventricle, left lateral ventricle, right unsegmented white matter, left unsegmented white matter, left choroid plexus, right choroid plexus, left inferior lateral ventricle, right inferior lateral ventricle, fourth ventricle, third ventricle, cerebral spinal fluid (CSF) and optic chiasm. The resulting preprocessed images, standardized through these rigorous methods, provided a robust dataset suitable for fine-tuning the pre-trained BioMedCLIP ViT model for accurate headache classification. Finally, 3D MRIs were converted into 2D sagittal slices for model fine-tuning and evaluation, ensuring consistency and precision in downstream analysis. To reduce computational complexity and facilitate transfer learning with ViT, we converted 3D MRI scans into 2D sagittal slices, as ViT models are optimized for 2D image inputs.
Headache classification and automatic biomarker extraction
Our method involves fine-tuning the pre-trained ViT image encoder from the BioMedCLIP foundation model, which is originally trained using a contrastive learning framework that leverages both image and text embeddings from its respective encoders. The model learns in a contrastive manner by maximizing the similarity score of the same image-text pair in the dataset and minimizing the similarity score for the unpaired image-texts in the dataset. This process is illustrated in (Fig. 1). In this study, we utilized the pre-trained vision transformer image encoder from the foundation model and fine-tuned it for the headache classification task. The fine-tuning process employs a cross-entropy loss function in a supervised learning framework. We fine-tuned the ViT model for three binary classification tasks: HC vs. Migraine, HC vs. APTH, and HC vs. PPTH, with the goal of extracting subtyping biomarkers. To prepare the data for fine-tuning and evaluation, we preprocessed the 3D MRI scans by slicing them into 2D sagittal slices for each patient.
Overview of the BioMedCLIP pre-training framework, utilizing a contrastive learning approach. The process involves fine-tuning a pre-trained vision transformer (ViT) as the image encoder, along with a parallel text encoder. During training, image and text embeddings are generated, and the similarity scores between matching image-text pairs (highlighted diagonally) are maximized, while the similarity scores of non-matching pairs (off-diagonal) are minimized. This strategy ensures the alignment of visual and textual representations. The image included in this figure is for illustrative purposes only and was not actually included in this study.
The dataset was randomly divided into three subsets: training, validation, and blind testing (Table 2). The validation set was used to identify the best-performing deep learning model, while the blind testing set was employed to evaluate the robustness and generalization of the model on unseen data. We executed five-fold cross-validation for each task and reported the average and standard deviation results. For each fold, we maintained the same number of patients for training, validation, and testing as specified in Table 2; however, each distinct fold utilized different test and validation samples, ensuring no overlap between the folds for model evaluation. For evaluation, we followed the multi-instance learning32,33 scheme, a variation of supervised learning. In this scheme, multiple patches of the subject’s image are considered as bag of instances and assigned to the same label. We employed a slice-based Multi-Instance Learning evaluation process where slice-level embeddings are extracted for each patient. We extracted each slice from the 3D MRI image, and for each slice, we derived the embeddings after passing the slices through the image encoder and converted them into probabilities, indicating the likelihood of being healthy or having headache. After that, we aggregated the slice-level scores by averaging them to make patient-level classification predictions. Notably, while the training set is imbalanced, we applied oversampling method to overcome this. The test set is kept balanced to facilitate a robust evaluation with five-fold cross validation. In addition to standard classification metrics, we report precision, specificity, and sensitivity to provide a comprehensive assessment of model performance and account for class imbalance.
