Abstract
Coronary microvascular dysfunction serves as one of the etiological factors for ischemic heart disease and represents a novel therapeutic direction for coronary artery diseases; however, the research on its pathogenesis remains inconsistent. This study aims to explore the single-cell gene expression profiles in rats with coronary microvascular dysfunction using single-cell RNA sequencing, with a particular focus on the in-depth analysis of endothelial cell gene expression characteristics. By establishing a rat model of coronary microvascular dysfunction, we collected cardiac apical tissue to prepare single-cell suspensions and further analyzed them using bioinformatics methods. From a total of 55,419 cells, we identified 28 distinct cell clusters, with endothelial cells and fibroblasts being the predominant cell types. Compared to the NC group, the proportion of endothelial cells in the CMD group was significantly reduced, while the number of fibroblasts was significantly increased. Through further analysis of the endothelial cells, we classified them into normal phenotype endothelial cells, mesenchymal phenotype endothelial cells, proliferative phenotype endothelial cells, and lymphatic endothelial cells, with mesenchymal and proliferative endothelial cells originating from normal phenotype endothelial cells. Additionally, the CMD group exhibited an increase in immune cells, enhanced inflammatory response, and increased oxidative stress. These findings may provide novel potential therapeutic targets for the treatment of Coronary microvascular dysfunction.
Similar content being viewed by others
Introduction
Coronary microvascular dysfunction is defined as impaired coronary flow reserve due to structural and/or functional abnormalities of the microvasculature, and is associated with adverse cardiovascular outcomes. CMD is considered one of the etiologies of ischemic heart disease, accompanied by signs and symptoms of myocardial ischemia, but without obstructive coronary artery disease1. Up to 70% of patients with angina pectoris have no obstructive coronary artery lesions detected after coronary angiography (epicardial vessel stenosis ≥ 50%), suggesting the presence of coronary microcirculatory dysfunction or non-obstructive coronary heart disease2. In recent years, multiple guidelines both domestically and internationally have highlighted the importance of CMD in cardiovascular diseases, making it a new direction in the treatment of coronary artery disease. However, the current diagnostic methods for CMD are limited, leading to missed and misdiagnoses. Due to the lack of targeted pharmacological treatment, traditional drugs for coronary heart disease have limited effects on CMD. Therefore, these patients continue to experience recurrent angina pectoris, impaired quality of life, leading to repeated hospitalizations and unnecessary coronary angiography, as well as short-term and long-term adverse cardiovascular outcomes3. Therefore, finding effective diagnostic and therapeutic methods for CMD is an urgent need to be addressed. Currently, single-cell sequencing technology, particularly single-cell RNA sequencing, has brought revolutionary changes to the research of cardiovascular diseases. It can identify different cell types and analyze the construction of single-cell trajectories to explore new drug targets4. Therefore, we collected apical heart tissue samples from CMD rats for single-cell RNA sequencing to reveal the dynamic changes of cells in the coronary microvasculature and surrounding tissues, seeking the etiology of the progression of endothelial cell (EC) dysfunction. The results presented allow us to explore the pathways and key genes involved in the interactions between different endothelial cell populations or with other cell types. By modulating their activities through inhibition or promotion, we can further prevent alterations in the structure and function of endothelial cells, thereby retarding the onset and progression of CMD. Thus providing potential new targets and theoretical basis for the diagnosis and treatment of CMD.
Materials and methods
Chemicals and reagents
Streptozotocin and Pentobarbital sodium were purchased from MedChemExpress, USA. Dobutamine was obtained from Zhejiang Ruixin Pharmaceutical Co., Ltd. PBS buffer was sourced from Solarbio Science & Technology Co., Ltd., China. The 10 × Genomics single-cell RNA sequencing kit v3 was procured from 10× Genomics, USA. Red cell lysate and Decaying cell kit were acquired from Miltenyi Biotec, Germany. Dissociating solvent was bought from Merck Group, Germany.
Animals
Twenty Sprague–Dawley rats (male, 4–6 weeks old) were obtained from the Animal Laboratory of Shanxi Medical University. All rats were housed in the laboratory of Shanxi Bethune Hospital. This study was approved by the Ethics Committee of Shanxi Bethune Hospital, and the ethics approval number was YXLL-2023–238. We numbered 20 rats and used a random number generator to randomly select rats, assigning the selected numbers to the NC group until there were 10 rats in this group. The remaining rats were then assigned to the CMD group. During the experimentation, the NC group (n = 10) rats were fed a normal diet, while the CMD group (n = 10) rats were fed a high-fat and high-sugar diet (67% rat maintenance feed + 10% lard + 20% sucrose + 2.5% cholesterol + 0.5% sodium cholate). After 4 weeks of high-fat and high-sugar diet feeding, the CMD group was fasted for 12 h and induced by intraperitoneal injection of streptozotocin (30 mg/kg bw). Meanwhile, age-matched NC group rats were fed a regular maintenance diet for 31 days. Three days after streptozotocin injection, the 12-h fasting blood glucose levels of all rats were measured.
Cardiology ultrasound
Two weeks later, rats were anesthetized with intraperitoneal injection of pentobarbital sodium (35 mg/kg) and placed in a supine position with chest hair removed. The room temperature was maintained at 37–38 °C, and heart rate was monitored using the Epiq software. Echocardiography was performed using a 13–24 MHz linear probe: the probe was positioned using a rail system to obtain a parasternal short-axis view, moved to locate the pulmonary artery, and slightly moved caudally to visualize the anterior descending artery and store the images. The anterior descending artery was displayed in color Doppler mode, and then the mode was switched to pulsed wave mode. The cine store was used to capture the resting blood flow velocity waveform at the peak diastolic phase of the anterior descending artery. After obtaining the resting velocity of the anterior descending artery, dobutamine liquid was administered via the tail vein at a dose of 20 μg/kg/min (observing changes in rat heart rate, with a maximum observation time of 5 min). The maximum velocity during the administration process was measured, and coronary flow reserve (calculated as the ratio of the peak velocity during stress to the resting velocity) was calculated. After the measurement, the infusion of dobutamine liquid was stopped, the tail vein infusion device was removed, hemostasis was performed, and the rats were allowed to recover from anesthesia.
