Abstract
The paradoxical impact of T-cell-derived circulating DNA (T-cirDNA) and prognostication in advanced non-small cell lung cancer (NSCLC) has been reported. Further exploration was conducted in 158 EGFR-mutated NSCLC participants who received EGFR inhibitors, correlated with tumor PD-L1 score, CD8 tumor-infiltrating lymphocytes (TILs), and bulk RNA sequencing. We categorized T-cirDNA levels into three groups based on a previous study: undetectable (26.8%), low (≤ 1% ratio; 36.6%), and high (> 1% ratio; 36.6%). Undetectable and high T-cirDNA groups were independent factors correlated with favorable outcomes. The presence of intra-tumoral CD8 TILs (≥ 1%) was also an independent unfavorable prognostic factor; however, it had the lowest proportion in the low T-cirDNA group (16%). Tumor-immune microenvironment (TIME)-related gene set enrichment analysis revealed an overlapped significant heme biosynthesis signature correlated with poor outcome and diverse T-cirDNA group. Despite a high heme biosynthesis signature score in the undetectable T-cirDNA group, inverse correlation with CIBERSORT-activated CD4 memory T-cells was found (R − 0.79, p-value 0.019). Those findings were contrary to the low and high T-cirDNA groups. The significant contribution of heme biosynthesis was the overexpression of CPOX. Crosstalk of EGFR and COPX function prohibits the activation of CD4 + memory T-cells or the spatial intra-tumoral CD8 + T-cells. Undetectable T-cirDNA represents inactivated naïve T-cells and solely active downstream EGFR signaling.
Similar content being viewed by others
Introduction
Circulating DNA (CirDNA) comprises both tumor-derived and non-tumor-derived cirDNA, originating from tumor cells, the tumor immune microenvironment, and normal cells via several mechanisms, including apoptosis, necrosis, phagocytosis, oncosis, and active secretion1,2,3,4. CirDNA helps monitor recurrent disease, inform treatment selection, facilitate post-treatment monitoring, and predict prognosis5,6. Only a few studies have evaluated the value of non-tumor-derived cirDNA and its utility in cancer care. A previous study demonstrated that the level of non-tumor-derived cirDNA was associated with survival outcomes7.
Since cancer development and progression result from an imbalance between cancer cells and host immunity. Evidence suggests that the tumor immune microenvironment (TIME), a potential source of non-tumor-derived cirDNA, is associated with immune checkpoint inhibitor efficacy and survival outcomes in advanced non-small cell lung cancers (NSCLC)8,9,10. We previously reported that paradoxical T-cell-derived circulating DNA (T-cirDNA) was associated with prognosis outcomes in advanced NSCLCs, with its level correlated to intra-tumoral tumor-infiltrating lymphocytes (TILs). We observed that undetectable T-cirDNA, indicating inactive T-cell TILs in a subgroup of participants with epidermal growth factor receptor (EGFR) alterations, resulted in durable disease control with EGFR TKIs11. Most epidermal growth factor receptor (EGFR) mutant NSCLCs exhibited low expression of T-cell TILs and tumor programmed cell death ligand-1 (PD-L1). It was associated with a poor response to immune checkpoint inhibitors9,12,13,14. However, the correlation between TIME and tyrosine kinase inhibitor (TKI) responses is limited and conflicting15,16,17. These inconsistencies underscore the need for more specific biomarkers that accurately reflect the interplay between the immune microenvironments. This study aimed to evaluate the impact of T-cirDNA in EGFR-mutant NSCLCs among subjects who received EGFR TKIs and assess treatment outcomes. Biological explanations for this impact were explored through TIME immunohistochemistry and bulk RNA sequencing. Part of this study was presented at the 2023 European Society for Medical Oncology (ESMO) congress (Abstract 172P).
Methods
Study participants
We conducted a prospective multicenter cohort study of 158 participants with advanced or recurrent EGFR-mutant NSCLC who were diagnosed and receiving EGFR TKIs treatment at two academic centers in Bangkok, Thailand, King Chulalongkorn Memorial Hospital and Siriraj Hospital, Mahidol University, between January 1, 2020, and May 31, 2022. Demographic data, including age, gender, Eastern Cooperative Oncology Group (ECOG) performance status, histological type, smoking status, and generation of EGFR TKIs, were extracted from the hospital’s electronic management system. EGFR testing was conducted as standard practice, which included the Cobas® mutation test v2, gene panel, or next-generation sequencing as determined by the provided physician. Response evaluations were assessed every two to three months, according to the standard protocol at individual institutions. All participants provided written informed consent. This study was approved by the Institutional Review Board of the Faculty of Medicine at Chulalongkorn University (IRB No. 894/63) and Mahidol University (IRB No. 373/2564).
Blood specimen collection
Blood samples were collected in EDTA containers from participants before the initiation of EGFR TKIs and 6–12 weeks after the start of treatment. Plasma samples were centrifuged at 1600g for 10 min and then stored at − 80 °C until analysis. The plasma samples were thawed and recentrifuged at 13,000g for 10 min before DNA extraction.