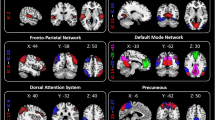
Once the classifier was trained for each task, to identify brain regions associated with different headache phenotypes, we utilized Gradient-weighted Class Activation Mapping (Grad-CAM) on the MRI images. Grad-CAM was applied to the final attention block of the fine-tuned ViT model. The resulting activation maps were aggregated across slices and projected onto anatomical regions to compute regional importance scores. For each patient, we first applied Grad-CAM to generate an activation map highlighting spatial locations contributing significantly to the classification decision. Next, we mapped the activation scores onto predefined brain regions by taking the maximum Grad-CAM activation score within each specific region, effectively assigning each region a representative activation value. We repeated this step for all patients in our dataset and then averaged these regional activation scores across patients to obtain patient-level consensus activation maps. Additionally, we executed this procedure separately for each of the five cross-validation folds, deriving regional activation maps from five independently trained models. Finally, we computed the average regional activation across these five folds, thereby ensuring robust and stable identification of regions consistently implicated in headache classification. The resulting regions were ranked according to their averaged activation scores, and the top activated regions were selected as the most salient biomarkers associated with headache conditions. The overall process is visualized in (Fig. 2).
Results
The fine-tuned model’s performance and the corresponding brain regions identified via Grad-CAM are detailed below. We summarize the three binary classification results, including accuracy, precision, specificity, and sensitivity in (Table 3). Each case is further discussed in the following subsections with the extracted biomarkers. We performed five-fold cross validation for each binary classification task, each with balanced non-overlapping testing and oversampled training sets and reported the mean along with the standard deviation of accuracy, precision, specificity and sensitivity.
Migraine classification
On the blind five-fold test sets, the model achieved an average accuracy of 89.96% (± 8.10%), with a precision of 85.71% (± 9.10%), sensitivity of 96.66% (± 6.60%), and specificity of 83.33% (± 10.54%) for the HC versus Migraine classification.
Key brain regions contributing to the model’s classification decisions were identified using Grad-CAM activation score rankings. These regions are visualized in (Fig. 3). Significant regions included the postcentral cortex, postcentral white matter, supramarginal cortex, supramarginal white matter, superior temporal cortex, superior temporal white matter, inferior parietal white matter, inferior parietal cortex, superior parietal cortex, superior parietal white matter, precuneus cortex, precuneus white matter, banks of the superior temporal sulcus white matter, hippocampus, middle temporal cortex, cerebellar cortex, and lingual white matter.
Acute post-traumatic headache classification
For the classification task distinguishing APTH from HC, we achieved an accuracy of 88.13% (± 6.10%), with 88.33% (± 9.00%) precision, 89.99% (± 7.50%) sensitivity, and 86.53% (± 11.58%) specificity on the five blind test sets.
Key brain regions contributing to the model’s classification decisions were identified using GradCAM activation scores. Significant regions included the rostral-middle frontal cortex, rostral-middle frontal white matter, precentral cortex, postcentral cortex, supramarginal cortex, superior temporal cortex, superior temporal white matter, middle temporal cortex, middle temporal white matter, lateral occipital cortex, lingual cortex, inferior parietal cortex, pars opercularis cortex, superior frontal cortex, inferior temporal cortex, inferior temporal white matter, fusiform cortex, cerebellar cortex, and cerebellar white matter. These regions are visualized in (Fig. 4).
Persistent post-traumatic headache classification
The model achieved an accuracy of 83.13% (± 5.20%), with 80.80% (± 10.36%) precision, specificity of 76.20% (± 13.60%) and sensitivity of 93.26% (± 8.20%) for HC versus PPTH classification task.
Significant brain regions contributing to the model’s classification decisions, as identified by GradCAM activation scores, included rostral middle frontal cortex, precentral cortex, precentral white matter, postcentral cortex, postcentral white matter, supramarginal white matter, supramarginal cortex, superior temporal cortex, inferior parietal cortex, superior parietal white matter, superior parietal cortex, lateral occipital cortex, lateral occipital white matter, precuneus cortex, middle temporal cortex, lingual cortex, cerebellar cortex. These regions are visualized in (Fig. 5).
