Abstract
Brain tumour identification, segmentation cataloguing from MRI images is most thought-provoking and is a very much essential for many medical image analysis applications. Every brain imaging modality provides information about various parts of the tumor. In current years deep learning systems have shown auspicious outcomes in medical image investigation tasks. Despite several recent works achieved a significant result on brain tumour segmentation and classification, they come with an improved performance at the expense of increased computational complexity to train and test the system. This exploration paper investigates the efficacy of popular deep learning architectures namely Xception Net, MobileNet for classification and DeepLab for segmentation of the cancerous region of brain tumor. Each architecture is trained using a BRATS 2018 dataset and evaluated for its performance in accurately classifying tumor presence and delineating tumor boundaries. The DeepLab models accomplished a best segmentation result with Pearson Correlation Coefficient values 0.50 respectively and the deep learning models Xception Net and MobileNet achieved an accuracy of 0.8921 and 0.9176respectively. The experimental results show that these architectures achieve high accuracy and precise segmentation. The findings of this study contribute to advancing the field of medical image analysis and hold implications for improving the analysis and dealing of brain tumors.
Similar content being viewed by others
Introduction
The growth of abnormal tissues is the major cause for brain Tumours. It affects or in some cases it halts the regular functions of the brain1. Malignant and benign are the two important classes of brain tumour2. The later class is curable and occur repeatedly and the former one is not curable and affects other regions of the brain rapidly3. The extensively applied modalities for detecting these tumours are CT, PET, and MRI4,5. Also the Medical Image Interpretation is popularly utilized for Research and Treatment in Medical community. This has several examples from image analysis6, medical bots7 to computer assisted diagnosis7. Among all the medical problems, brain tumour is the consistently explored and very fascinating medical challenge among all other challenges reported in the literature. The accurate early detection of these tumour very much essential to provide the quick treatment to the affected patients. The research community has proposed several models with respect to the brain tumour detection and classification in the literature. Despite huge effort from the research community the precise detection, dissection and cataloguing of brain tumour remain as a challenge. This may be due to Brain Tumor (BT) is a great healthcare challenge, with its diagnosis and treatment heavily reliant on accurate image analysis. Manual analysis of MRI scans is tedious, and practitioners are in need for automated methods to assist clinicians in interpreting medical images. Recently Deep Learning (DL) has sought huge attention for medical image analysis offering promising results for automating problems like Tumor Segmentation and Classification (TSC). But convolutional neural networks (CNNs) are proficient of learning multifaceted patterns from large scale data surpassing baseline systems in several medical imaging tasks. During past, several models have been contributed by researchers in the literature as a solution for brain tumour related problems8,9.
The brain tumour diagnosis consists of pre-processing, parameterization of images, segmentation, and classification steps.
Pre-processing phase is focused with the noise reduction, enhancement and skull–tripping11. Texture, shape and colour identities are recorded in parameterized co-efficient’s. Classification is segregating the test image into diseased or non-diseased image. This paper investigates the application of DL models for brain tumor dissection problem. In this article, our main focus is on enhancing the performance of four CNN architectures: Xception, MobileNet for classification DeepLab for segmentation of cancerous region in the tumours.
-
The proposed model’s segmentation performance was evaluated using Pearson Correlation Coefficient (0.51), Bland–Altman mean (0.13 mm2), and CDC (80% error < 1.25 mm2).
-
Classification performance was assessed using accuracy and loss metrics: Xception achieved 89.21% accuracy, and MobileNet reached 91.76%.
-
Peak-and-valley filtering enhanced image quality with 92.32% noise elimination.
-
Modified Xception and MobileNet architectures improved classification efficiency via a custom head with GAP, Dense, Dropout, and Sigmoid layers.
-
The framework combines accurate segmentation and robust classification for effective brain tumor diagnosis
Literature review on model contributions for solving brain tumour problem
To organize a short survey on the conventional models proposed in the literature for diagnosing the brain tumours following strategy is made.
-
To provide an extensive analysis of available datasets, software tools, performance measures, and drawback of the existing models.
-
To offer the remarkable research space and drawbacks of the baseline models for motivating the aspiring researchers.
It highlights the effectiveness of deep learning methods in addressing this issue, citing their success in various computer vision tasks. The survey reviews over 150 scientific papers, discussing topics like model architecture, performing segmentation in various conditions, and multimodality processes. the paper offers insights into future directions for development in this field12. Another work addresses the critical need for early detection of brain tumours, which significantly impacts patient survival rates.
It reviews recent Artificial Intelligence (AI) models for detecting brain tumours in MRI scanned images categorizing them into Supervised, Unsupervised, and DL techniques. The study highlights MRIs roles as a non-invasive imaging method for brain tumor detection, but also notes the challenge of limited medical staff proficient in employing advanced technologies. It emphasizes the importance of developing robust automated brain tumor detection methods. The work includes the performance analysis of modern methods and summarizes various image segmentation techniques, while also suggesting directions for future research13. A system for addressing the issue of brain tumor detection and the significance of early detection for improved cure rates is proposed. The model utilized a hybrid combination of saliency map with Efficient Net DL model. Deep transfer learning and an improved fusion approach are explored. Finally, classification task is done using ELM. The results derived from the investigation on three public datasets demonstrated significant performance improvement over number of neural nets14.
An investigation of generative models for medical image analysis starting with a detailed overview of DL fundamentals is proposed and the work then covers the baseline systems applied brain tumor detection and Covid-19 lung pneumonia diagnosis. The work interpreted the effectiveness of generative adversarial networks and autoencoders for semantic segmentation, data augmentation, and improved classification algorithms. The work highlights the best models for handling medical applications, challenges present in the domain and future research guidelines15. A model that addressed the need for automatic classification of brain tumor’s is proposed. It introduces a novel optimization driven model for tumor classification, utilizing a hybrid segmentation approach that combines U-Net model, CFP Net and Squeeze Net. The optimization is achieved using the algorithms like ASMO, SMO, AO and FC. The results convey that the system is capable of achieving effective classification16.