Cell-free DNA extraction and measurement of rearranged TCRβ loci and RPP30
Two milliliters of plasma were used for DNA extraction using QIAamp® DNA Blood Mini Kit (QIAGEN), according to the manufacturer’s instructions. The plasma was eluted to 50 µL, dried with a speed vac without heating, and resuspended to a final volume of 10 µL. Quantitative measurement of total cirDNA and T-cirDNA was analyzed by multiplex real-time polymerase chain reaction assay in duplicate fashion using the ABI PRISM® 7500 system (Applied Biosystems, Carlsbad, CA, USA). 5 µL of plasma-extracted DNA was mixed with 10µL of 2X TaqMan™ Fast Advanced Master Mix and primers and probes for RPP30, representing total cirDNA, and TCRβ, representing T-cirDNA, as previously described protocol11. Initial denaturation was performed at 95 °C for 10 min, followed by 60 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 1 min, and extension at 72 °C for 30 s. An average of two replicates was used to represent the measured value of each patient. Standard curve of CD3 sorted T-lymphocyte from flow cytometry (BD FACS Aris II cell sorter; BD Biosciences, Singapore) was applied to cover the sensitivity points of 100, 10, 1, 0.1, 0.01 ng per reaction, which represented the lowest amplified thresholds of five copies per reaction. The T-cirDNA/cirDNA %ratio was calculated from total cirDNA and T-cirDNA values as follows:
Tumor-immune microenvironment evaluation
Pretreatment formalin-fixed paraffin-embedded tissue samples were evaluated for TIME, which included PD-L1, CD8 and FOXP3, using the immunohistochemistry method. Three-micron-thick sections were deparaffinized and stained by the Autostainer Link 48 (Dako, United States) by ready-to-use PD-L1 clone 22C3 Primary Monoclonal Mouse Anti-Human (cat. No. 22C3 pharmaDx; Dako, United States), CD8 clone C8/144B Primary Monoclonal Mouse AntiHuman (cat.No. IR623; Dako, United States), and FOXP3 (236 A/E7, Abcam) (dilution 1:100), as previous described14. PD-L1 was evaluated by tumor proportion score (TPS) as recommended18. CD8 TILs were scored into stromal CD8 TILs and intra-tumoral CD8 TILs. Intra-tumoral CD8 TILs were defined as CD8 TILs with direct cell-to-cell contact with cancer cells. Stromal CD8 TILs were defined as CD8 TILs dispersed in the stroma and do not directly contact cancer cells. Stromal CD8 TILs were scored according to the international TILs Working Group 2014 recommendation19. FOXP3, which represents regulatory T cells (Treg), was evaluated comprehensively. All slides were reviewed by one of the authors (S.S.), who was blinded to patient treatment, clinical outcome, and measured circulating DNA and T-cirDNA levels.
Bulk RNA sequencing
RNA was extracted from paraffin-embedded tissue, using the High Pure RNA paraffin Kit (Roche®) following the manufacturer’s protocol. After quality control, forty-one qualified samples proceeded to library construction using Agilent SureSelect XT HS2 Human All Exon V8 library preparation and target enrichment (San Diego, CA, USA). The sequencing was conducted using NovaSeq 6000 to generate 150 paired-end reads of at least 60Gb at Molecular Genomics Pte Ltd (Singapore). The FASTQ data was adjusted for quality control, trimmed with a sequence adapter, and aligned to the human reference genome (GRCh38) using the STAR aligner20. The transcript was quantified using the Salmon workflow21. We analyzed the TIME proportion via the deconvoluted CIBERSORT workflow22. A single-sample gene set enrichment score (ssGSEA), which provides more power to detect subtle pathway activity, was generated23. The estimated significant signatures from a total of 322 TIME-related gene signatures were analyzed using the R package (IOBR version 2.0)24.
Statistical analysis
The chi-square test, the Mann–Whitney U test, and the Kruskal-–Wallis test were used to test for mean differences in categorical data and non-parametric continuous data, respectively. Overall survival (OS) was defined as the time from recurrence and metastasis of disease until death from any cause. EGFR TKIs Progression-free survival (PFS), defined as the time from TKI initiation until disease progression, was assessed by study physicians. Both PFS and OS were estimated by the Kaplan-Meier method and log-rank test. Hazard ratios and 95% confidence intervals were calculated by Cox proportional hazard analysis. Univariate and multivariate analyses were conducted to identify factors influencing PFS and OS. Participants who did not develop the event were censored as of December 31, 2023. The statistical significance level was defined as a two-sided p-value less than 0.05. All statistical analyses were performed by STATA version 17.0, R-package version 3.6.3, or GraphPad Prism 9.4 (GraphPad Software, San Diego, CA, USA).
Results
Demographic characteristics
A total of 158 participants were enrolled in the study cohort (Fig. 1). Demographic characteristics were female predominance (67.1%), age ≥ 60 years (69.0%), never smokers (75.3%), and adenocarcinoma subtype (92.4%). The EGFR exon 19 deletion was the most common of EGFR alterations in our study (58.9%). Based on the reimbursement schedule for coverage in Thailand, first-generation EGFR TKIs were predominantly represented in our study (76.6%). Upfront third-generation EGFR TKIs, the current standard treatment, were found in 18.3% of our cohort. The baseline characteristics of this cohort are shown in Table 1. At the cutoff date, 121 (76.6%) participants progressed on first-line EGFR TKIs with a median follow-up of 22.4 months [IQR 13.5–31.2]. The median PFS of upfront EGFR TKIs was 12.8 months [IQR 10.9–14.2]. Subsequently, Osimertinib, a third-generation EGFR TKI, was given after disease progression in 33.9% of study participants. A total of 56% of participants received at least two treatment regimens, including EGFR TKIs and/or chemotherapy. Seventy-nine participants (50.3%) were still alive at the censored date, resulting in a median overall survival of 30.5 months [IQR 22.9–39.1].