Discussion
One of the primary challenges in medical imaging-based downstream tasks, e.g., biomarker extraction, is the scarcity of large, annotated imaging datasets. Most previous deep learning methods require task-specific labeled training data to extract useful image features. However, a single-modal image model trained from scratch on a relatively small dataset could be limited in extracting robust disease-related image features. Multi-modal models trained on large-scale image-text pairs34 have been shown to learn a robust understanding of visual concepts guided by natural language descriptions, allowing them to discriminate between categories they haven’t explicitly seen before. This task-agnostic large-scale training to extract semantically meaningful image representations allows the model to adapt to different downstream applications with fine-tuning. In this study, we used headache disorders as a case example for testing our methods. Prior studies had demonstrated differences in brain structure amongst those with migraine or PTH compared to HC, making our headache dataset useful for such testing. In headache classification, since access to labeled datasets and clinically verified biomarkers is rare, we hypothesized that leveraging a pre-trained multi-modal model may be a viable solution for extracting robust headache biomarkers.
The effectiveness of BiomedCLIP stems from its large-scale training on image-text pairs tailored for biomedical applications. It captures a more complete picture of the data by integrating complementary information from various sources (text and images here), which also loosely mimics how humans perceive the world using multiple senses. With the rationale of localizing biomarkers using imaging data only, we used only the image encoder of this foundation model for headache classification.
We identified brain regions utilizing the Grad-CAM activation scores that were important for differentiating three types of headaches from HC. As expected, based on prior imaging studies of headache and headache-associated symptoms that localize to many brain regions, there were numerous brain regions contributing to classification of each headache type. It is beyond the scope of this manuscript to discuss the potential role of every identified brain region in migraine or PTH pathophysiology or symptomatology. However, regions identified in our analyses have been previously implicated in migraine and/or PTH, supporting the validity of our findings. For example, the hippocampus has been highlighted as an area associated with migraine and other chronic pain, perhaps related to stress associated with pain35,36. Atypical functional connectivity of the supramarginal gyrus has been identified amongst those with migraine, likely due to its role as a somatosensory association region. The postcentral gyrus, a key region of the pain matrix given its involvement in primary somatosensation, was identified as significant for migraine in prior research37. Several of the identified regions are part of the default mode network, a network that has previously identified to be abnormal in migraine38. Amongst those with PTH, several regions that we found to differ compared to HC have been previously identified as regions associated with PTH39. For the regions found significant for PPTH, we found similar regions in one of our prior analyses (Siddique et al.5, such as the lingual, supramarginal, middle temporal, lateral occipital, postcentral, and cerebellar cortex. Chong et al.40. showed that there are cortical thickness differences for PPTH patients in precuneus, precentral, supramarginal inferior and superior parietal regions and from our Grad-CAM activation scoring, we also found these regions to be significant for the classification prediction. Differentiating participants with headache from HC provides potentially important information about brain regions that might participate in generating each headache type or be impacted by recurrent headaches. There could eventually be clinical utility for brain MRI-based headache classification models as well. Theoretically, “normalization” of structure or function in brain regions identified as important for differentiating those with headache from HC could be used as biomarkers of headache preventive treatment response, perhaps especially useful when developing new headache therapies. Furthermore, differentiating patients with headaches from HC is an important step toward developing classification models that differentiate between headache types with overlapping features, such as migraine and PTH. Although we do not see a future in which brain MRI is required for all headache diagnoses, it could be used to supplement diagnostic decision making when the diagnosis is difficult to determine based on patient symptoms alone.