Another novel approach for brain tumor classification (BTC) using a HieDNN classifier framework, that utilizes input images from the Brats dataset and preprocesses them with SG denoising method is proposed. Texture features are removed using GLCM, and the processed dataset is fed into the HieDNN for classification. The proposed method demonstrated improved accuracy for benign, malignant, and normal brain tumor classifications17. A research work addressed the importance of early-stage diagnosis of brain tumors and the limitations of existing classification schemes, such as long execution times and uncertain predictions. The low complexity hybrid model is a combination of K-means and SSO RBNN algorithm. The wavelet transforms is applied for feature extraction. The projected method is authenticated on customary datasets and paralleled with standing schemes, demonstrating higher classification accuracies of 96%, 92%, and 94% on three datasets18.
The research work focused on cancerous tumor detection using ML and DL techniques. The study covered the interpretation of works published during the past five years19. The work effectively identified the best models, metrics for tumor identification20. The work also highlights the higher mortality rates associated with these specific cancers and identifies open research challenges in each category providing opportunities for future research endeavours21.
Review based on chronology
The percentage contributions from the previous works related to brain tumour research are graphically represented in Fig. 1. The number shown in Fig. 1 from the year 2011 to 2021 are taken from the work22 and the remaining numbers from the year 2022 to 2024 are collected by the authors.
Review by considering the various modalities
Figure 2 shows Different imaging modalities are utilized for detecting the brain tumours. Among all the modalities the usage of MRI is 93% and use of remaining modalities is less than 4.1%
Datasets, segmentation, parameterization, and classification methods for brain tumour diagnosis
Datasets
BRATS is a series of state-of-the-art BTS Benchmarking Datasets 23,24,25. Many datasets are proposed for performing the research related to Brain Tumor during the past26,27,29. A detailed classification of datasets proposed in the literature for brain related research are shown in Fig. 3. In this work BRATS 2018 dataset is used to perform the BTSC tasks29,30,31. Structured classification of medical imaging datasets commonly used in brain tumor detection, segmentation, and classification. These datasets are organized according to the imaging modality they originate from, namely 2D Ultrasonic Images, CT (Computed Tomography), MRI (Magnetic Resonance Imaging), and combined CT and MRI modalities. Each modality serves a specific diagnostic purpose and supports different research objectives in medical image analysis.
Under 2D Ultrasonic Images, the diagram lists the Transrectal ultrasound image analysis dataset, which is frequently utilized for analyzing soft tissue structures, particularly in the pelvic and prostate regions. This modality is beneficial for real-time and non-invasive imaging, although it provides limited spatial resolution compared to MRI or CT.
In the case of CT imaging, the referenced category includes a general medical image database comprising 3D volumetric scans. CT data is vital for detecting calcifications, hemorrhages, and bone structures and is widely used in neurosurgical planning and emergency assessments.
MRI datasets, however, dominate the list due to their superior soft tissue contrast and relevance in brain tumor analysis. The MRI section includes a rich array of datasets such as the BRATS (Brain Tumor Segmentation) challenge datasets from various years (2012, 2013, 2015, and 2018), which are standard benchmarks for evaluating brain tumor segmentation algorithms. Additional resources include real-time MR images, 3D DICOM real images, and simulated Brain Web datasets. The Digital Imaging and Communications in Medicine (DICOM) standard and the Internet Brain Segmentation Repository (IBSR) provide clinically annotated datasets. The Montreal Neurological Institute (MNI) and datasets from UZ Gent and UZ Leuven hospitals support multi-institutional research by offering multiparametric MR images. These datasets enable the development and evaluation of machine learning and deep learning techniques across different tumor types, grades, and modalities.
Tumour segmentation algorithms
Segmentation in image processing is very crucial for reducing the noise from images and processed image is used for detecting edges and boundaries. The categorization of brain tumours is illustrated in Fig. 4.The image illustrates a comprehensive taxonomy of block-based segmentation techniques commonly used in medical image analysis, particularly for applications such as brain tumor segmentation. The taxonomy is organized into three primary categories: soft computing-based techniques, region-based segmentation techniques, and other advanced techniques, each comprising specific algorithms tailored to improve accuracy, adaptability, and robustness.
The soft computing-based techniques employ artificial intelligence and machine learning models to perform intelligent segmentation. These include improved fuzzy connected algorithms that enhance region-growing strategies, and deep learning models such as Deep Neural Networks (DNN) and Convolutional Neural Networks (CNN) that learn complex spatial patterns from data. Kernel-based CNNs coupled with multiclass Support Vector Machines (M-SVM) offer powerful classification post-feature extraction. Additional techniques in this category include single image super-resolution approaches, fuzzy entropy-based methods for optimizing uncertainty during segmentation, and biologically inspired neural architectures like SK-TPCNN and HDCNet. Classic machine learning methods like Support Vector Machines (SVM) are also utilized as classifiers within hybrid segmentation pipelines.
The second category, region-based segmentation techniques, is further divided into three subtypes: region-based, clustering-based, and thresholding-based methods. Region-based techniques such as Gabor filter-based Region of Interest (ROI) detection and Local Statistical Based Level Approximation (LSBLA) are employed to identify spatially homogeneous areas. Clustering methods—such as Fuzzy C-Means (FCM), Rough Set Constrained FCM (RSCFCM), and Bayesian fuzzy clustering—group pixels with similar features into segments. More advanced variants like BAT-IT2FCM and WMMFCM integrate swarm intelligence or weighted membership functions to improve segmentation robustness and noise tolerance. Thresholding methods like multi-level thresholding divide the image histogram into distinct classes to isolate pathological regions, while improved thresholding algorithms refine segmentation accuracy by adjusting thresholds dynamically.