Pretreatment circulating T-cirDNA/cirDNA correlated with treatment outcomes
We were able to evaluate circulating DNA in 153 participants, representing a 96.8% feasibility of the assay. The median total cirDNA, T-cirDNA, and T-cirDNA/cirDNA %ratio was 7.48 ng/mL [IQR 3.30–17.67 ng/mL], 34.59 pg/mL [IQR 0–401.02 pg/mL], and 0.19% [IQR 0–4.64%], respectively. Based on our previous study of T-cirDNA as a prognostic factor for advanced NSCLC11, participants were classified into three groups according to the %ratio of T-cirDNA/cirDNA: undetectable (41 participants, 26.8%), low T-cirDNA/cirDNA %ratio (≤ 1%) (56 participants, 36.6%), and high T-cirDNA/cirDNA %ratio (> 1%) (56 participants, 36.6%). No statistically significant differences were noted in baseline characteristics between the groups.
The median PFS in undetectable, low, and high T-cirDNA groups was 13.1 months [95% CI 8.8–18.3], 9.9 months [95% CI 7.4–13.0], and 15.5 months [95% CI 11.3–19.3], respectively (Fig. 2A). Compared to the low T-cirDNA group, the high T-cirDNA group was an independent factor correlated with longer PFS, with an HR of 0.63 [95% CI 0.40–0.99, p-value 0.046]. The undetectable T-cirDNA group was also an independent factor correlated with longer PFS, with an HR of 0.61 [95% CI 0.38–0.99, p-value 0.047] (Table 2). Performance status (ECOG 0–1 vs. 1–2) was an independent factor correlated with longer PFS with an HR of 0.34 [95% CI 0.18–0.63, p-value < 0.001]. The generation of EGFR TKIs (1st–2nd vs. 3rd ) was also correlated with shorter PFS, with an HR of 2.78 [95% CI 1.56–4.94, p-value < 0.001].
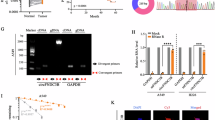
Progression-free survival of EGFR TKI treatment stratified by T-cirDNA/cirDNA category. Paradoxical correlation, which revealed unfavorable PFS in the low T-cirDNA group with a median of 9.9 months [95% CI 7.4–13]. As compared to the low T-cirDNA group, the hazard ratio of PFS in undetectable and high T-cirDNA groups was 0.72 [95% CI 0.46–1.14, log-rank p-value 0.17] and 0.65 [95% CI 0.42–0.99, log-rank p-value 0.045], respectively (A). Consistent with overall survival, unfavorable overall survival (OS) was observed in the low T-cirDNA group, with a median OS of 18.7 months [95% CI 13.5−23.6]. Compared to the low T-cirDNA group, the hazard ratios of the undetectable and high T-cirDNA groups were 0.41 [95% CI 0.24–0.72, log-rank p-value 0.001] and 0.34 [95% CI 0.19–0.59, log-rank p-value < 0.001], respectively (B). Forest plot of a significant TIME-related gene signature score correlated with EGFRi HR of PFS. CD8 T-cell signature was the most considerable signature score with the HR of PFS 3.18 [95% CI 1.3–7.7] (C). Heatmap of those significant TIME-related gene signatures from individual single-sample gene enrichment analysis (GSEA) correlated according to T-cirDNA group (D).
The median OS in the undetectable, low, and high T-cirDNA groups was 46.3 months [95% CI 25.2–NR], 18.7 months [95% CI 13.5–23.6], and NR [95% CI 32.4–NR], respectively (Fig. 2B). Compared to the low T-cirDNA group, both undetectable and high T-cirDNA groups were independent factors correlated with longer OS, with an HR of 0.38 [95% CI 0.21–0.70, p-value 0.002] and 0.32 [95% CI 0.18–0.59, p-value < 0.001], respectively (Table S1). Generation of EGFR TKIs treatment (1st -2nd vs. 3rd generation) and pattern of EGFR mutation (exon 21 L828R vs. exon 19 del) were independent factors correlated with shortened OS with an HR of 2.29 [95% CI 1.11–4.70, p-value 0.02] and 2.20 [95% CI 1.28–3.78, p-value 0.004], respectively. Performance status (ECOG 0–1 vs. 2–3) and histologic subtype (adenocarcinoma vs. others) were also independent factors correlated with longer OS with an HR of 0.28 [95% CI 0.14–0.56, p-value < 0.001] and 0.36 [95% CI 0.16–0.83, p-value 0.02], respectively (Table S1). Among known prognostic factors such as performance status, generation of EGFR TKI, and mutation pattern consistent with other studies, T-cirDNA was a novel independent prognostic factor that could be applied to both EGFR TKIs disease-free survival and overall survival.
The dynamic changes of T-cirDNA 6–12 weeks after starting EGFRi were minimal (Table S2, Fig. S1) and did not affect the outcomes of EGFR TKIs (Fig. S2). The details of the dynamic change of T-cirDNA are described in the supplementary results.