To the best of our knowledge, the only deep learning-based approach for headache classification reported in the literature is from Siddiquee et al.5 which used the same dataset as ours. The regions identified as significant for migraine using the proposed method slightly overlap with those identified by Siddiquee et al.., including the hippocampus and precentral white matter. Additionally, we found other regions that contribute to migraine, including the supramarginal cortex, superior temporal cortex, supramarginal white matter, and precuneus. We also found multiple overlapping significant regions for APTH with those identified by Siddiquee et al.., including the cerebellar cortex, lateral occipital cortex, inferior parietal, and lingual. We found some new regions that are significant for APTH, including superior temporal, middle temporal, rostral middle frontal cortex, postcentral cortex, and fusiform. In our analysis of PPTH, we identified additional overlapping significant regions consistent with our prior study, including the cerebellum, lateral occipital white matter, precuneus, postcentral cortex, and inferior parietal cortex. Furthermore, we discovered new regions, such as the precentral cortex, rostral middle frontal cortex, supramarginal cortex, and superior temporal cortex, contributing to PPTH classification. The classification accuracy of the proposed method is significantly improved for migraine and APTH classification. For migraine, we achieved an average accuracy of 89.96% for five-folds, which is significantly improved compared to the 75% accuracy achieved in our previous method. We saw similar improvements for APTH, where our new method achieved 88.13% average accuracy compared to 75% accuracy in the previous method. We achieved an accuracy of 83.13% for PPTH, which is lower than the 91.70% accuracy reported in the prior method. However, it is crucial to highlight that our evaluation utilized a more rigorous five-fold cross-validation approach, in contrast to the prior work, which assessed only a single model on a limited dataset. Consequently, while our accuracy may be lower for PPTH classification, it is indicative of a more robust and generalizable performance owing to the comprehensive evaluation methodology.
Although the classification accuracy for Migraine and APTH were higher with the current method, we found that some of the significant brain regions identified by our current method differed from those identified in our previous study. This discrepancy could be attributed to the following reasons: (i) Our current method employs a pretrained model trained on PMC-15 M, a dataset comprising 15 million figure-caption pairs extracted from biomedical research articles in PubMed Central. As a result, our current model is likely more generalized compared to the one used in the previous study. (ii) We adopted a different model architecture and training approach, specifically ViT with pretraining, which may capture features more effectively than the traditional ResNet trained from scratch. While this study focused on binary classification tasks (e.g., HC vs. each headache type), future work will explore multi-class classification (e.g., Migraine vs. APTH vs. PPTH vs. HC) to better reflect real-world diagnostic needs. We also plan to evaluate cross-site generalization and model calibration in future clinical validation studies.
Limitations of this study include: (i) Although we used a pretrained ViT model, the validation and testing sets are still small. (ii) MRIs from those with headache and HCs were obtained using various scanners and acquisition parameters. While this variability might be viewed as a limitation, it can also be considered a strength of our study. The heterogeneity in the dataset may lower classification accuracy but increases the likelihood that the classification results will generalize to new patient populations. (iii) The consistency observed in the brain regions that most contributed to classification in this study and the previous studies may partly be attributed to participant overlap between the studies. (iv) Although the participants in the IXI dataset are classified as healthy, it is possible that they were not screened for conditions such as migraine, a history of mTBI, or PTH. (v) All participants with migraine and most participants with PTH were enrolled at the Mayo Clinic. (vi) As this study relies solely on structural MRI, factors such as sex, handedness, medication use, aura presence, and white matter hyperintensities were not controlled for. This broad inclusion may have introduced variability and impacted the robustness of the model. (vii) We used six samples from each class in the test set to enable a one-to-one comparison with the only existing method in the literature, but this small sample size may lead to potential statistical instability in the reported accuracy metrics. Although the participant demographics, headache characteristics, and mTBI characteristics are consistent with expectations, it is possible that the enrolled patient population differs from the general population of individuals with migraine and PTH, which could limit generalizability of study results. In the future, we will explore other experimental designs, such as a different data split, with a larger set for testing and a smaller set for fine tuning.