Feature vectors generation
Many features extraction techniques, classification architectures and software tools have been invented and implemented in the collected works for pursuing brain tumor research. Figure 5 illustrates the detailed categorization of feature extraction methods based on shape, texture and others. A detailed categorization of features, classification architectures and software tools are used in the brain tumor research are depicted in Fig. 6, 7 respectively.
Figure 5 presents a comprehensive classification of feature extraction methods used in medical image analysis, especially for tasks like tumor segmentation and classification. These methods are grouped into three categories: shape-based, texture-based, and other advanced techniques. Shape-based feature extraction focuses on structural and geometric properties using methods such as histogram analysis, active contour models via level set techniques, image binarization, and region-based level set models to delineate object boundaries. Texture-based methods capture spatial intensity variations and include wavelet transforms, PCA, Gabor wavelets and textons, and statistical texture descriptors like GLCM, GLDM, GLRLM, GLSZM, SGLDM, SVD, and LDP—particularly useful for identifying subtle patterns in tissues. The third category comprises advanced or hybrid approaches such as SPDM, CMC, genetic algorithms (GA), Gaussian filtering, self-organizing maps (SOM), DAPP, and multi-modal feature kernels. It also includes deep learning methods like convolutional neural networks (CNNs) and the overlapping patches method, which enhance context-aware feature learning. Collectively, these feature extraction strategies enable robust, accurate, and modality-adaptive analysis of complex medical images for improved diagnostic decision-making.
The Fig. 7 describes the various software tools available for brain tumour research.
A summary of the methods with corresponding approach related to brain tumor research are listed in Table 1.
Methodology
The projected framework in the Fig. 8 presents a method In BT detection from MRI images, the process starts with image acquisition, where MRI scans and corresponding manually segmented ground truth masks outlining tumor regions are collected. Pre-processing follows, involving techniques like noise reduction and contrast enhancement to improve image quality for dissection. CNN-based frameworks like are then used for segmentation, where the pre-processed images and ground truth masks serve as training data to distinguish tumor from healthy tissue. Post-training, model enactment is gaged by means of metrics such as Dice coefficient, Jaccard Index, and Bland–Altman plots. Finally, the segmented tumor is visualized with highlighted regions, along with heatmaps and cumulative distribution curves to further analyze intensity distributions in the tumor area.
Functional requirements
Functional Requirements refer to the specific functions that a software framework must perform and how it should respond to particular inputs or situations. These functions may include data management, calculations, handing out, and other precise jobs.
Dataset collection
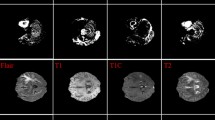
The BRATS 2018 dataset, cited in Menze et al.32, is a substantial collection developed for segmenting brain tumours in multimodal MRI images The dataset consists of a 210 are diagnosed with High-Grade Glioma (HGG) and 75 with Low-Grade Glioma (LGG), thereby ensuring a balanced representation of both malignant and benign tumor cases. Each subject in the dataset includes four MRI modalities: T1-weighted (T1), contrast-enhanced T1-weighted (T1c), T2-weighted (T2), and Fluid Attenuated Inversion Recovery (FLAIR). These modalities provide complementary anatomical and pathological information about the brain tissue and tumor structure. All MRI volumes are preprocessed by the BRATS organizing team and are skull-stripped, co-registered to a common anatomical space, and resampled to an isotropic resolution of 1 mm3. The dataset also includes high-quality manual annotations by expert radiologists marking the whole tumor (WT), tumor core (TC), and enhancing tumor (ET), which serve as the ground truth for segmentation tasks. This rich, well-curated dataset forms a robust foundation for training and evaluating both segmentation and classification models. The revised manuscript now includes this detailed description under the methodology section to clarify the dataset’s scope and structure.
In this study, the BRATS 2018 dataset was selected over BRATS 2020 and BRATS 2021 due to its suitability for the research objectives. BRATS 2018 offers high-quality, expert-annotated ground truth, which ensures reliable training and evaluation of segmentation models as shown in Table 2. Additionally, it provides a balanced distribution of High-Grade Gliomas (HGG) and Low-Grade Gliomas (LGG), allowing for effective learning across different tumor types. The dataset also includes consistently preprocessed, skull-stripped, and modality-aligned images, minimizing the need for extensive preprocessing. In contrast, BRATS 2020 and 2021, while larger in size, contain raw or partially processed data that would require significant additional preprocessing effort, making them less practical for the focused scope of this study. To ensure effective model training and evaluation, the dataset was partitioned into training (80%), validation (10%), and testing (10%) sets.Importantly, we employed stratified sampling during the splitting process to maintain a balanced representation of both classes across all three subsets. This step was crucial due to the class imbalance inherent in the BraTS 2018 dataset. Stratification ensured that the model learned from a proportionate distribution of tumor , minimizing bias and enhancing generalization. For performance evaluation, we implemented k-fold cross-validation (with k = 5) during the training phase to validate the robustness and stability of the model across different data folds.
The peak-and-valley filter in image processing
It is a non-linear filtering technique designed to address impulsive noise, particularly in gray-level profiles and two-dimensional (2-D) images. In this comprehensive explanation, we delve into its operation, equations, and distinctions from morphological filters.
Operation on gray level profiles (1-D)
In its one-dimensional (1-D) application, the peak-and valley filter aims to eliminate narrow “peaks” and “valleys” in gray-level profiles through a series of iterative cutting and filling operations. A crucial aspect is the consideration of a three-pixel neighborhood, where “r” and “s” are variables with values of − 1, 0, or 1, ensuring that “r” and “s” are not both 0.