Tumor immune microenvironment and treatment outcomes
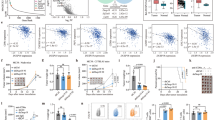
Eighty-two of 158 participants had available tissue blocks and adequate tumor portions for evaluating TIME (Fig. 1). We used the previously defined optimal cut-off of PD-L1 TPS in EGFR-mutated NSCLC receiving EGFRi, specifically the cutoff level of ≥ 15%, to determine levels of expression (high vs. low) and enable us to discriminate between prognostic outcomes25. Only nine of 82 participants (11.0%) had high PD-L1 expression. The spatial expression of CD8 + TILs and the cutoff threshold were based on our prior study11which reported meaningful results. The presence of intra-tumoral CD8 + TILs (≥ 1%), high stromal CD8 + TILs expression (≥ 5%), and high Treg/FOXP3 expression (≥ 5%) were 30.5%, 48.8%, and 37.8%, respectively. The TIME correlation and regression analysis information are in the supplementary results and Figure S3.
The median PFS of high and low PD-L1 expression was 5.3 months [95% CI 0.7–18.0] and 15.6 months [95% CI 12.5–21.6], respectively, with an HR of 2.79 [95% CI 1.31–5.95, p-value 0.006] (Fig. S4a, Table S3). The median OS in high and low PD-L1 expression was 14.8 months [95% CI 4.0–20.4] and 46.3 months [95% CI 30.5–NR], respectively, with HR of 3.65, 95% CI 1.56–8.56, p-value 0.001 (Fig. S4b, Table S4). These findings are consistent with previously published studies; however, tumor PD-L1 expression level was not an independent factor in multivariate analysis when including CD8 TILs. The presence of intra-tumoral CD8 TILs was an independent factor correlated with shorter PFS and OS, with HR of PFS 2.35 [95% CI 1.24–4.48, p-value 0.009] and HR of OS 5.13 [95% CI 2.10–12.50, p-value 0.003] (Fig. S5). This impact was contrary to stromal CD8 TILs (Fig. S6). High expression of FOXP3 was also an independent factor correlated with shorter OS with HR of 2.55 [95% CI 1.07–6.07, p-value 0.03] (Table 3, Fig. S7).
Correlation between circulating T-cirDNA/cirDNA %ratio and tumor immune microenvironment
Seventy-seven participants with overlapping tissue blocks and available T-cirDNA results were analyzed for interaction and correlation with clinical factors. The PD-L1, intra-tumoral CD8 TILs, stromal CD8 TILs, and Treg/FOXP3 expression level by T-cirDNA groups are shown in Table S5 and Figure S8. High PD-L1 expression proportionately increased with T-cirDNA groups: 4.0%, 12.0%, and 18.5% in undetectable, low T-cirDNA, and high T-cirDNA groups, respectively. The presence of intra-tumoral CD8 TILs and stromal CD8 TILs ≥ 5% was lowest in the low T-cirDNA group (16% and 36%, respectively), in contrast to undetectable (36% and 52%, respectively) and high T-cirDNA group (44.4% and 63%, respectively). FOXP3 expression was generally consistent among the three groups (36–40%).
When combining clinical data, T-cirDNA groups, and TIME IHC into the Cox regression analysis of PFS and OS, we found that undetectable T-cirDNA and high T-cirDNA groups were independent factors of OS with an HR of 0.19 [95% CI 0.07–0.58, p-value 0.003] and 0.14 [95% CI 0.04–0.46, p-value 0.001], respectively. The prognostic prediction of disease control (PFS) of EGFR TKIs was reduced when intra-tumoral CD8 TILs were incorporated in the model (Table 3).
Deconvolute the TIME proportion and related signatures
Bulk tumor RNA sequencing was conducted to explore the pathogenesis behind the paradoxical prognostic phenomenon of T-cirDNA. A total of 322 TIME-related gene signature scores were extracted (Table S6). Eleven significant TIME-related gene signatures correlated with EGFR TKI progression-free survival were retrieved. Among them, the CD8 T-cell signature was the most important, with an HR of 3.18 [95% CI 1.3–7.7] for PFS (Fig. 2C). Twelve TIME-related gene signatures correlated with T-cirDNA groups. Only the heme biosynthesis signature showed significant overlap between the hazard ratio of PFS EGFRi and T-cirDNA groups (Fig. 3A). Heme biosynthesis signature score was in 0.31, 0.19, and − 0.51 (p-value 0.03) in undetectable, low, and high T-cirDNA groups (Fig. 3B). The details of the TIME-related gene signature score related to the EGFRi hazard ratio of PFS and three T-cirDNA groups are shown in Tables S7 and S8.
A significant number of TIME-related signatures were observed in EGFRi PFS and T-cirDNA groups. The heme biosynthesis signature was the only one that overlapped with these outcomes (A). The boxplot of the heme biosynthesis signature score in three T-cirDNA groups, 0.31, 0.19, and − 0.51 in undetectable, low, and high T-cirDNA, with a Kruskal–Wallis p-value of 0.037 (B). Negative correlation was observed between CPOX gene expression and CIBERSORT-activated CD4 memory T-cells in the undetectable group (R -0.71, p-value 0.049). Neither low nor high detectable T-cirDNA was correlated with CIBERSORT-activated CD4 memory T-cells (C). Spearman correlation plot between a significant TIME-related signature and CIBERSORT-TIME fracture. The size of the dot represents − log10(p-value) (D). Heatmap of individual nine gene expressions of heme biosynthesis signature: ALAD, ALAS1, ALAS2, CPOX, FECH, HMBS, PPOX, UROD, and UROS expressions according to T-cirDNA group (E).