Conclusion
This study, using headaches as a case example, demonstrates the potential of the pretrained CLIP model for disease classification and biomarker classification, particularly in scenarios with limited training data. Compared to the literature using deep learning (e.g., ResNet-based model) trained from scratch, the CLIP-based approach showed improved classification performance. Notably, the evaluation of our model using a more robust five-fold cross-validation process ensures that the extracted biomarkers are derived from multiple models, enhancing their reliability compared to those generated in previous work. These findings highlight the promise of pretrained models in advancing clinical diagnostic tools and underscore the importance of rigorous evaluation methodologies.
Data availability
Data from the two studies sponsored by the United States Department of Defense (DOD) and one of the studies sponsored by the National Institutes of Health (NIH) will be made available through the Federal Interagency Traumatic Brain Injury Research (FITBIR) Informatics System in accordance with the rules and regulations of the DOD and NIH funding contracts. Patient consent for the other NIH-sponsored study and from the Amgen-sponsored study did not include a data sharing agreement. The IXI data set can be obtained from https://brain-development.org/ixi-dataset/.
Abbreviations
- APTH:
-
Acute post-traumatic headache
- DL:
-
Deep learning
- HC:
-
Healthy control
- Mig:
-
Migraine
- ML:
-
Machine learning
- PPTH:
-
Persistent post-traumatic headache
- PTH:
-
Post-traumatic headache
References
Stovner, L. J., Hagen, K., Linde, M. & Steiner, T. J. The global prevalence of headache: an update, with analysis of the influences of methodological factors on prevalence estimates. J. Headache Pain. 23 (1), 34. https://doi.org/10.1186/s10194-022-01402-2 (2022).
Khan, L. et al. Migraine headache (MH) classification using machine learning methods with data augmentation. Sci. Rep. 14 (1), 5180. https://doi.org/10.1038/s41598-024-55874-0 (2024).
Messina, R. & Filippi, M. What we gain from machine learning studies in headache patients. Front. Neurol. 11, 221. https://doi.org/10.3389/fneur.2020.00221 (2020).
Chong, C. D. et al. Migraine classification using magnetic resonance imaging resting-state functional connectivity data. Cephalalgia 37 (9), 828–844. https://doi.org/10.1177/0333102416652091 (2017).
Rahman Siddiquee, M. M. et al. Headache classification and automatic biomarker extraction from structural MRIs using deep learning. Brain Commun. 5 (1), fcac311. https://doi.org/10.1093/braincomms/fcac311 (2022).
Bouhafra, S. & El Bahi, H. Deep learning approaches for brain tumor detection and classification using MRI images (2020 to 2024): A systematic review. J. Imaging Inf. Med. 30 https://doi.org/10.1007/s10278-024-01283-8 (2024).
Biradar, S. & Virupakshappa. AG-MSTLN-EL A Multi-source transfer learning approach to brain tumor detection. J. Imaging Inf. Med. 38 (1), 245–261. https://doi.org/10.1007/s10278-024-01199-3 (2024).
Che, Y. et al. Anomaly Detection with Forward Process of Diffusion Models for Brain MRI. In Proceedings of the Winter Conference on Applications of Computer Vision 1113–1122. (2025).
Kaplan, E. et al. PFP-HOG: pyramid and Fixed-Size Patch-Based HOG technique for automated brain abnormality classification with MRI. J. Digit. Imaging. 36 (6), 2441–2460. https://doi.org/10.1007/s10278-023-00889-8 (2023).
Felefly, T. et al. A 3D convolutional neural network based on non-enhanced brain CT to identify patients with brain metastases. J. Imaging Inform. Med. https://doi.org/10.1007/s10278-024-01240-5 (2024).
Yang, H., Zhang, J., Liu, Q. & Wang, Y. Multimodal MRI-based classification of migraine: using deep learning convolutional neural network. Biomed. Eng. OnLine. 17 (1), 138. https://doi.org/10.1186/s12938-018-0587-0 (2018).
Schramm, S. et al. Functional magnetic resonance imaging in migraine: A systematic review. Cephalalgia 43 (2), 03331024221128278. https://doi.org/10.1177/03331024221128278 (2023).