Sample collection
After completing the ultrasound measurements, one week later, we used a random number generator to randomly select three rats from each of the NC and CMD groups for the study. These rats were anesthetized and euthanized by cervical dislocation, and their hearts were promptly removed and placed in ice-cold saline at 4 °C. The apical tissue of the hearts was then minced using ophthalmic scissors and stored in PBS solution at 4 °C.
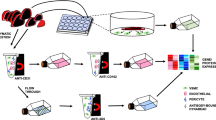
Preparation of single-cell suspension
The tissue was transferred to a culture dish pre-filled with 1 × PBS (free of RNase and Ca, Mg ions) and placed on wet ice. The tissue was then cut into small pieces of 0.5 mm2 and repeatedly washed with 1 × PBS to remove surface blood stains, fats, and other adherent substances. The cleaned tissue pieces were then added to a dissociation solution containing 0.35% collagenase IV, 2 mg/ml papain, and 120 Units/ml DNase I, and incubated at 37 °C on a shaking water bath (100 rpm) for 20 min. The dissociation was terminated by adding 1 × PBS containing 10% fetal bovine serum (FBS). The cell suspension was then pipetted up and down 5–10 times with a pipette (gentle operation to prevent shear force-induced cell death). The cell suspension was filtered through a 70–30 μm cell strainer and centrifuged at 300 g for 5 min at 4 °C. The cell pellet was collected after centrifugation and resuspended in 100 μl of 1 × PBS (0.04% BSA). To remove red blood cells, 1 ml of red blood cell lysis buffer (MACS 130–094,183, 10 ×) was added and incubated on wet ice for 2–10 min. After lysis, the cells were centrifuged at 300 g for 5 min at room temperature, and the cell pellet was collected. The cell pellet was then resuspended in 100 μl of dead cell removal reagent: Dead Cell Removal MicroBeads (MACS 130-090-101) and dead cells were removed using the Miltenyi® Dead Cell Removal Kit (MACS 130-090-101). After the reaction, the reagent was removed by centrifugation. The cells were then resuspended in 1 × PBS (0.04% BSA) and centrifuged at 300 g for 3 min at 4 °C (repeated twice). The resulting cell suspension, which had undergone tissue dissociation, red blood cell lysis, and dead cell removal, was then adjusted to 50 μl of 1 × PBS (0.04% BSA). Cell viability was assessed using the trypan blue staining method, with a requirement of > 85% viability. The cell count was performed using a Countess II Automated Cell Counter, with a required cell concentration of 700–1200 cells/μl.
Library construction and sequencing
The cell suspension obtained after dissociation was loaded into the 10× Chromium instrument, and cell capture (targeting 5,000 cells) was performed according to the manufacturer’s instructions for the library preparation kit (10X Genomics Chromium Single-Cell 3’ kit, V3). Following this, cDNA amplification and library construction were conducted. Upon completion of library construction, the NovaSeq 6000 sequencing platform was utilized for paired-end multiplexing sequencing with a read length of 150 bp, with a required sequencing depth of 20,000 reads per cell.
ScRNAseq analysis
Sequencing data were processed using the Cell Ranger single-cell software suite v7.1.0 (10 × Genomics) to handle single-cell expression data, including data filtering, genome alignment, transcript quantification, and cell identification, ultimately yielding gene expression matrices for individual cells. The gene barcode matrix and UMI count data were integrated using Seurat (V4.1.1)5. The output was then imported into Seurat (V4.1.1) R for subsequent quality control and downstream analysis of single-cell RNA sequencing data6. The PercentFeatureSet function of the Seurat software package was employed to calculate mitochondrial gene expression7. Cells with > 25% UMI in the mitochondrial genome and fewer than 500 genes were removed through quality control. The integrated matrix was scaled, and principal component analysis (PCA) was performed to identify 20 principal components for downstream analysis. The FindNeighbors function was used to cluster cells with similar expression patterns, and the FindClusters function was applied to delineate distinct cell subpopulations, which were visualized using the Uniform Manifold Approximation and Projection (UMAP) algorithm. Subsequently, the FindAllMarkers function in Seurat was utilized to identify differentially expressed genes in each cluster for subsequent cell annotation and classification. Cell clusters were manually annotated into major cell types based on known markers. To further delineate EC subpopulations, the entire EC subclass was extracted using Seurat, and the FindClusters function was applied again with a resolution of 0.3 for clustering analysis, followed by manual annotation of cell types. Additionally, the FindMarkers function in Seurat was used to screen for differentially expressed genes between the CMD and NC groups. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes (DEGs) were performed using the ClusterProfiler (v 3.14.0) software package.
Pseudotemporal analysis
We utilized the Monocle 2 software package to perform machine learning based on the expression patterns of key genes and simulate the dynamic changes over time. Initially, we selected genes with substantial intercellular expression differences and performed spatial dimensionality reduction according to their expression profiles. Subsequently, we constructed a minimum spanning tree and identified the longest path that represents the differentiation trajectory of cells with similar transcriptional features.
