Abstract
Alzheimer’s Disease (AD) is a progressive condition of a neurological brain disorder recognized by symptoms such as dementia, memory loss, alterations in behaviour, and impaired reasoning abilities. Recently, many researchers have been working to develop an effective AD recognition system using deep learning (DL) based convolutional neural network (CNN) model aiming to deploy the automatic medical image diagnosis system. The existing system is still facing difficulties in achieving satisfactory performance in terms of accuracy and efficiency because of the lack of feature ineffectiveness. This study proposes a lightweight Stacked Convolutional Neural Network with a Channel Attention Network (SCCAN) for MRI based on AD classification to overcome the challenges. In the procedure, we sequentially integrate 5 CNN modules, which form a stack CNN aiming to generate a hierarchical understanding of features through multi-level extraction, effectively reducing noise and enhancing the weight’s efficacy. This feature is then fed into a channel attention module to select the practical features based on the channel dimension, facilitating the selection of influential features. . Consequently, the model exhibits reduced parameters, making it suitable for training on smaller datasets. Addressing the class imbalance in the Kaggle MRI dataset, a balanced distribution of samples among classes is emphasized. Extensive experiments of the proposed model with the ADNI1 Complete 1Yr 1.5T, Kaggle, and OASIS-1 datasets showed 99.58%, 99.22%, and 99.70% accuracy, respectively. The proposed model’s high performance surpassed state-of-the-art (SOTA) models and proved its excellence as a significant advancement in AD classification using MRI images.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a neurodegenerative brain disease that stands out as the most prevalent form of dementia, requiring significant medical attention1,2,3. Early and accurate diagnosis of AD is crucial to initiate timely therapeutic interventions and ensure effective patient care1,4. As per research findings, in 2018, approximately 50 million people new cases of dementia were recorded annually5. According to the World Health Organization (WHO), many people will be affected by AD in 2023, and the number is projected to exceed 152 million worldwide by 2050, surpassing global cancer patient statistics. Additionally, AD ranks fifth in terms of the worldwide mortality ratio, causing the death of numerous individuals globally5,6,7. AD is characterized as a persistent neurological brain disorder that progressively damages brain cells, leading to memory loss and cognitive difficulties. Ultimately, it accelerates the decline in the ability to carry out everyday activities in real-life scenarios5,6. This state leads to the shrinkage of the human brain, affecting memory and leading to the decline of behavioral , social, and reasoning abilities. Alzheimer’s is thought to be caused by the build-up of protein fragments in the brain. This leads to the formation of plaques and tangles around nerve cells. As a result, it badly impacts the lobes and hippocampus in terms of changing shape or shrinkage or enlarged ventricles 8,9,10.
It is true that AD is a most deathful, incurable, and fatal disease that mainly causes the patient to suffer a lifetime. Consequently, it brings numerous burdens on the patient’s family, including financial, mental, and physical issues11,12. Researchers still can not find the origin of AD, and there are no effective therapies or medications to protect people from this disease. Consequently, doctors and researchers are not able to reverse the dementia of AD-affected people. However, researchers found some stages of AD disease, such as the pre-clinical stage of AD, can be defined by Mild Cognitive Impairment (MCI), recognized as the pre-clinical stage of AD. This invention signifies a transitional state between normal aging and the onset of AD6. Early AD detection and assessment of its risk and severity are crucial13,14. In contrast, many traditional diagnostic centers are available in various countries where doctors use computer-assisted or neuro-imaging systems. Researchers found some drawbacks with low-performance accuracy during the initial stage of AD. Computed tomography (CT) and Positron emission tomography (PET) scans are medical imaging techniques that provide detailed information about the internal structures and functions of the body. CT scans use X-rays to create cross-sectional images, while PET scans involve the injection of a radioactive tracer to visualize metabolic activity. These scans help diagnose and monitor various medical conditions, revolutionizing the medical diagnosis of AD6. In addition, MRI is a practical, non-invasive method for collecting valuable information about the human body. This information is continuously used in the medical diagnosis of AD, and this has become a significant recent trend in computed-aided AD diagnosis8,9.