Alzubaidi, L. et al. A survey on deep learning tools dealing with data scarcity: definitions, challenges, solutions, tips, and applications. J. Big Data. 10 (1), 46. https://doi.org/10.1186/s40537-023-00727-2 (2023).
Deng, J. et al. Li Fei-Fei. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 https://doi.org/10.1109/CVPR.2009.5206848 (IEEE, 2009).
Wen, Y., Chen, L., Deng, Y. & Zhou, C. Rethinking pre-training on medical imaging. J. Vis. Commun. Image Represent. 78, 103145. https://doi.org/10.1016/j.jvcir.2021.103145 (2021).
Lu, Z., Xie, H., Liu, C. & Zhang, Y. Bridging the gap between vision transformers and convolutional neural networks on small datasets. https://doi.org/10.48550/ARXIV.2210.05958 (2022).
Zhao, Z. et al. CLIP in medical imaging: A comprehensive survey. https://doi.org/10.48550/ARXIV.2312.07353 (2023).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. (accessed 21 November 2024); http://arxiv.org/abs/2010.11929
Vaswani, A. et al. Attention is all you need. https://doi.org/10.48550/ARXIV.1706.03762 (2017).
Zhang, S. et al. BiomedCLIP: a multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs. (accessed 21 November 2024); http://arxiv.org/abs/2303.00915
Gu, Y. et al. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. https://doi.org/10.48550/ARXIV.2007.15779 (2020).
Patel, T., El-Sayed, H. & Sarker, M. K. Evaluating vision-language models for hematology image classification: Performance analysis of CLIP and its biomedical AI variants. In 36th Conference of Open Innovations Association (FRUCT) 578–584 https://doi.org/10.23919/FRUCT64283.2024.10749850 (IEEE, 2024).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630 (8015), 181–188. https://doi.org/10.1038/s41586-024-07441-w (2024).
Steinmetz, J. D. et al. Global, regional, and National burden of disorders affecting the nervous system, 1990–2021: a systematic analysis for the global burden of disease study 2021. Lancet Neurol. 23 (4), 344–381. https://doi.org/10.1016/S1474-4422(24)00038-3 (2024).
Safiri, S. et al. Global, regional, and National burden of migraine in 204 countries and territories, 1990 to 2019. Pain 163 (2), e293–e309. https://doi.org/10.1097/j.pain.0000000000002275 (2022).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via Gradient-Based localization. Int. J. Comput. Vis. 128 (2), 336–359. https://doi.org/10.1007/s11263-019-01228-7 (2020).
Headache Classification Committee of the International Headache Society (IHS) The international classification of headache disorders, 3rd edition (beta version). Cephalalgia 33 (9), 629–808. https://doi.org/10.1177/0333102413485658 (2013).
Zhang, Y. et al. International classification of headache disorders 3rd edition beta-based field testing of vestibular migraine in china: demographic, clinical characteristics, audiometric findings and diagnosis statues. Cephalalgia 36 (3), 240–248. https://doi.org/10.1177/0333102415587704 (2016).
Brain Development.org. IXI Dataset. http://brain-development.org/ixi-dataset/
Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17 (3), 143–155. https://doi.org/10.1002/hbm.10062 (2002).
Jenkinson, M. & Smith, S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5 (2), 143–156. https://doi.org/10.1016/S1361-8415(01)00036-6 (2001).
Barbosa, D., Ferreira, M., Junior, G. B., Salgado, M. & Cunha, A. Multiple instance learning in medical images: A systematic review. IEEE Access. 12, 78409–78422. https://doi.org/10.1109/ACCESS.2024.3403538 (2024).
Quellec, G., Cazuguel, G., Cochener, B. & Lamard, M. Multiple-Instance learning for medical image and video analysis. IEEE Rev. Biomed. Eng. 10, 213–234. https://doi.org/10.1109/RBME.2017.2651164 (2017).