GSVA analysis
Gene set variation analysis (GSVA) was performed using theOmicStudio tools at https://www.omicstudio.cn/tool.The plots was drawn based on the R version 4.1.3 (2022-03-10) on the Omicstudio platform (https://www.omicstudio.cn).
Cell-to-cell communication network analysis
We employed the CellChat package, which is based on the R programming language, to analyze intercellular communication. Overexpressed ligands or receptors identified within the cell population were mapped onto the protein–protein interaction (PPI) network. CellChat infers biologically significant intercellular communication by assigning probability values to each interaction and conducting permutation tests. Communication networks and signaling pathways were visualized using circular plots and heatmaps.
Results
Animals and ultrasound
After 31 days of high-fat and high-sugar feeding and 3 days following intraperitoneal injection of streptozotocin (STZ), blood glucose levels were measured. The results indicated that all rats in the CMD group had blood glucose levels above 11.1 mmol/L, whereas those in the normal control (NC) group had levels below 11.1 mmol/L (P < 0.05) (Table 1). Subsequently, echocardiographic examinations were performed on both groups of rats. The findings (Fig. 1A-D) revealed a significant reduction in the coronary flow reserve in the CMD group compared to the NC group (P < 0.05) (Table 2). For further investigation, rats from the NC group with the IDs 2, 5, and 10, and rats from the CMD group with the IDs 1, 4, and 7 were randomly selected as study subjects and designated as NC1, NC2, NC3, CMD1, CMD2, and CMD3, respectively, for subsequent analyses.
The transcriptomic profiles of the rats in the NC group and CMD group
After filtering cells with low RNA content or high mitochondrial RNA content, a total of 55,419 cells were annotated (ranging from 7,558 to 11,010 cells per sample). Unsupervised clustering of gene expression revealed 28 clusters (Fig. 2A). The segregation of these cell subclusters was well-illustrated by the top 10 marker genes for each subcluster (Fig. 2B). Upon reviewing the pertinent literature, we have observed the presence of various cell types, including endothelial cells, fibroblasts, and immune cells, within coronary microvascular and the surrounding tissue8. Under ischemic conditions due to myocardial infarction, endothelial cells may interact with immune cells and potentially undergo transient mesenchymal activation, leading to endothelial cell dysfunction9. Consequently, we speculate that these cells may also play a crucial role in the pathogenesis of CMD. Therefore, by referring to relevant literature, we manually annotated 10 cell types: endothelial cells (ECs) (Cdh5, Vwf, Sox17, Cldn5, and Pecam1), fibroblasts (Col1a1, Col3a1, Pdgfrb, and Dcn), vascular smooth muscle cells (VSMCs) (Acta2, Tagln, Cald1, and Myl9), pericytes (Rgs5 and Abcc9), T cells (Cd3g, Cd3e, Cd3c), B cells (Ms4a1 and Cd79a), natural killer (NK) cells (Gzma, Ctsw, Nkg7, Xcl1, and Ccl5), macrophages (Cd68, Adgre1, C1qa, Csf1r, and C1qc), monocytes (Ccr2, Csf1r, Fcgr3, Plac8, Mal, and Fcnb), and neutrophils (S100a8, G0s2, and Alox5ap) (Fig. 2C)10. We observed that the predominant cell populations were ECs (25,119, 45.33%) and fibroblasts (15,793, 28.50%), with the remaining cell types occupying varying proportions: VSMCs (2,356, 4.25%), pericytes (2,548, 4.60%), T cells (2,447, 4.41%), macrophages (2,345, 4.23%), NK cells (2,023, 3.65%), monocytes (1,015, 1.83%), neutrophils (980, 1.77%), and B cells (793, 1.43%). The bubble plots of marker genes for different cell types demonstrated a clear separation between the various cell types (Fig. 2E). A further comparison of the cell percentages between the CMD and NC groups (Fig. 2D) revealed that, compared to the NC group, the CMD group exhibited the most significant decrease in the number and proportion of ECs, while the number and proportion of fibroblasts increased the most. Additionally, the number and proportion of VSMCs did not change significantly, while the number and proportion of pericytes relatively decreased. In contrast, the number and proportion of immune cells, such as macrophages, T cells, NK cells, monocytes, B cells, and neutrophils, relatively increased. The UMAP plot also illustrated the changes in different cell types between the two groups (Fig. 2F).
(A) The UMAP plot of all sample clusters revealed 28 clusters. (B) Heatmap of the top 10 marker genes for the 28 cell clusters. (C) The UMAP plot of all samples manually clustered into 10 cell subclasses. (D) Proportion of the 10 cell types between the CMD group and the NC group. (E) Bubble plot of marker genes for the 10 cell subclasses. (F) UMAP plot of the 10 cell subclasses between the CMD group and the NC group.
Differential phenotypes and functions of endothelial cells
We further investigated the primary cell types contributing to the progression of coronary microcirculatory dysfunction. Unsupervised clustering analysis using Seurat identified 11 ECs subclusters (Fig. 3A). The top 10 marker genes of these ECs subclusters demonstrated good separation between different subclusters (Fig. 3B), and these 11 types of ECs were distributed in each sample (Fig. 3C) (excluding cluster 10, which was cell debris, for subsequent analysis). Based on the functional differences of their expressed genes, we classified clusters 0–5 and 9 as normal endothelial phenotype (EC1), cluster 6 as mesenchymal endothelial phenotype (EC2), cluster 8 as proliferative endothelial phenotype (EC3), and cluster 7 as lymphatic endothelial phenotype (EC4) (Fig. 3D). The separation of these four EC types was well illustrated by the bubble plot of marker genes for each cell type (Fig. 3E). The identification rationale is as follows:
(A) The UMAP plot of all sample endothelial cell clusters revealed 11 clusters. (B) Heatmap of the top 10 marker genes for the 11 endothelial cell clusters. (C) UMAP plot of the distribution of 11 endothelial cell types across all samples. (D) The UMAP plot of all samples manually clustered into 4 endothelial cell subclasses. (E) Bubble plot of marker genes for the 4 endothelial cell subclasses. (F) Gene Ontology (GO) enrichment analysis for EC2. (G) GO enrichment analysis for EC3.