In recent years, researchers worldwide have developed numerous ML15 and DL16 algorithms to identify and categorize AD. However, while some research has achieved the best results using DL algorithms, there is still room for improvement. A sequence of DL models has been introduced in this domain, including a hybrid CNN that incorporates slice selection and integrating histogram stretching17,18,19, respectively. Other researchers proposed a CNN architecture that incorporates skull stripping techniques20. Additionally, a CNN model that includes slicing samples for pre-processing is introduced in21. Nevertheless, these deep models primarily emphasize classification tasks, which can be attributed to the inherent black-box nature of CNNs. Several datasets are available online, and the Alzheimer’s Disease Neuroimaging Initiative dataset stands out as one of the most challenging among them. Additionally, the ADNI dataset is widely used due to its publically available and comprehensive collection of clinical, imaging, and genetic data from individuals with AD, MCI, and health control. Numerous researchers have been diligently working to develop an AD recognition system using various DL technologies, aiming to enhance the performance accuracy and efficiency of the system with the ADNI dataset.
Kamal et al. develop a CNN-SVC system to improve the performance accuracy with the Kaggle dataset22. In the same way, researchers developed DEMNET23, CNN24, Triad25, and FDCT-WR26, where they reported performance accuracy compared to the previous model with kaggle dataset. Moreover, Sharma et al. used hybrid systems by combining transfer learning and machine learning algorithms such as DenseNet201-SVM27, and they reported 91.00% accuracy with the Kaggel dataset. Fareed et al.1 employed image augmentation techniques to overcome the challenges of balancing class labels among AD disease classes. They utilized a CNN-based feature extraction and classification module, namely ADD-Net, reporting a 97.05% accuracy with the Kaggle dataset. Al-Alhadia et al.28 explored the pre-trained models such as AlexNet and ResNet50 for AD diagnosis and reported an accuracy of 94.53%. The AlexNet-ResNet5028, ADD-Net1, DEMNET23, and Bi-Vision Transformer (BiViT)29 models try to improve performance accuracy, but they still face computational complexity problems and performance accuracy. Additionally, these models require more parameters for training as compared to our model. In this scenario, it is crucial to develop an Alzheimer’s disease detection module to overcome performance accuracy and efficiency challenges. To overcome the problem, we proposed a simple lightweight stack CNN with a channel attention network (SCCAN) module where we employed a series of conventional blocks with diverse deep layers, which later integrated with the channel attention module to achieve outstanding classification results in terms of the accuracy and efficiency, particularly in the early stages of AD. Our research makes significant contributions to the field, advancing the SOTA in AD detection through the following key contributions:
-
Innovative Model Architecture: We introduced the efficacy of a Stacked Convolutional Neural Network with a Channel Attention Network (SCCAN) explicitly designed for AD detection. Our model comprises two stages of the feature extraction technique: stack CNN and channel attention module after applying the data augmentation technique. Stack CNN comprises integrated 5 CNN modules, which form a stack CNN aiming to generate a hierarchical understanding of features through multi-level extraction, effectively reducing noise and enhancing the weight’s efficacy.
-
Channel-wise Feature Extraction: We incorporated a channel attention module to address the nuances of channel-wise features from the stack CNN spatial features. This module enhances the model’s capability to extract relevant features based on channel dimensions, aligning with the intricacies of AD classification from MRI images. Finally, we employed a classification module.
-
Comparative Evaluation: We conducted an extensive comparative evaluation of the proposed SCCAN model with the ADNI1 Complete 1Yr 1.5, ADNI Kaggle, and OASIS-1 datasets. The results unequivocally demonstrate the superior performance of our proposed SCCAN approach, marking a substantial advancement in AD classification using MRI images. Our code and other materials for preprocessing, feature extraction selection, and classification are available in the GitHub link (https://github.com/najm-h/Alzheimer/.).
The organization of our paper is outlined as follows: Section 2 delves into the related work. Datasets are described in the Section 3. Section 4 presents the proposed methodology including dataset preprocessing, feature extraction procedure, and classification method. Section 5 provides detailed results, including optimal parameter values and a comparative analysis. Finally, Section 5 (5.6) and Section 6 presented the discussion and conclusion of this paper respectively.