Cutting Operation: When applied to a pixel, the cutting operation replaces the pixel’s gray level with the maximum of the gray levels of its neighbors if the central pixel’s value is higher. Filling Operation: The filling operation replaces a pixel’s gray level with the minimum of the gray levels of its neighbors if the central pixel’s value is lower. These operations are applied iteratively until no peaks or valleys remain. The equation for 2-D Peak-and-Valley Filter: In the two-dimensional (2-D) version, the filter operates on a 3 × 3 pixel neighborhood. The expression for the central pixel’s new value is as follows:
where: V ′(x, y) is the new gray value of the central pixel at coordinates, (x, y).V (x, y) characterises the original gray level value of the vital pixel. V (x ± 1, y ± 1) represents the gray values of the surrounding eight pixels in the 3 × 3 neighbourhood. The function f implements the cutting and filling logic to adjust the central pixel’s gray level value based on the gray levels of its neighbouring pixels. The peak-and-valley filter exhibits certain similarities with morphological filters but is fundamentally distinct. Unlike morphological filters, which often alter the shape of image elements through erosion or dilation, the peak-and valley filter preserves the shape of image elements. While the peak-and-valley filter operates iteratively, not recursively, making it non-self-replicating, some morphological filters may use iterations but can also be non-recursive. Due to its iterative nature, applying the peak-and-valley filter more than once does not provide additional benefits, contrasting with certain morphological filters that may benefit from multiple applications.
DeepLab for tumor segmentation
DeepLab utilizes atrous convolution to expand the receptive field of convolutional layers without increasing the number of parameters or reducing spatial resolution. This allows the network to extract multi-scale features and capture more global context, which is particularly beneficial in segmenting objects that appear at different scales or are embedded within cluttered scenes.ASPP (Atrous Spatial Pyramid Pooling): ASPP is a key module in DeepLab that applies multiple parallel atrous convolutions with different dilation rates. This facilitates multi-scale feature extraction, enabling the model to concurrently capture both fine and coarse features. By aggregating these parallel outputs, ASPP enhances the model’s ability to discriminate between foreground and complex background regions, which is essential in medical imaging and real-world applications with high variability in texture and appearance.After feature extraction and classification, DeepLab employs bilinear upsampling to recover the original image resolution. Unlike nearest-neighbor methods, bilinear interpolation preserves edge smoothness and reduces aliasing artifacts. This step ensures that the final segmentation masks are spatially coherent and accurately aligned with the original input, which is critical for delineating object boundaries—especially in domains like brain tumor segmentation where boundary precision is crucial.
Figure 9 shows the architecture of DeepLab that uses ResNet, a deep convolutional neural network (DCNN), as its backbone. The ResNet architecture is defined by its use of residual blocks, which are described mathematically as follows: For a residual block, the transformation F(x) is applied to the input xxx, which includes convolutions, batch regularisation (BR), and a non-linear activation (ReLU). The residual connection allows the network to bypass these transformations:
where: x is the input to the outstanding unit. F(x) is the result of the transformations within the chunk. y is the output of the outstanding chunk. In the case of bottleneck residual blocks, the transformation F(x) involves a series of convolutions, as follows:
where: W1,W2,W3 are the weights for the 1*1,3*3,1*1convolutions, respectively. σ is the ReLU activation function. Batch normalization is applied after each convolution and before ReLU. Thus, the final output from the bottleneck block can be written as:
-
1.
Atrous Convolutions: To detention topographies, DeepLab modifies the standard convolutional blocks by introducing atrous (dilated) convolutions. An atrous convolution is defined by a dilation rate rrr, which enlarges the sympathetic field of the filter without aggregate the factors or computational cost. For a given convolution operation with filter WWW and dilation rate rrr, the output is computed as:
$$y[i] = \sum {kW[k] \cdot x[i + r \cdot k]}$$(5)where i is the spatial position of the yield. k is the index for the convolution filter. r is the dilation rate, which controls the spacing between filter elements. As a result, the effective receptive field of a 3 × 3 atrous convolution with distention rate r = 2 becomes equivalent to a 5 × 5 receptive field, while maintaining only 9 parameters.
-
2.
Atrous Spatial Pyramid Pooling (ASPP): The key innovation of DeepLab is the Atrous Spatial Pyramid Pooling (ASPP). Mathematically, the output of the ASPP module is a concatenation of features obtained from parallel atrous convolutions:
$$YASPP = [F1(x),F2(x),F3(x),GAP(x)]$$(6)where: F1 (x) is the result of a 1 × 11 times 11 × 1 convolution. F2 (x),F3 (x),F4 (x) are the results of 3 × 3 convolutions with dilation rates 6,12,18, respectively. GAP(x) represents the Global Average Pooling of the input x.Each convolution captures features at different scales.The result of ASPP is upsampled using bilinear interpolation to match the resolution of the input image:
$${\text{Youtput}} = {\text{Upsample}}\left( {{\text{YASPP}},{\text{scale}} {\text{factor}}} \right)$$(7) -
3.
Output Stride and Feature Map Size: The output stride (OS) controls the degree of down sampling applied to the input image throughout the network. The output stride OS is defined as:
$${\text{OS}} = {\text{input}} {\text{size}}/{\text{output}} {\text{feature}} {\text{map}} {\text{size}}$$(8)For example, if the output stride is 16, an input image of size 224 × 224 will produce a feature map of size 14 × 14.By controlling the output stride, DeepLab avoids excessive decimation, ensuring higher-resolution feature maps for dense prediction tasks like semantic segmentation.
-
4.
Multi-grid Dilations in Block 4: DeepLab also introduces the multi-grid method in the final ResNet block (Block 4) to further capture multi-scale context. The dilation rates for Block 4’s convolutions are given by:
$${\text{rBlock}} {4} = {2} \cdot \left( {{1},{2},{4}} \right) = \left( {{2},{4},{8}} \right)$$(9) -
5.
Complete Model Equation:The full output of the DeepLab model can be summarized as:
$${\text{YDeepLab}} = {\text{Upsample}}\left( {\left[ {{\text{ASPP}}\left( {{\text{FBlock}} {4}\left( {\text{x}} \right)} \right)} \right],{\text{scale}} {\text{factor}}} \right)$$(10)where:x is the input image. FBlock 4(x) is the output from ResNet Block 4 with atrous convolutions. ASPP aggregates multi-scale features from FBlock 4 (x). The final output is up sampled to match the input resolution.