TIME proportion analysis via bulk RNAseq was also conducted using the deconvolute CIBERSORT-TIME workflow. This method has been reported to yield consistent and accurate results, and its findings have been correlated with those from flow cytometry26,27. The details of all CIBERSORT-TIME correlated with T-cirDNA and PFS ICIs are shown in Tables S9 and S10. We found no significant overlap with those parameters using CIBERSORT-TIME. However, correlations between the CIBERSORT-CD8 + proportion from RNA-seq and stromal/total CD8 + IHC were demonstrated by Pearson correlation coefficients; R 0.32–0.34 (p-value 0.05–0.07) (Fig. S9). Spearman correlation analysis between significant TIME-related gene signature scores and CIBERSORT TIME was conducted. Heme biosynthesis signature was also significantly correlated with CIBERSORT-activated CD4 memory T-cell, R − 0.04, p-value < 0.001 (Fig. 3D), which was mainly contributed by the undetectable T-cirDNA group, R − 0.79, p-value 0.019 (Fig. S10). In detail, the heme biosynthesis signature was composed of nine genes: ALAD, ALAS1, ALAS2, CPOX, FECH, HMBS, PPOX, UROD, and UROS. We found that only CPOX (coproporphyrinogen oxidase) gene expression was significantly diverse among T-cirDNA groups (Table S11). The average fold-change of CPOX gene expression was 0.61, 0.03, and − 0.64 in the undetectable, low, and high T-cirDNA groups, respectively (p-value 0.014). Furthermore, a negative correlation was observed between CPOX gene expression and CIBERSORT-activated CD4 memory T-cells in the undetectable group (R − 0.71, p-value 0.049). Neither low nor high detectable T-cirDNA was correlated with CIBERSORT-activated CD4 memory T-cells (Fig. 3C).
Discussion
We previously reported the impact of circulating T-cell-derived DNA and prognostic survival outcomes in advanced-stage non-small cell lung cancer. Our results showed that intra-tumoral T-lymphocytes might be a potential source of T-cell-derived DNA11. In this study, we explored only advanced-stage EGFR-mutated NSCLC participants who received EGFR TKIs. Pretreatment circulating T-cell-derived genomic material could be detected in 73% of the cohort. The paradoxical effects of T-cirDNA were consistent with those of a previous study. Undetectable and high T-cirDNA groups were independent factors correlated with PFS and OS, with multivariate HRs of PFS 0.61 [95% CI 0.38–0.99, p-value 0.047] and OS 0.63 [95% CI 0.40–0.99, p-value 0.046]. Clinical factors correlated with prognosis outcomes, such as the 3rd generation of EGFR TKIs, ECOG, and EGFR mutation subtypes, were consistent with several previous publications28,29,30,31. We performed a dynamic change of T-cirDNA in the 2nd timepoint after 6–12 weeks of EGFR TKIs treatment according to the usual time-to-treatment response of EGFR TKIs28,32. The dynamic change in T-cirDNA did not affect the outcomes of EGFR TKIs. These findings were contradictory to circulating tumor-derived genomic material, which mostly achieved clearance after a short duration of EGFR TKIs and corresponded with radiologic tumor responses33,34,35,36. Circulating immune-derived DNA more accurately represents perturbations of immune homeostasis37.
EGFR-mutant NSCLCs were generally associated with low PD-L1 and low CD8 TILs tumor immune microenvironment (TIME)12,13,14. EGFR mutation crosstalk with PD-L1 expression in lung cancer cell line studies was also reported38,39. PD-L1 expression by tumor cells also reduced the activity of effector T-cells40. Several studies demonstrated a poor response and shorter PFS with EGFR TKI treatment in EGFR-mutated lung cancer participants with high tumor PD-L1 expression17,25,41. We explored tumor PD-L1 together with CD8 and regulatory T-cell (Treg/FOXP3) TILs involved in tumor growth and development42. The non-immunological effects of EGFR mutation were strongly associated with the prevention of recruitment of effector CD8 TILs by down-regulation of CXCL10 through IRF143. We found that negative (< 1%) intra-tumor CD8 TILs were an independent factor correlated with longer PFS and OS. This implied that the absence of CD8 TILs represented activity solely downstream of EGFR signaling. It also appeared to be the reason for the greater durability of EGFR TKIs treatment and better survival outcomes. Treg/FOXP3 expression was not significantly different between the detected and undetected T-cirDNA groups. However, high Treg/FOXP3 expression (≥ 5%) in our study was significantly correlated with shortened survival outcomes (multivariate HR of OS 2.55 [95% CI 1.07–6.07, p-value 0.03) and potentially shortened disease control with EGFR TKIs (multivariate HR of PFS 1.79 [95% CI 0.96–3.32, p-value 0.067]. Consistent findings in resected NSCLCs have already been reported44,45,46.
We extracted the effect of tumor PD-L1, CD8 TILs, and Treg/FOXP3 TILs expression on T-cirDNA groups. An incrementally high PD-L1 expression (TPS score ≥ 15%) was observed in 4.0%, 12.0%, and 18.5% corresponding to the undetectable, low, and high T-cirDNA groups, respectively. The PD-L1 signaling is an essential component of tumor immunosuppression47. Therefore, despite the presence of intra-tumoral CD8 TILs (36%), these were inactive and led to consequences of undetected T-cell-derived genomic material in the undetectable T-cirDNA group. Treg/FOXP3 expression moderately correlated with stromal CD8 + TILs expression (R 0.5821, p-value < 0.0001). This might explain the prognostic phenomenon of stromal CD8 + TILs48. Combining the T-cirDNA and TIME expression, we could define the active function of CD8 + TILs, a novel finding, and explain the paradoxical prognostic effects of circulating T-cell-derived genomic material.