All four types of ECs expressed pan-endothelial genes Sox17, Cldn5, Pecam1, Cdh5, Vwf, Kdr, but EC1 had relatively higher expression levels of these pan-endothelial genes compared to other types of ECs. These genes collaborate in the development, maintenance, and function of the vascular system, playing a crucial role in maintaining the stability, adaptability, and normal function of blood vessels. Therefore, EC1 was identified as the normal endothelial phenotype.
EC2 also expressed pan-endothelial genes, but at relatively lower levels, and significantly expressed genes related to Endothelial-to-mesenchymal transition (EndoMT), such as Mfap5, Bgn, Timp1, Fn1, Col5a2, Col3a1, and Serpine19. EndoMT refers to the process of ECs transforming into mesenchymal cells. During this process, ECs, mainly vascular ECs, lose their typical endothelial characteristics and gradually acquire mesenchymal cell features11. This is a phenomenon of cell transformation, similar to Epithelial-to-Mesenchymal Transition, but occurring in ECs. Therefore, EC2 was identified as the mesenchymal endothelial phenotype. The Gene Ontology (GO) analysis of these ECs mainly enriched in extracellular matrix organization, response to endoplasmic reticulum stress, cell adhesion, regulation of collagen fibril organization, response to oxidative stress, positive regulation of cell migration, and Transforming Growth Factor-β receptor signaling pathway (Fig. 3F).
EC3 also expressed pan-endothelial genes at relatively lower levels and significantly expressed genes such as HMGB2, MKI67, CENPF, and Top2a, which are associated with tumors. Overexpression of HMGB2 and CENPF can regulate the cell cycle and promote cell proliferation12,13. Ki-67 is commonly used as a biomarker for cell proliferation. Histologically, the expression of Ki-67 can estimate the proliferation status of cells, and high Ki-67 levels are usually associated with more active cell proliferation14. Top2a plays a key role in cell division, helping to unwind the supercoiled structure of DNA and allowing chromosomes to separate. Therefore, the overexpression of Top2a may be related to excessive cell proliferation15. In summary, the genes significantly expressed by EC3 are related to cell proliferation, so we identified it as the proliferative endothelial phenotype. The GO analysis of these ECs mainly enriched in cell division, mitotic cell cycle, cell cycle regulation, DNA replication initiation, DNA template-dependent transcription, cell growth, positive regulation of telomerase maintenance of telomeres, regulation of cell population proliferation, and positive regulation of cell migration (Fig. 3G). This indicates that the genes significantly upregulated in EC3 regulate cell division and proliferation compared to other types of ECs.
EC4 also expressed pan-endothelial genes, but in addition, it expressed lymphatic endothelial cell-related genes such as Pdpn, Prox1, Lyve1, and Itga2b9. Lymphatic ECs are key components of the lymphatic vascular system, closely related to the circulatory system, responsible for transporting interstitial fluid and fat metabolic products back to the circulatory system, and also participate in the transport of immune cells and inflammatory responses. Therefore, EC4 was identified as the lymphatic endothelial phenotype.
Endothelial cell pseudotime analysis
In a comparative analysis, we observed a significant decrease in the quantity and proportion of EC1 cells, while a notable increase in the number and proportion of EC2 and EC3 cells was evident in the CMD group relative to the NC group (Fig. 4A-B). Given that EC4 cells do not originate from the same lineage as vascular ECs, we excluded EC4 and employed pseudotime analysis to identify gene expression trajectories associated with functional changes, thereby delving into the transformation relationships among EC1, EC2, and EC3. Pseudotime analysis of ECs revealed that EC1 primarily occupies the starting point of the developmental trajectory, branching into EC2 and EC3 during differentiation. EC2 and EC3 are positioned at the terminal ends of the two trajectories in the pseudotime analysis (Fig. 4C). ECs from the NC group were predominantly distributed near the starting point and branching stages of the developmental trajectory, with a small number located at the terminal end. Conversely, ECs from the CMD group were mainly distributed at the terminal ends of the two developmental trajectories, with a minority found at the starting point and branching stages (Fig. 4D). Finally, we analyzed the markers of mesenchymal phenotype ECs and proliferative phenotype ECs to observe their specific expression patterns along the pseudotime trajectory (Fig. 4E, F). The results suggest that the expression of mesenchymal phenotype EC markers such as Bgn, Fn1, Col3a1, and Timp1 gradually increases throughout the trajectory, peaking at the location of EC2 distribution. Conversely, proliferative phenotype EC markers such as HMGB2, MKI67, CENPF, and Top2a were barely expressed at the onset of the trajectory, with expression gradually increasing as the trajectory progresses, reaching its peak at the location of EC3 distribution.
(A) Proportion of the four endothelial cell types in the CMD group compared to the NC group. (B) UMAP plot of the four endothelial cell types in the CMD group versus the NC group. (C) Pseudotemporal analysis after excluding lymphatic endothelial cells. (D) Pseudotemporal analysis of endothelial cells in the CMD group versus the NC group. (E) Pseudotemporal analysis of marker genes for mesenchymal phenotype endothelial cells. (F) Pseudotemporal analysis of marker genes for proliferative phenotype endothelial cells.