Xception Net
Xception Net invented by François Chollet in 2017 is as shown in Fig. 10. It is inspired by the Inception architecture but adopts a different approach to feature extraction. Instead of using traditional convolutional layers Xception replaces them with depthwise separable convolutions. These convolutions include point wise convolutions post depth wise convolutions. This separation of spatial and channel wise convolutions reduces the computational cost while maintaining expressive power, leading to improved efficiency and performance. The depthwise separable convolution can be represented as:
Depthwise convolution
Given an input feature map X of size \(H \times W \times D_{in}\) where \(H{ }is{ }the{ }height,{ }W{ }is{ }the{ }width,{ }and{ }D_{in} { }is{ }the{ }number{ }of{ }input{ }channels\) and a depthwise convolutional filter Fd of size K × K × 1 where K is the kernel size:
Depthwise convolution can be expressed as:
where: where i and j iterate over the spatial dimensions H and W, and k iterates over the depth dimension \(D_{in}\).
Pointwise convolution
Pointwise convolution can be expressed as Given the output feature map \(Y_{d}\) from the depthwise convolution and a pointwise convolutional filter \(F_{p}\) of size 1 × 1 × Dout where \(D_{out}\) is the desired number of output canals. The output feature map Yp is computed as:
where i and j iterate over the spatial dimensions H and W, and l iterates over the depth dimension Dout, and the end output of the depth wise divisible convolution is determined by augmenting the production of the depth wise convolution \(Y_{d}\) and the pointwise convolution \(Y_{p}\).
A Global Average Pooling (GAP) layer was used to reduce spatial dimensions and aggregate feature maps.This was followed by a fully connected Dense layer with 256 units and ReLU activation, enabling non-linear learning of discriminative tumor features.To prevent overfitting, a Dropout layer with a rate of 0.5 was included after the dense layer. Dense output layer with a single neuron and Sigmoid activation was added to generate probability scores for binary classification (tumor vs. no tumor). This design supports the use of binary cross-entropy loss during training. For input compatibility:The MRI volumes from the BRATS 2018 dataset (240 × 240 × 155 voxels) were processed by extracting representative axial slices, which were then resized to 224 × 224 (or 240 × 240) pixels to align with model input requirements. Where needed, grayscale images were converted to 3-channel format by duplication or stacking across modalities This configuration ensures optimal feature extraction, maintains compatibility with pretrained weights, and enables efficient learning of tumor-specific patterns from MRI slices.”
MobileNet
MobileNet is another lightweight CNN variant designed for mobile devices with limited computational resources whose architecture is as shown in Fig. 11. It was proposed by Andrew G. Howard et al. in 201733. MobileNet employs depthwise separable convolutions similar to Xception but introduces depthwise convolution with depth multiplier and pointwise convolution. The depth multiplier parameter controls the number of output channels in the depthwise convolution effectively reducing the model size and computational cost.
The architectural choices in MobileNet, particularly the use of depthwise separable convolutions, represent a significant advancement in CNN design. As a result, MobileNet is widely used in real-time applications, including mobile devices and embedded systems, where computational resources are limited. Its lightweight nature makes it highly suitable for tasks such as medical image analysis and other vision-related applications that require low latency and low power consumption while maintaining strong performance in image recognition. For an input feature map X with dimensions W × H × Din, whereW and H are the spatial dimensions and Din is the quantity of input canals, the depthwise convolution uses Din filters of size K × K × 1. The output feature map Ydepth is computed as Eq. 13:
where Wdepth represents the depthwise convolutional filters, and bdepth is the bias term. Here, i and j iterate over the spatial dimensions, and k iterates over the input channels.
Following the depthwise convolution, pointwise convolution (1 × 1 convolution) is applied to combine features across channels. This step uses Dout filters of size 1 × 1 × Din, where Dout is the desired number of output channels. The pointwise onvolution output feature map Ypoint is calculated as Eq. 14:
where Wpoint denotes the pointwise convolutional filters, and bpoint is the bias term. The variable l iterates over the output channels.
The hyper parameters listed in Table 3 were selected based on empirical experimentation and validation performance monitoring. The learning rate of 0.001 allowed efficient gradient descent, while the Adam optimizer provided robustness in handling sparse gradients common in medical images. A batch size of 16 offered a practical trade-off between training speed and generalization. Training for 30 epochs, with early stopping based on validation loss, avoided overfitting.
Results
This segment undertakes a thorough examination of the outcomes and their interpretation, shedding light on the significance of our study’s findings. Here, we systematically present, analyze, and contextualize the data obtained from rigorous experiments, surveys, or analyses.
Figure 12 and 13 shows the sample input image for the proposed system and enhanced image using Peak and Valley 1D that removes impulse noise and preserves edges and fine details. It’s particularly effective in applications where edge preservation is critical.