Lastly, integrating bulk RNA sequencing enables us to elucidate the biological significance of T-cirDNA. Heme biosynthesis, which plays a central role in mitochondrial respiration by supplying the prosthetic group to oxidative phosphorylation49 and is correlated with outcome and treatment response in NSCLC49,50,51. It also correlated with the immunosuppressive condition52 and was recently reported as an underlying mechanism of acquired osimertinib resistance53. The detectable T-cirDNA group exhibited active heme biosynthesis and T-cell function. In contrast to the undetectable T-cirDNA group, despite active heme biosynthesis, inactivated T-cells were present. CPOX, as part of heme biosynthesis, is essential for T-cell development and is involved in the process of T-cell lineage commitment to either CD4 + or CD8 + cells54. Overexpression of CPOX in undetectable T-cirDNA is crucial for the inactivation of T-cell effector function. Crosstalk of EGFR and CPOX prohibits the activation of CD4 + memory T-cells or the spatial intra-tumoral CD8 + T-cells. The biological explanation is summarized in Fig. 4.
Summary of the biological explanation of paradoxical prognostic effect of undetectable T-cirDNA vs. detectable T-cirDNA in mutated-EGFR NSCLC who received EGFRi. Active heme biosynthesis pathway via CPOX overexpression depended on the active function of intra-tumoral CD8 TILs, which subsequently develops an immunosuppressive TIME condition. Solely active EGFR signaling with active heme biosynthesis but without active intra-tumoral CD8 TILs led to a favorable outcome of EGFRi. More active heme biosynthesis and a less immune response against cancer cells in low-detectable T-cirDNA, despite EGFRi treatment, are associated with the worst prognosis outcome and shortest disease control. https://app.biorender.com/illustrations/6888922da1291bd128698c30 “Created with BioRender.com”.
In conclusion, the paradoxical prognostic effect of T-cirDNA and its biological significance have been reported. Undetectable T-cirDNA, representing inactivated naïve T-cells by crosstalk of CPOX, part of heme biosynthesis pathway. It was solely active downstream of EGFR signaling, which correlated with a favorable prognosis. While the high-T-cirDNA, which represents active T-cells, also had favorable outcomes via a less active heme biosynthesis pathway, the presence of intra-tumoral CD8 + and Treg/FOXP3 TILs was an independent factor impacting both PFS and OS.
Data availability
The RNA sequencing datasets generated and/or analyzed in this study were submitted to the NCBI database; Bioproject ID PRJNA1275863.
References
Cisneros-Villanueva, M. et al. Cell-free DNA analysis in current cancer clinical trials: a review. Br. J. Cancer. 126, 391–400. https://doi.org/10.1038/s41416-021-01696-0 (2022).
Jahr, S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001).
Thierry, A. R., Messaoudi, E., Gahan, S., Anker, P. B., Stroun, M. & P. & Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 35, 347–376. https://doi.org/10.1007/s10555-016-9629-x (2016).
Aucamp, J., Bronkhorst, A. J., Badenhorst, C. P. S. & Pretorius, P. J. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc. 93, 1649–1683. https://doi.org/10.1111/brv.12413 (2018).
Song, P. et al. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 6, 232–245. https://doi.org/10.1038/s41551-021-00837-3 (2022).
Cheng, M. L. et al. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J. Clin. 71, 176–190. https://doi.org/10.3322/caac.21650 (2021).
Vinayanuwattikun, C., Winayanuwattikun, P., Chantranuwat, P., Mutirangura, A. & Sriuranpong, V. The impact of non-tumor-derived circulating nucleic acids implicates the prognosis of non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 139, 67–76. https://doi.org/10.1007/s00432-012-1300-5 (2013).
Shirasawa, M. et al. Differential immune-related microenvironment determines programmed cell death Protein-1/Programmed death-Ligand 1 Blockade efficacy in patients with advanced NSCLC. J. Thorac. Oncol. 16, 2078–2090. https://doi.org/10.1016/j.jtho.2021.07.027 (2021).
Jin, R. et al. Molecular heterogeneity of anti-PD-1/PD-L1 immunotherapy efficacy is correlated with tumor immune microenvironment in East Asian patients with non-small cell lung cancer. Cancer Biol. Med. 17, 768–781. https://doi.org/10.20892/j.issn.2095-3941.2020.0121 (2020).
Lin, Z. et al. Deciphering microenvironment of NSCLC based on CD8 + TIL density and PD-1/PD-L1 expression. J. Cancer. 10, 211–222. https://doi.org/10.7150/jca.26444 (2019).
Sitthideatphaiboon, P. et al. Paradoxical prognostic phenomenon of plasma T-cell-derived circulating DNA level in advanced non-small cell lung cancer. Clin. Transl. Oncol. 22, 1117–1125. https://doi.org/10.1007/s12094-019-02238-0 (2020).
Dong, Z. Y. et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology. 6, e1356145. https://doi.org/10.1080/2162402X.2017.1356145 (2017).
Gainor, J. F. et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway Blockade in non-small cell lung cancer: A retrospective analysis. Clin. Cancer Res. 22, 4585–4593. https://doi.org/10.1158/1078-0432.CCR-15-3101 (2016).