Radford, A. et al. Learning transferable visual models from natural Language supervision. https://doi.org/10.48550/ARXIV.2103.00020 (2021).
Wilcox, S. L. et al. Hippocampal volume changes across developmental periods in female migraineurs. Neurobiol. Pain. 14, 100137. https://doi.org/10.1016/j.ynpai.2023.100137 (2023).
Neumann, N., Domin, M., Schmidt, C. & Lotze, M. Chronic pain is associated with less grey matter volume in the anterior cingulum, anterior and posterior Insula and hippocampus across three different chronic pain conditions. Eur. J. Pain. 27 (10), 1239–1248. https://doi.org/10.1002/ejp.2153 (2023).
Tolner, E. A., Chen, S. P. & Eikermann-Haerter, K. Current Understanding of cortical structure and function in migraine. Cephalalgia 39 (13), 1683–1699. https://doi.org/10.1177/0333102419840643 (2019).
Hu, S. et al. Resting-state abnormalities in functional connectivity of the default mode network in migraine: A meta-analysis. Front. Neurosci. 17, 1136790. https://doi.org/10.3389/fnins.2023.1136790 (2023).
Xu, H. et al. Abnormal longitudinal changes of structural covariance networks of cortical thickness in mild traumatic brain injury with posttraumatic headache. Prog Neuropsychopharmacol. Biol. Psychiatry. 133, 111012. https://doi.org/10.1016/j.pnpbp.2024.111012 (2024).
Chong, C. D., Berisha, V., Chiang, C., Ross, K. & Schwedt, T. J. Less cortical thickness in patients with persistent Post-Traumatic headache compared with healthy controls: an MRI study. Headache J. Head Face Pain. 58 (1), 53–61. https://doi.org/10.1111/head.13223 (2018).
Acknowledgements
The authors are thankful to Arizona State University Research Computing (ASURC) for hosting and maintaining their computational nodes.
Funding
This work was supported by the United States Department of Defense W81XWH-15-1-0286 and W81XWH1910534, National Institutes of Health K23NS070891, National Institutes of Health—National Institute of Neurological Disorders and Stroke, Award Number 1R61NS113315-01, and Amgen Investigator Sponsored Study 20187183.
Author information
Authors and Affiliations
Contributions
Fazle Rafsani, Devam Sheth, Yiming Che, Jay Shah, Md Mahfuzur Rahman Siddiquee and Teresa Wu contributed to the study conception and design. Implementation and coding were done by Fazle Rafsani and Devam Sheth. Material preparation, data collection and analysis were performed by Catherine D. Chong, Simona Nikolova, Katherine Ross, Gina Dumkrieger and Todd J. Schwedt. The first draft of the manuscript was written by Fazle Rafsani and all authors commented on previous versions of the manuscript. All authors read and approved of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Todd Schwedt, within the prior 24 months, has received consulting fees from AbbVie, Amgen, Linpharma, Lundbeck, Salvia BioElectronics, and Scilex, and royalties from UpToDate. He holds stock options in Allevalux and Nocira. He has received research funding from the American Heart Association, Flinn Foundation, Henry Jackson Foundation, National Headache Foundation, National Institutes of Health, Patient Centered Outcomes Research Institute, Pfizer, Spark Neuro, and United States Department of Defense. Catherine Chong has received research funding from the National Institutes of Health, the American Heart Association and United States Department of Defense.
Ethics approval and consent to participate
This study was approved by the Mayo Clinic Institutional Review Board (IRB), the Phoenix Veterans Administration IRB, and the United States Department of Defense Human Research Protection Office, and all participants provided written informed consent for their participation.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rafsani, F., Sheth, D., Che, Y. et al. Leveraging multi-modal foundation model image encoders to enhance brain MRI-based headache classification. Sci Rep 15, 33256 (2025). https://doi.org/10.1038/s41598-025-18507-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-18507-8