Table 4 illustrates the comparative evaluation between the Peak-and-Valley filter and the Gaussian filter reveals that the Peak-and-Valley approach significantly outperforms the Gaussian method across multiple critical metrics relevant to tumor MRI image preprocessing. Specifically, the Peak-and-Valley filter achieved a noise elimination rate of 92.32%, substantially higher than the 83.15% attained by the Gaussian filter. This indicates its superior ability to suppress impulsive noise without compromising image structure. Additionally, the noise attraction percentage—which measures the filter’s tendency to misclassify actual signal as noise—was markedly lower for the Peak-and-Valley filter (0.25%) compared to the Gaussian filter (1.85%), highlighting its precision in preserving clinically relevant features. The image spoilage percentage, reflecting the extent of structural degradation due to filtering, was also significantly lower for the Peak-and-Valley filter (0.30%) than for the Gaussian filter (0.85%), confirming its edge-preserving capability. In terms of average attenuation, which denotes the loss of signal intensity, the Peak-and-Valley filter recorded a lower value (0.041) than the Gaussian filter (0.093), suggesting minimal distortion of the original tissue contrast. While both filters demonstrated similar average spoilage times (0.5132 s for Peak-and-Valley vs. 0.4973 s for Gaussian), the slight increase in processing time for the Peak-and-Valley filter is negligible when weighed against the substantial quality improvement. the Peak Signal-to-Noise Ratio (PSNR), a key indicator of overall image quality, was considerably higher for the Peak-and-Valley filter (42.85 dB) compared to the Gaussian filter (36.42 dB). This confirms the Peak-and-Valley filter’s effectiveness in enhancing image clarity while preserving diagnostic detail Fig. 14 illustrates the Correlation Analysis between DeepLab and GT: The Pearson correlation coefficient (CC) is commonly utilized to assess the association between the area estimated by artificial intelligence (AI) and the ground truth (GT) area for two models: DeepLab. The CC assessment for the DeepLab model is 0.51,
Figure 15 illustrates BA Plot for DeepLab and GT Area: This plot shows the discrepancy between the ground truth (GT) and DeepLab areas, with the y-axis signifying the difference and the x-axis signifying the mean of the DeepLab and GT areas. Lower values of mean and standard deviation (SD) indicate superior performance. Figure 13 depicts the Bland Altman plots for both the DeepLab. The mean and SD values for DeepLab model are 0.13 mm2 and 1.35 mm2 correspondingly.
Cumulative Distribution Curves (CDC) for Area Error among DeepLab and GT: The area error, measured in square millimeters (mm2), is a critical metric for evaluating model performance. It represents the disparity between the areas estimated by the DeepLab and the ground truth (GT). It is determined by transforming ground truth mask and predicted areas from pixels to millimeter dimensions. Then a resolution factor of 0.0625 mm per pixel is applied. Lesser area errors indicate superior model performance.
In Fig. 16, CDC illustrate the area error among the GT and DeepLab-estimated masks for the DeepLab models. For the DeepLab model, 80% of the scans exhibited an area error of less than 1.25 mm2.
From the Table 5, its evident that DeepLab perform across all the parameters assessed. It achieves a higher Pearson correlation coefficient, indicating a stronger correlation between DeepLab and ground truth areas. Additionally, DeepLab exhibits lower mean and standard deviation values in the Bland Altman (BA) plot, signifying better agreement among DeepLab and ground truth areas. Moreover, DeepLab demonstrates lower area errors in the cumulative distribution analysis, highlighting its superior performance in accurately estimating tumor areas. Figure 17 shows how the model predicts the mask after every epoch.
Figure 17, depicts the progression of our segmentation model after 30 epochs. We train the model for each epoch, predicting masks for images as we progress. The displayed images highlight the DeepLab models gradual improvement in segmentation accuracy over time providing valuable insights into its evolving performance.
.
Table 6 illustrates the 30 epoch values of the The accuracy and loss values achieved by Xception Net Deep learning architecture are 0.8921 and 0.2118 respectively.
The above curve in the Fig. 18 is the training and validation results of Xception for brain tumour classification reveal a similar trend of improvement over epochs, albeit with slightly different performance metrics compared to MobileNet. Beginning with a higher loss and moderate accuracy in the initial epochs, Xception gradually reduces its loss and increases accuracy, showcasing its ability to learn complex patterns from the dataset. By the conclusion of training, Xception achieves a respectable validation loss of 0.2118 and an accuracy of 0.8921, demonstrating its efficacy in accurately classifying BT from MRI metaphors. Despite minor fluctuations in performance during training, Xception demonstrates robust learning capabilities and holds promise for further optimization and deployment in medical imaging applications.
Table 7 illustrates the30 epoch information for The accuracy and loss values achieved by Mobile Net Deep learning architecture are 0.9176 and 0.1381 respectively.
Based on the training and validation results of MobileNet for BT cataloguing shown in Fig. 19 (a), it is obvious that the model demonstrates a steady improvement in both loss reduction and accuracy enhancement across epochs. Beginning with a relatively high loss and moderate accuracy, the model converges towards lower loss values and higher accuracies with increasing epochs. By the end of training, MobileNet achieves a satisfactory performance with a final validation loss of 0.1381 and accuracy of 0.9176. These results convey the effectiveness of MobileNet for BTC tasks, showcasing its capability to learn and generalize well on the given dataset. Further fine tuning or optimization strategies may lead to even better performance, but the current outcomes highlight the promise of MobileNet in aiding medical professionals in accurate brain tumor diagnosis.
The Fig. 19 b and c shows the output obtained for No tumor, the first racket shown in the original enhanced image the second racket is the mask predicted by the mobile net model. It shows the region which is having the most probability of identified muscles with different heat signature.
The Fig. 20 a and b shows the output obtained for tumor, the first racket shown in the original enhanced image the second racket is the mask predicted by the mobile net model. It shows the region which is having the most probability of cancer.
Discussion
The study inspects the performance of DL models—DeepLab, Xception, and MobileNet—in the context of BT concealment and dissection using MRI images. The image enhancement process employing a peak and valley filter significantly improved image quality, achieving a 92.32% noise elimination rate while preserving essential details. DeepLab demonstrated a moderate Pearson Correlation Coefficient (CC) of 0.51 in tumor area segmentation, indicating room for improvement, but still achieving a mean area error of 0.13 mm2. In contrast, Xception showed steady performance improvements, culminating in a validation accuracy of 0.8921. MobileNet outperformed both models, achieving a validation accuracy of 0.9176 and exhibiting strong generalization capabilities.