Toki, M. I. et al. Immune marker profiling and programmed death ligand 1 expression across NSCLC mutations. J. Thorac. Oncol. 13, 1884–1896. https://doi.org/10.1016/j.jtho.2018.09.012 (2018).
Lin, C. et al. Programmed Death-Ligand 1 expression predicts tyrosine kinase inhibitor response and better prognosis in a cohort of patients with epidermal growth factor receptor mutation-positive lung adenocarcinoma. Clin. Lung Cancer. 16, e25–35. https://doi.org/10.1016/j.cllc.2015.02.002 (2015).
Bai, Y. et al. PD-L1 expression and its effect on clinical outcomes of EGFR-mutant NSCLC patients treated with EGFR-TKIs. Cancer Biol. Med. 15, 434–442. https://doi.org/10.20892/j.issn.2095-3941.2018.0223 (2018).
Matsumoto, Y. et al. Impact of tumor microenvironment on the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with EGFR-mutant non-small cell lung cancer. Cancer Sci. 110, 3244–3254. https://doi.org/10.1111/cas.14156 (2019).
PD-L1 IHC 22C3 pharmDx Interpretation Manual—NSCLC. https://www.agilent.com/cs/library/usermanuals/public/29158_pd-l1-ihc-22C3-pharmdx-nsclc-interpretation-manual.pdf.
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs working group 2014. Ann. Oncol. 26, 259–271. https://doi.org/10.1093/annonc/mdu450 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14, 417–419. https://doi.org/10.1038/nmeth.4197 (2017).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7. https://doi.org/10.1186/1471-2105-14-7 (2013).
Zeng, D. et al. Enhancing immuno-oncology investigations through multidimensional decoding of tumor microenvironment with IOBR 2.0. Cell. Rep. Methods. 4, 100910. https://doi.org/10.1016/j.crmeth.2024.100910 (2024).
Cruz-Rico, G. et al. Association of lung adenocarcinoma subtypes according to the IASLC/ATS/ERS classification and programmed cell death ligand 1 (PD-L1) expression in tumor cells. Pathol. Oncol. Res. 27, 597499. https://doi.org/10.3389/pore.2021.597499 (2021).
Saito, N. et al. Selection of RNA-based evaluation methods for tumor microenvironment by comparing with histochemical and flow cytometric analyses in gastric cancer. Sci. Rep. 12, 8576. https://doi.org/10.1038/s41598-022-12610-w (2022).
Nadel, B. B. et al. Systematic evaluation of transcriptomics-based Deconvolution methods and references using thousands of clinical samples. Brief. Bioinform. 22 https://doi.org/10.1093/bib/bbab265 (2021).
Soria, J. C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125. https://doi.org/10.1056/NEJMoa1713137 (2018).
Ramalingam, S. S. et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 382, 41–50. https://doi.org/10.1056/NEJMoa1913662 (2020).
Lin, J. H. et al. The association between clinical prognostic factors and epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) efficacy in advanced non-small-cell lung cancer patients: a retrospective assessment of 94 cases with EGFR mutations. Oncotarget 8, 3412–3421. https://doi.org/10.18632/oncotarget.13787 (2017).
Choi, Y. W. et al. EGFR exon 19 deletion is associated with favorable overall survival after First-line gefitinib therapy in advanced Non-Small cell lung cancer patients. Am. J. Clin. Oncol. 41, 385–390. https://doi.org/10.1097/COC.0000000000000282 (2018).
Wu, Y. L. et al. Tumor response and health-related quality of life in clinically selected patients from Asia with advanced non-small-cell lung cancer treated with first-line gefitinib: post hoc analyses from the IPASS study. Lung Cancer. 81, 280–287. https://doi.org/10.1016/j.lungcan.2013.03.004 (2013).
Ku, B. M. et al. Role of circulating tumor DNA profiling in patients with non-small cell lung cancer treated with EGFR inhibitor. Oncology. 100, 228–237. https://doi.org/10.1159/000516813 (2022).
Li, Y. et al. Disease monitoring of epidermal growth factor receptor (EGFR)-mutated non-small-cell lung cancer patients treated with tyrosine kinase inhibitors via EGFR status in circulating tumor DNA. Thorac. Cancer. 13, 2201–2209. https://doi.org/10.1111/1759-7714.14545 (2022).
Boysen, F. et al. Clearing of circulating tumour DNA predicts clinical response to osimertinib in EGFR mutated lung cancer patients. Lung Cancer. 143, 67–72. https://doi.org/10.1016/j.lungcan.2020.03.020 (2020).
Moiseenko, F. V. et al. Monitoring of the presence of EGFR-mutated DNA during EGFR-targeted therapy May assist in the prediction of treatment outcome. Cancer Treat. Res. Commun. 31, 100524. https://doi.org/10.1016/j.ctarc.2022.100524 (2022).
Fox-Fisher, I. et al. Remote immune processes revealed by immune-derived circulating cell-free DNA. Elife. 10 https://doi.org/10.7554/eLife.70520 (2021).
Chen, N. et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-Driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J. Thorac. Oncol. 10, 910–923. https://doi.org/10.1097/JTO.0000000000000500 (2015).
Zhang, N. et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int. J. Oncol. 49, 1360–1368. https://doi.org/10.3892/ijo.2016.3632 (2016).
Topalian, S. L., Drake, C. G. & Pardoll, D. M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 27, 450–461. https://doi.org/10.1016/j.ccell.2015.03.001 (2015).