Transcriptional landscape of macrophages
In this analysis of macrophages, we utilized unsupervised clustering based on the Seurat platform to identify macrophage subclusters, revealing seven distinct macrophage subclusters (Fig. 5A). The top 10 marker genes of these macrophage subclusters demonstrated clear separation between different cell types (Fig. 5B), with these seven macrophage subtypes distributed across all samples (Fig. 5C). When comparing the percentage of cells in the CMD group versus the NC group, we observed a significant increase in the overall number and proportion of macrophages in the CMD group compared to the NC group (Fig. 2D). Further subdivision showed an increase in the proportions of clusters 0, 2, 3, 4, and 6, with clusters 0 and 2 showing the most pronounced increase, while the proportions of clusters 1 and 5 decreased relatively (Fig. 5D). The UMAP plots between the two groups also illustrated the changes in different macrophage clusters (Fig. 5C). To investigate the functions of different macrophage subclusters, we conducted GSVA analysis, which revealed that inflammatory signaling pathways such as TNF signaling pathway, NF-kappa B signaling pathway, NOD-like receptor signaling pathway, IL-17 signaling pathway, Apoptosis, and Fc gamma R-mediated phagocytosis were upregulated in clusters 0, 2, and 6, while they were relatively downregulated in clusters 1, 3, and 4; pathways promoting angiogenesis and cell proliferation, such as VEGF signaling pathway, TGF-beta signaling pathway, PI3K-Akt signaling pathway, and ErbB signaling pathway, and anti-inflammatory and antioxidant pathways, such as FoxO signaling pathway and PPAR signaling pathway, were upregulated in cluster 5 (Fig. 5E).
(A) UMAP plot of macrophage clustering across all samples, revealing seven distinct clusters. (B) Heatmap of the top 10 marker genes for the seven endothelial cell clusters. (C) UMAP plot showing the distribution of seven macrophage types in the CMD group versus the NC group. (D) Bar graph illustrating the proportion of seven macrophage types in the CMD group compared to the NC group. (E) Heatmap of GSVA (Gene Set Variation Analysis) for the seven macrophage types.
Endothelial-macrophage communication network analysis
To investigate whether EndoMT influences the interactions between ECs and other cells in the heart, we constructed a cell–cell communication network based on CellChat, with a particular focus on the communication network between ECs and macrophages.
The overall analysis of intercellular communication between CMD and NC samples was similar; therefore, we concentrated on intercellular communication within CMD (Fig. 6A, B). The primary signaling pathways detected by CellChat included MIF, CCL, PTN, CXCL, SPP1, GALECTIN, MK, CSF, IGF, TNF, TGFb, GAS, OSM, and VEGF (Fig. 6C-D). Among these, MIF exhibited the highest enrichment level, followed by CCL, PTN, CXCL, and SPP1. These signaling pathways are primarily associated with inflammatory responses, immune cell recruitment, cell proliferation, differentiation, and migration.
(A) Network visualization of potential specific interactions in CMD rats, where nodes represent clusters and edges denote the number of significant ligand-receptor pairs. (B) Network visualization of potential specific interactions in NC rats, with nodes representing clusters and edges indicating the number of significant ligand-receptor pairs. (C) Ingress signaling patterns in CMD rats. (D) Egress signaling patterns in CMD rats.
Discussion
In this study, we characterized the single-cell gene expression profiles of rats with CMD. Previous research has primarily focused on how various risk factors lead to EC dysfunction through oxidative stress and endothelial nitric oxide synthase (eNOS), subsequently affecting coronary microcirculatory perfusion. However, these studies have not addressed the cellular heterogeneity. In our investigation, we analyzed 55,419 cells through single-cell RNA sequencing and identified 28 cell clusters and 10 cell subtypes, including ECs, fibroblasts, vascular smooth muscle cells, pericytes, T cells, B cells, NK cells, macrophages, monocytes, and neutrophils. In healthy rats, ECs were the most abundant, but their proportion significantly decreased in CMD rats, while fibroblasts and immune cells increased. Studies have shown that under stress conditions (such as hypoxia, hyperglycemia, and high levels of free fatty acids), reactive oxygen species and their downstream signaling pathways are activated, leading to reduced eNOS signaling, mitochondrial damage, upregulation of the NLRP3 inflammasome, accelerated degradation of NO, and decreased utilization, resulting in impaired EC functions such as proliferation, adhesion, migration, and apoptosis. This dysfunction also causes microvascular constriction and a decrease in coronary flow reserve fraction16. Our study further demonstrates that during disease progression, an increase in immune cells, enhanced inflammation and oxidative stress, and a decrease in the number of ECs affect their function.
Furthermore, due to the significant role of ECs in the development of CMD and their marked reduction, we conducted a further analysis of their gene expression characteristics. Through re-clustering, we identified 11 EC subclusters and categorized them into four EC subtypes. Our data indicate that genes related to extracellular matrix organization, endoplasmic reticulum stress response, cell adhesion, regulation of collagen fiber organization, response to oxidative stress, positive regulation of cell migration, and TGF-β receptor signaling pathways are highly enriched in EC2. Studies have shown that under pathologically pro-inflammatory conditions associated with high TGF-β levels, ECs can undergo EndoMT during adulthood, acquiring characteristics of mesenchymal cells, such as extracellular matrix remodeling, cytoskeletal rearrangement, and enhanced migratory capacity17,18. In EC3, tumor-related genes were upregulated, which are associated with biological processes such as cell division, cell cycle regulation, DNA replication initiation, cell growth, and regulation of cell population proliferation, promoting cell proliferation and migration through interactions with other cells. Studies have shown that in certain pathological states, EC proliferation may become abnormal, potentially leading to vascular stenosis or obstruction and vascular remodeling19,20. Electron microscopy revealed swollen ec nuclei, irregular narrowing of arteriolar lumens, and hypertrophy and deformation of the media smooth muscle cells21, which is consistent with our findings.