Our study demonstrates superior classification performance compared to previous works as depicted in Table 8, leveraging advanced deep learning architectures for brain tumor classification. Abiwinanda et al.35, S. A. Yazdan et al.36 used a basic CNN model, achieving 88.86% accuracy, whereas our Xception model (89.21%) and MobileNet (91.76%) provide improved results due to deeper feature extraction. Our results, based on the BRATS 2018 dataset and a four-class classification setup, demonstrate that attention-based MobileNet significantly enhances classification accuracy, making it a promising approach for medical image analysis.
While the results are promising, limitations such as dataset size and computational constraints for real-time deployment remain challenges. MobileNet’s superior performance in identifying various tumor types underscores its potential for clinical applications. Overall, the findings support the usage of advanced DL techniques to enhance diagnostic accuracy and decision-making in medical imaging.
Conclusion
In this work, we explored the performance of DL architectures for BT segmentation and classification, leveraging MRI data across four modalities. Each modality contributes unique characteristics that help the network distinguish between tumor classes effectively. The DeepLab model exhibited a robust correlation, showcasing better agreement with ground truth (as indicated by lower mean and standard deviation in the Bland Altman plot) and reduced area errors. Furthermore, both Xception Net and Mobile Net demonstrated commendable accuracy in brain tumor classification, achieving final validation accuracies of 0.8921 and 0.9176, respectively. These results highlight the significant prospective of DL models to improve brain tumor diagnosis, while also revealing avenues for further research in medical image analysis.
Despite these encouraging findings, the study identifies certain limitations. The relatively small dataset, especially for rare tumor types, poses challenges for model generalization. Additionally, deploying these models in real-time scenarios may face computational hurdles, particularly in settings with limited resources. Future research should prioritize optimizing model architectures to accommodate larger datasets and enhance real-time segmentation capabilities. In summary, this study illustrates the promise of deep learning models, such as MobileNet and Xception Net, in medical imaging. The findings underscore the necessity of selecting models tailored to the specific demands of tumor detection tasks, encompassing accuracy, processing speed, and computational efficiency. With continued refinement and optimization, these models have the impending to significantly enhance scientific decision-making in tumor diagnosis.
7. References
Popuri, K., Cobzas, D., Murtha, A. & Jägersand, M. 3D variational brain tumor segmentation using Dirichlet priors on a clustered feature set. Int J CARS. 7(4), 493–506. https://doi.org/10.1007/s11548-011-0649-2 (2012).
Njeh, I. et al. 3D multimodal MRI brain glioma tumor and edema segmentation: A graph cut distribution matching approach. Comput. Med. Imaging Graph.. 40, 108–111. https://doi.org/10.1016/j.compmedimag.2014.10.009 (2015).
Rangaiah, P. K. B. & Augustine, R. Enhancing medical image reclamation for chest samples using B-coefficients, DT-CWT and EPS algorithm. IEEE Access 11, 113360–113375. https://doi.org/10.1109/ACCESS.2023.3322205 (2023).
Manoj, H. M. Comparative assessment of machine learning models for predicting glucose intolerance risk. SN Comput. Sci. 5, 894. https://doi.org/10.1007/s42979-024-03259-5 (2024).
Rajendran, A. & Dhanasekaran, R. Fuzzy clustering and deformable model for tumor segmentation on MRI brain image: A combined approach. Procedia Eng. 30, 327–333. https://doi.org/10.1016/j.proeng.2012.01.868 (2012).
Xia, Y., Eberl, S., Wen, L., Fulham, M. & Feng, D. D. Dual-modality brain PET-CT image segmentation based on adaptive use of functional and anatomical information. Comput. Med. Imaging Graph. 36(1), 47–53. https://doi.org/10.1016/j.compmedimag.2011.06.004 (2012).
Darshan, S. L. S. et al. Design of chest visual based image reclamation method using dual tree complex wavelet transform and edge preservation smoothing algorithm. SN Comput. Sci. 5, 352. https://doi.org/10.1007/s42979-024-02742-3 (2024).
Srinidhi, N. N. et al. Design of cost efficient VBIR technique using ICA and IVCA. SN Comput. Sci. 5, 560. https://doi.org/10.1007/s42979-024-02936-9 (2024).
Giridhar R. B., Ofori M., Liu J., Ambati L. S., Early public outlook on the coronavirus disease (COVID-19): A social media study. Social Media Analysis on Coronavirus (COVID-19) 2020.
Rangaiah, Pramod and Augustine, Robin, Improving Liver Cancer Diagnosis: A Multifaceted Approach to Automated Liver Tumor Identification in Ultrasound Scans. Available at SSRN: https://ssrn.com/abstract=4646452 or https://doi.org/10.2139/ssrn.4646452
Robin Augustine Improving burn diagnosis in medical image retrieval from grafting burn samples using B-coefficients and the CLAHE algorithm, Biomedical Signal Processing and Control, Volume 99, 2025, 106814, ISSN 1746–8094, https://doi.org/10.1016/j.bspc.2024.106814.
Sachdeva, J., Kumar, V., Gupta, I., Khandelwal, N. & Ahuja, C. K. Segmentation, feature extraction, and multiclass brain tumor classification. J. Digit Imaging. 26(6), 1141–1150. https://doi.org/10.1007/s10278-013-9600-0 (2013).
Augustine, Robin, Vbir-Based Assessment of Radiographic-Divergence Agent Attention in Prostate Melanoma Patients. Available at SSRN: https://ssrn.com/abstract=4752359 or https://doi.org/10.2139/ssrn.4752359
Augustine, Robin, Enhanced Glaucoma Detection Using U-Net and U-Net+ Architectures Using Deep Learning Techniques. Available at SSRN: https://ssrn.com/abstract=4831407 or https://doi.org/10.2139/ssrn.4831407
Parisot, S., Wells, I. W., Chemouny, S., Duffau, H. & Paragios, N. Concurrent tumor segmentation and registration with uncertainty-based sparse non-uniform graphs. Med. Image Anal. 18(4), 647–659. https://doi.org/10.1016/j.media.2014.02.006 (2014).