Shiozawa, T. et al. Prognostic implication of PD-L1 expression on osimertinib treatment for EGFR-mutated non-small cell lung cancer. Anticancer Res. 42, 2583–2590. https://doi.org/10.21873/anticanres.15736 (2022).
Madeddu, C. et al. EGFR-mutated non-small cell lung cancer and resistance to immunotherapy: role of the tumor microenvironment. Int. J. Mol. Sci. 23 https://doi.org/10.3390/ijms23126489 (2022).
Sugiyama, E. et al. Blockade of EGFR improves responsiveness to PD-1 Blockade in EGFR-mutated non-small cell lung cancer. Sci. Immunol. 5 https://doi.org/10.1126/sciimmunol.aav3937 (2020).
Zhao, S. et al. Clinicopathological and prognostic significance of regulatory T cells in patients with non-small cell lung cancer: A systematic review with meta-analysis. Oncotarget. 7, 36065–36073. https://doi.org/10.18632/oncotarget.9130 (2016).
O’Callaghan, D. S. et al. Tumour islet Foxp3 + T-cell infiltration predicts poor outcome in nonsmall cell lung cancer. Eur. Respir. J. 46, 1762–1772. https://doi.org/10.1183/13993003.00176-2014 (2015).
Shimizu, K. et al. Tumor-infiltrating Foxp3 + regulatory T cells are correlated with cyclooxygenase-2 expression and are associated with recurrence in resected non-small cell lung cancer. J. Thorac. Oncol. 5, 585–590. https://doi.org/10.1097/JTO.0b013e3181d60fd7 (2010).
Jiang, X. et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer. 18, 10. https://doi.org/10.1186/s12943-018-0928-4 (2019).
Hashemi, S. et al. Surprising impact of stromal til’s on immunotherapy efficacy in a real-world lung cancer study. Lung Cancer. 153, 81–89. https://doi.org/10.1016/j.lungcan.2021.01.013 (2021).
Kalainayakan, S. P., FitzGerald, K. E., Konduri, P. C., Vidal, C. & Zhang, L. Essential roles of mitochondrial and Heme function in lung cancer bioenergetics and tumorigenesis. Cell. Biosci. 8, 56. https://doi.org/10.1186/s13578-018-0257-8 (2018).
Bo, T., Van Wijk, K. & Nakajima, O. Heme biosynthesis is crucial for cell survival and mitochondrial OXPHOS after X irradiation. Radiat. Res. 201, 48–54. https://doi.org/10.1667/RADE-23-00035.1 (2024).
Hooda, J. et al. Enhanced Heme function and mitochondrial respiration promote the progression of lung cancer cells. PLoS One. 8, e63402. https://doi.org/10.1371/journal.pone.0063402 (2013).
Akter, Z., Salamat, N., Ali, M. Y. & Zhang, L. The promise of targeting Heme and mitochondrial respiration in normalizing tumor microenvironment and potentiating immunotherapy. Front. Oncol. 12, 1072739. https://doi.org/10.3389/fonc.2022.1072739 (2022).
Wang, H. et al. Ferritinophagy mediates adaptive resistance to EGFR tyrosine kinase inhibitors in non-small cell lung cancer. Nat. Commun. 15, 4195. https://doi.org/10.1038/s41467-024-48433-8 (2024).
Kumar, A. et al. Iron regulates the quiescence of Naive CD4 T cells by controlling mitochondria and cellular metabolism. Proc. Natl. Acad. Sci. U. S. A. 121, e2318420121. https://doi.org/10.1073/pnas.2318420121 (2024).
Acknowledgements
Acknowledgments: This project is funded by the Health Systems Research Institute (Thailand) (Grant number 66-153) to NZ, PS, CV, VS, and NH, and Rachadapisek Sompote Matching fund [RA-MF-17/67] to NH. HN is a member of the Thailand Hub of Talents in Cancer Immunotherapy (TTCI). The academic endeavors of TTCI received support from the National Research Council of Thailand [Grant number N N34E670096]. Biobank, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, supported the biospecimen collection.
Author information
Authors and Affiliations
Contributions
N.Z. contributed to the investigation, analysis, and drafting of the manuscript. S.S. contributed to investigation and data acquisition. C.N., P.S. and N.L. and K.K. contributed specimen collection and data acquisition. C.A. contributed to the data analysis and drafted the manuscript. C.V. contributed to the design of the research, conception, analysis of the data, funding acquisition, and drafting of the manuscript. P.C. and V.S. provided consultation. N.H. contributed to funding acquisition and provided consultation. All authors critically revised the manuscript, agreed to be fully accountable for ensuring the integrity and accuracy of the work, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
The authors are accountable for all aspects of the work and ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients provided written informed consent. This study was approved by the Institutional Review Board of the Faculty of Medicine at Chulalongkorn University (IRB No. 894/63) and Mahidol University (IRB No. 373/2564) and performed in accordance with the Health Insurance Portability and Accountability Act and the Declaration of Helsinki (as revised in 2013).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zungsontiporn, N., Ngamchokwathana, C., Santisukwongchote, S. et al. The paradoxical prognostic effects of T-cell-derived circulating DNA in EGFR-mutated advanced non-small-cell lung cancer involve the crosstalk between heme biosynthesis and immune microenvironment. Sci Rep 15, 36434 (2025). https://doi.org/10.1038/s41598-025-20307-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-20307-z