Pseudotemporal analysis suggests that during disease progression, ECs transition from a normal phenotype to a mesenchymal and proliferative phenotype. EndoMT is a biological process in which ECs lose their endothelial characteristics and acquire mesenchymal cell features. EndoMT has been reported in tumors and is believed to promote atherosclerosis22,23 and myocardial fibrosis24,25 after cardiac stress or injury. Although fibroblasts significantly increased during disease progression in our study, recent fibroblast lineage tracing analyses suggest that the overall contribution of ECs to the cardiac fibroblast population is limited26. Partial mesenchymal transformation or activation may also promote pathological vascular growth through the enhancement of pro-migratory and pro-invasive EC phenotypes27. Additionally, studies have shown that during myocardial infarction, ECs undergo transient mesenchymal activation, which is related to metabolic adaptation. After 10 days of recovery following vascular recanalization, mesenchymal transcripts return to levels detected under steady-state conditions, indicating they experience transient EndoMT rather than complete EndoMT9. Recent single-cell RNA sequencing studies have also failed to detect a large number of cells undergoing complete mesenchymal transformation after myocardial infarction28. Therefore, we hypothesize that ECs can promote mesenchymal cell transformation and abnormal proliferation through oxidative stress and cytokine action, leading to vascular remodeling and EC dysfunction. Although we have not yet studied the reversibility of the transformation of ECs to mesenchymal and proliferative phenotypes in CMD rats, we speculate that this transformation may be persistent due to the non-transient nature of myocardial ischemia in CMD rats. Further research is needed to validate this hypothesis.
Macrophages and fibroblasts have become the main drivers of angiogenic responses following myocardial ischemia29. In our research findings, a marked increase in the ratio of macrophages to fibroblasts is observed during the progression of CMD. Furthermore, correlative studies have demonstrated that macrophages play a decisive role in the vascular remodeling essential for arterial formation.30. Studies have shown that pro-inflammatory lymphocytes are mostly derived from the blood, while the myocardium primarily retains macrophages, which play a key role in lymphocyte recruitment and cardiac fibrosis8. Therefore, we conducted an in-depth analysis of macrophages. Macrophage re-clustering and GSVA results showed that inflammation-related pathways were relatively upregulated in clusters 0, 2, and 6, which also increased in proportion during CMD progression. In contrast, pathways related to neovascularization, cell proliferation, and anti-inflammatory and antioxidant responses were relatively upregulated in cluster 5, which decreased in proportion during CMD progression. As central cellular components of the human myocardium, ECs not only regulate angiogenesis and neovascularization during tissue remodeling but also modulate leukocyte infiltration to regulate local inflammation31. Therefore, we explored the communication network between ECs and macrophages. The key signaling pathways identified in our investigation encompass MIF,CCLs,PTN, and CXCLs.MIF, an important pro-inflammatory factor in atherosclerosis, has been reported to promote atherogenic leukocyte recruitment and pathological inflammation through interactions with chemokine receptors CXCR2 and CXCR432,33. PTN is a cytokine belonging to the growth factor family that affects various cell types. The PTN signaling pathway refers to a series of biological processes mediated by PTN molecules, involving cell proliferation, differentiation, migration, and survival34. The CXCL signaling pathway is a class of important cell signaling pathways primarily involved in regulating cell migration, inflammatory responses, and immune responses. Members of the CXCL family include several different chemokines that can attract immune cells, such as T cells and neutrophils, to migrate towards inflammatory areas35. The CCL signaling pathway is another important pathway in regulating cell migration, inflammatory responses, and immune responses, similar to the CXCL signaling pathway. It regulates cell behavior through the binding of chemokines to receptors on the cell surface36. In summary, we believe that the interaction between ECs and macrophages plays a crucial role. When vascular ECs are damaged, macrophages are recruited to the site of injury and participate in inflammatory responses and vascular remodeling through the release of various inflammatory and growth factors, leading to microvascular dysfunction in coronary arteries.
In conclusion, through single-cell RNA sequencing and bioinformatics analysis of rat myocardial tissue, we found that under conditions of myocardial ischemia, there is an increase in immune cells, enhanced inflammation and oxidative stress, and a decrease in the number of ECs. Coronary microvascular ECs can transform into mesenchymal cells and proliferative phenotypes. During this process, macrophages are recruited to the site of injury, promoting coronary microvascular remodeling through inflammation, immune cell recruitment, cell proliferation, migration, and differentiation, leading to luminal narrowing or obstruction.
Therefore, on one hand, we can manipulate the pathways associated with macrophages that are most closely related to endothelial cells, such as MIF, CCLs, PTN, and CXCLs. Through further experimentation, we can find common hub genes within these pathways for intervention, thereby inhibiting inflammation and oxidative stress, and reducing the recruitment of immune cells. On the other hand, we can regulate the pathways and genes related to EndoMT and cell proliferation, such as the TGF-β pathway and cancer-associated genes, to suppress or even reverse the transformation of normal endothelial cells to other phenotypes. By employing these methods to modulate inflammation and prevent vascular remodeling, we can effectively alleviate endothelial dysfunction, aiding in the identification of therapeutic targets for CMD. Additionally, by measuring changes in related genes and proteins, we can develop CMD biomarkers based on endothelial cells or macrophages, which will facilitate early detection of CMD and enable the implementation of appropriate treatment measures to improve patient prognosis.