Cordova, J. S. et al. Quantitative tumor segmentation for evaluation of extent of glioblastoma resection to facilitate multisite clinical trials 1,2. Transl. Oncol. 7(1), 40–47 (2014).
Naresh, E. et al. Autonomous garbage accumulation robot using internet of things. J Mach. Comput. 4(2), 431–440. https://doi.org/10.53759/7669/jmc202404041 (2024).
Swaminathan, S. V., Surendiran, J., Design and implementation of Kogge Stone adder using CMOS and GDI design: VLSI based. Int. J. Eng. Adv. Technol. (IJEAT), 8(6S3) 2019
Chen, Y., Zhao, B., Zhang, J. & Zheng, Y. Automatic segmentation for brain MR images via a convex optimized segmentation and bias field correction coupled model. Magn. Reson. Imaging. 32(7), 941–955. https://doi.org/10.1016/j.mri.2014.05.003 (2014).
Tabjula, J., Kalyani, S., Rajagopal, P. & Srinivasan, B. Statistics-based baseline-free approach for rapid inspection of delamination in composite structures using ultrasonic guided waves. Struct. Health Monit. https://doi.org/10.1177/14759217211053776 (2021).
Imani, F., Boada, F. E., Lieberman, F. S., Davis, D. K. & Mountz, J. M. Molecular and metabolic pattern classification for detection of brain glioma progression. Eur. J. Radiol. 83(2), e100–e105. https://doi.org/10.1016/j.ejrad.2013.06.033 (2014).
Liu, Z. et al. Deep learning based brain tumor segmentation: A survey. Complex and Intell. Syst. https://doi.org/10.1007/s40747-022-00815-5 (2023).
Manjunatha, M. B. Design and Development of ASL Recognition by Kinect Using Bag of Features. In International Proceedings on Advances in Soft Computing, Intelligent Systems and Applications Advances in Intelligent Systems and Computing Vol. 628 (eds Reddy, M. & Viswanath, K. K. M. S.) (Springer, Singapore, 2018). https://doi.org/10.1007/978-981-10-5272-9_31.
Prasad, S., Samimalai, A., Rani, S. R., Kumar, H. & N., Banu, S., Information Security and Privacy in Smart Cities, Smart Agriculture, Industry 4.0, Smart Medicine, and Smart Healthcare. In IoT Based Control Networks and Intelligent Systems. Lecture Notes in Networks and Systems Vol. 528 (eds Joby, P. P. et al.) (Springer, Singapore, 2023).
Ranjbarzadeh, R., Caputo, A., Tirkolaee, E. B., Jafarzadeh Ghoushchi, S. & Bendechache, M. Brain tumor segmentation of MRI images: A comprehensive review on the application of artificial intelligence tools. Comput. Biol. Med. https://doi.org/10.1016/j.compbiomed.2022.106405 (2023).
Prathap C and M. Swamy, “Design and analysis of UHF BJT feedback oscillator using linear and non-linear simulation,” 2013 International Conference on Emerging Trends in Communication, Control, Signal Processing and Computing Applications (C2SPCA), Bangalore, India, 2013, pp. 1–6, https://doi.org/10.1109/C2SPCA.2013.6749386.
Khan, M. A. et al. Multimodal brain tumor detection and classification using deep saliency map and improved dragonfly optimization algorithm. Int. J. Imaging Syst. Technol. 33(2), 572–587 (2023).
Celard, P. et al. A survey on deep learning applied to medical images: From simple artificial neural networks to generative models. Neural Comput. Appl. https://doi.org/10.1007/s00521-022-07953-4 (2023).
Nirmalapriya, G., Agalya, V., Regunathan, R. & Ananth, M. B. J. Fractional Aquila spider monkey optimization based deep learning network for classification of brain tumor. Biomed Signal Process Control https://doi.org/10.1016/j.bspc.2022.104017 (2023).
Shajin, F. H., Rajesh, S. P. P. & Nagoji Rao, V. K. Efficient framework for brain tumour classification using hierarchical deep learning neural network classifier. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. https://doi.org/10.1080/21681163.2022.2111719 (2023).
Nanda, A., Barik, R. C. & Bakshi, S. SSO-RBNN driven brain tumor classification with saliency-K-means segmentation technique. Biomed. Signal Process. Control https://doi.org/10.1016/j.bspc.2022.104356 (2023).
Menze, B. H. et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024. https://doi.org/10.1109/TMI.2014.2377694 (2015).
Howard, A.G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., Andreetto, M., & Adam, H. MobileNets: EfficientConvolutional Neural Networks for Mobile Vision Applications. ArXiv:abs/1704.04861 (2017).
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, ,M., Crimi, A., et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. Preprint https://arxiv.org/abs/1811.02629 (2018).
Abiwinanda N., Hanif M., Hesaputra S. T., Handayani A., Mengko T. R. (2019). “Brain tumor classification using convolutional neural network,” in World Congress on Medical Physics and Biomedical Engineering 2018: June 3–8, 2018, Prague, Czech Republic (Springer), 183–189. https://doi.org/10.1007/978-981-10-9035-6_33
Yazdan, S. A. et al. ‘An efficient multi-scale convolutional neural network based multi-class brain MRI classification for SaMD’. Tomography 8(4), 1905–1927. https://doi.org/10.3390/tomography8040161 (2022).
Funding
Open access funding provided by B.M.S. College of Engineering.
Author information
Authors and Affiliations
Contributions
All authors are contributed equally.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Data availability
The datasets generated during and/or analysed during the current study are available from pradi14cta@gmail.com on reasonable request.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pradeep Kumar, B.P., Naresh, E., Raghavendra, C.K. et al. Comprehensive brain tumour concealment utilizing peak valley filtering and deeplab segmentation. Sci Rep 15, 34780 (2025). https://doi.org/10.1038/s41598-025-18574-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-18574-x