However, this study has some limitations. First, the sample size is small, and the use of rats as the research object may not fully reflect the molecular characteristics of human CMD. Second, although single-cell RNA sequencing technology can provide gene expression information at the single-cell level, its results may be influenced by cellular heterogeneity and individual differences. Therefore, in future research, we need to expand the sample size and combine animal experiments, in vitro cell experiments, and other methods to validate our research findings and predictions, further exploring the molecular changes in CMD and the specific pathophysiological mechanisms of disease occurrence.
Conclusions
To investigate the single-cell gene expression profiles of cardiac microvascular disease (CMD) and the specific characteristics of endothelial cell gene expression, we conducted a comparative analysis using single-cell RNA sequencing on apex cardiac tissues from CMD rats and control groups. Our findings revealed a significant reduction in both the number and proportion of coronary microvascular endothelial cells in CMD rats. Additionally, we observed the occurrence of endothelial-to-mesenchymal transition and a transformation towards a proliferative endothelial phenotype, accompanied by heightened inflammatory responses and oxidative stress levels. Furthermore, there was an evident recruitment of macrophages, which interacted with the endothelial cells, leading to dysfunction of these cells—a primary cause of coronary microvascular pathology in CMD.Our research not only provides a comprehensive reference for the cellular types and interaction networks involved in the progression of CMD but also offers novel insights and theoretical foundations for understanding the disease mechanism and advancing future therapeutic strategies.
Data availability
Our data was sequenced by Lianchuang Company. Due to the involvement of these raw data in other related topics, it cannot be made public at the moment. We have uploaded the raw data to the GSA database as requested, You can download the raw data within 3 months at the following link: https://ngdc.cncb.ac.cn/gsa/s/nU9i9m0J, and the raw data will be officially released in 2 years.
Abbreviations
- CMD:
-
Coronary microcirculation dysfunction
- ECs:
-
Endothelial cells
- VSMCs:
-
Vascular smooth muscle cells
- EndoMT:
-
Endothelial-to-mesenchymal transition
- eNOS:
-
Endothelial nitric oxide synthase
References
Dean, J., Cruz, S. D., Mehta, P. K. & Merz, C. N. Coronary microvascular dysfunction: Sex-specific risk, diagnosis, and therapy. Nat. Rev. Cardiol. 12, 406–414. https://doi.org/10.1038/nrcardio.2015.72 (2015).
Chinese Society of Cardiology, C. M. A. & Editorial Board of Chinese Journal of, C. [Chinese expert consensus on diagnosis and management on patients with ischemia and non-obstructive coronary artery disease]. Zhonghua Xin Xue Guan Bing Za Zhi, 50, 1148–1160, https://doi.org/10.3760/cma.j.cn112148-20220908-00682 (2022).
Jespersen, L., Abildstrom, S. Z., Hvelplund, A. & Prescott, E. Persistent angina: Highly prevalent and associated with long-term anxiety, depression, low physical functioning, and quality of life in stable angina pectoris. Clin. Res. Cardiol. 102, 571–581. https://doi.org/10.1007/s00392-013-0568-z (2013).
Viswanathan, G. et al. Single-cell analysis reveals distinct immune and smooth muscle cell populations that contribute to chronic thromboembolic pulmonary hypertension. Am. J. Respir. Crit. Care Med. 207, 1358–1375. https://doi.org/10.1164/rccm.202203-0441OC (2023).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413. https://doi.org/10.1126/science.aan6733 (2017).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. https://doi.org/10.1038/nbt.3192 (2015).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177(7), 1888–1902-e1821. https://doi.org/10.1016/j.cell.2019.05.031 (2019).
Rao, M. et al. Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res. Cardiol. 116, 55. https://doi.org/10.1007/s00395-021-00897-1 (2021).
Tombor, L. S. et al. Single cell sequencing reveals endothelial plasticity with transient mesenchymal activation after myocardial infarction. Nat. Commun. 12, 681. https://doi.org/10.1038/s41467-021-20905-1 (2021).
Gao, X. F. et al. Single-cell RNA sequencing of the rat carotid arteries uncovers potential cellular targets of neointimal hyperplasia. Front. Cardiovasc. Med. 8, 751525. https://doi.org/10.3389/fcvm.2021.751525 (2021).
Cheng, W. H. et al. Amphiregulin induces CCN2 and fibronectin expression by TGF-beta through EGFR-dependent pathway in lung epithelial cells. Respir. Res. 23, 381. https://doi.org/10.1186/s12931-022-02285-2 (2022).
Cui, G., Cai, F., Ding, Z. & Gao, L. HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum. Pathol. 84, 133–141. https://doi.org/10.1016/j.humpath.2018.09.017 (2019).
Xu, P. et al. N6-methyladenosine modification of CENPF mRNA facilitates gastric cancer metastasis via regulating FAK nuclear export. Canc. Commun. (Lond) 43, 685–705. https://doi.org/10.1002/cac2.12443 (2023).
Uxa, S. et al. Ki-67 gene expression. Cell Death Differ. 28, 3357–3370. https://doi.org/10.1038/s41418-021-00823-x (2021).
Uuskula-Reimand, L. & Wilson, M. D. Untangling the roles of TOP2A and TOP2B in transcription and cancer. Sci. Adv. 8, eadd4920. https://doi.org/10.1126/sciadv.add4920 (2022).
Deng, J. Research progress on the molecular mechanism of coronary microvascular endothelial cell dysfunction. Int. J. Cardiol. Heart Vasc. 34, 100777. https://doi.org/10.1016/j.ijcha.2021.100777 (2021).
Fan, M. et al. Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Sci. Adv. 9, eadc9465. https://doi.org/10.1126/sciadv.adc9465 (2023).
Del Buono, M. G. et al. Coronary microvascular dysfunction across the spectrum of cardiovascular diseases: JACC state-of-the-art review. J. Am. Coll. Cardiol. 78, 1352–1371. https://doi.org/10.1016/j.jacc.2021.07.042 (2021).
Su, M. et al. Ufmylation is activated in vascular remodeling and lipopolysaccharide-induced endothelial cell injury. DNA Cell Biol. 37, 426–431. https://doi.org/10.1089/dna.2017.4073 (2018).
Kadomatsu, K. et al. Therapeutic potential of midkine in cardiovascular disease. Br. J. Pharmacol. 171, 936–944. https://doi.org/10.1111/bph.12537 (2014).
Suzuki, H., Takeyama, Y., Koba, S., Suwa, Y. & Katagiri, T. Small vessel pathology and coronary hemodynamics in patients with microvascular angina. Int. J. Cardiol. 43, 139–150. https://doi.org/10.1016/0167-5273(94)90003-5 (1994).
Chen, P. Y. et al. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J. Clin. Invest. 125, 4514–4528. https://doi.org/10.1172/JCI82719 (2015).
Evrard, S. M. et al. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat. Commun. 7, 11853. https://doi.org/10.1038/ncomms11853 (2016).
Zeisberg, E. M. et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 13, 952–961. https://doi.org/10.1038/nm1613 (2007).
Piera-Velazquez, S. & Jimenez, S. A. Endothelial to mesenchymal transition: Role in physiology and in the pathogenesis of human diseases. Physiol. Rev. 99, 1281–1324. https://doi.org/10.1152/physrev.00021.2018 (2019).
Fu, X. et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J. Clin. Invest. 128, 2127–2143. https://doi.org/10.1172/JCI98215 (2018).
Welch-Reardon, K. M., Wu, N. & Hughes, C. C. A role for partial endothelial-mesenchymal transitions in angiogenesis?. Arterioscler. Thromb. Vasc. Biol. 35, 303–308. https://doi.org/10.1161/ATVBAHA.114.303220 (2015).
Li, Z. et al. Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur. Heart J. 40, 2507–2520. https://doi.org/10.1093/eurheartj/ehz305 (2019).
Wu, X., Reboll, M. R., Korf-Klingebiel, M. & Wollert, K. C. Angiogenesis after acute myocardial infarction. Cardiovasc. Res. 117, 1257–1273. https://doi.org/10.1093/cvr/cvaa287 (2021).
Cooke, J. P. & Lai, L. Role of angiogenic transdifferentiation in vascular recovery. Front Cardiovasc. Med. 10, 1155835. https://doi.org/10.3389/fcvm.2023.1155835 (2023).
Swirski, F. K. & Nahrendorf, M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science 339, 161–166. https://doi.org/10.1126/science.1230719 (2013).
van der Vorst, E. P., Doring, Y. & Weber, C. MIF and CXCL12 in cardiovascular diseases: Functional differences and similarities. Front. Immunol. 6, 373. https://doi.org/10.3389/fimmu.2015.00373 (2015).
Lue, H., Dewor, M., Leng, L., Bucala, R. & Bernhagen, J. Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell Signal 23, 135–144. https://doi.org/10.1016/j.cellsig.2010.08.013 (2011).
Papadimitriou, E. et al. On the role of pleiotrophin and its receptors in development and angiogenesis. Int. J. Dev. Biol. 66, 115–124. https://doi.org/10.1387/ijdb.210122ep (2022).
Zhou, C., Gao, Y., Ding, P., Wu, T. & Ji, G. The role of CXCL family members in different diseases. Cell Death Discov. 9, 212. https://doi.org/10.1038/s41420-023-01524-9 (2023).
Atri, C., Guerfali, F. Z. & Laouini, D. Role of human macrophage polarization in inflammation during infectious diseases. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19061801 (2018).
Acknowledgements
We acknowledge the substantial support provided by the members of our research group during the process of animal modeling and experimental procedures. We also extend our appreciation to Hangzhou LianChuan Biotechnology Company for their technical assistance in our study.
Funding
This work was supported by grants from NSFC-82102104 (L.Y.Y), China Postdoctoral Science Foundation,2021M702054 (L.Y.Y) and Fundamental Research Program of Shanxi Province, 20210302123485 (L.Y.Y), Shanxi Province Higher Education “Billion Project” Science and Technology Guidance Project, BYJL065(L.Y.Y and Z.L).
Author information
Authors and Affiliations
Contributions
Hao Li: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. Yuping Gao: Data curation, Formal Analysis, Writing–review and editing. Yuanyuan Lin and Lin Zuo: Funding acquisition, Project administration, Supervision, Writing–review and editing. Yiding jia, Zelin Chen, Luqun Yang, Lin Ni, Yuchen Cao, Rong Fan, Zitong Yuan, Kaiyi Zhu, Zhijun Zhang, Lin Zuo, Ping Wu: Animal rearing, data recording, specimen acquisition.All authors have approved the final version of the editorial.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
The animal study was approved by the Animal Use Ethics Committee of Shanxi Medical University. The study was conducted in accordance with the local legislation and institutional requirements. The study was conducted in accordance with the ARRIVE guidelines requirements.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, H., Jia, Y., Chen, Z. et al. Bioinformatics analysis of coronary microvascular dysfunction in rats based on single-cell RNA sequencing. Sci Rep 15, 5050 (2025). https://doi.org/10.1038/s41598-025-85318-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-85318-2