Related Work
The accurate detection and classification of medical images pose a challenging task due to the complex nature of acquiring medical datasets30. Therefore, institutions and organizations such as the ADNI31 and the Open Access Series of Imaging Studies (OASIS)32, which provide medical datasets, implement a stringent screening process. Recently there are many researchers has been working to develop an Alzheimer’s disease detection system utilizing the ADNI31 and OASIS32 datasets. Islam et al.33 utilized the OASIS dataset, which comprised 416 3D data samples, to construct a several-layer CNN model. They assessed the accuracy of their model by comparing it with two distinct pre-trained architecture models, namely Inception V434 and ResNet34. Khan et al.35 used a twelve-layer CNN structure to integrate convolution and pooling operations with the OASIS data set in a similar manner. To solve the gradient vanishing issue, other researchers used the Leaky Rectified Linear Unit ReLU36 with MaxPooling as the activation function while staying away from ReLU37. Recently, many researchers employed pre-trained models, including MobileNetV2, VGG19, InceptionV3, and Xception, to enhance the performance accuracy and efficiency of the AD system38,39,40. Ebrahimighahnavieh et al.16 proposed a hybrid framework including ResnetV2 with InceptionV4 and residual connections to provide the skip connection35. They reported 79.12% accuracy for the OASIS dataset. Similarly, Pradhan et al.41 employed VGG19 and DenseNet169 models for comparative analysis of AD classification42, and they reported accuracy of 88% and 87%, respectively, for these two transfer learning methods for the OASIS dataset. Meanwhile, Battineni et al.43 implemented a five-layer CNN model using the OASIS-3 dataset, focusing on classifying three distinct early stages of AD.
El-aal et al.44 selected potential features among the whole DL models to reduce the computational cost and reported better accuracy. To enhance the feature selection concept of DL, some researchers used refined genetic algorithm (RGA)45 and probability binary particle swarm optimization (PBPSO)46 to select the effective features. Additionally, some existing works presented on attention mechanism-based approach47,48,49,50,51 for the MRI image diagnosis. Several recent studies incorporate attention mechanisms to improve feature extraction in AD detection models. For example, Illakiya et al.47 developed an adaptive hybrid attention network, achieving 98.53% accuracy on the ADNI dataset; however, their model suffers from high computational demands, making it less practical for real-time deployment. Attention-based methods like these are effective but require substantial resources, which limits their utility in clinical settings where efficiency is essential. Yen et al.52 developed a model based on attention mechanisms for the classification of AD and reported an accuracy of 85.24%. Besides the computational complexity, many researchers have been working to improve the performance accuracy using Deep learning scratch22,23,24,26,27. Kamal et al. proposed an AD detection system using SpinalNet with a CNN module, and they reported 97.60% accuracy for the Kaggle MRI AD dataset22. Chabib et al. utilized a cruvlet transformer-based CNN module, DeepCurvMRI, achieving 98.62% accuracy26 with the same dataset. Kim et al. employed an ensemble CNN, specifically 1D CNN, to classify AD. They combined ten 1D CNNs and reported a 98.60% accuracy and a 0.0386% loss for their model. Murugan et al.23 proposed a CNN network to build a framework for AD detection using the Kaggle dataset. Sharma et al.27 introduced a hybrid AI-based model that includes the combination of permutation-based ML and transfer learning-based voting classifier in two phases of feature extraction. Finally, they employed several machine learning algorithms for classification, reporting 91.75% accuracy and a 96.50% F1 score.
Transformer-based models like Vision Transformers (ViT) have shown promise due to their ability to handle large-scale features53,54. As evidenced by Dhinaagar et al.54 achieving 82.69% accuracy. Fareed et al.1 employed image augmentation techniques to balance class labels among AD disease classes. They utilized a CNN-based feature extraction and classification module, reporting a 97.05% accuracy with the Kaggle dataset.
However, the above-mentioned method often lacks the interpretability and efficiency of CNNs, especially when applied to smaller datasets, where overfitting and generalization issues may arise. However, the mentioned existing AD recognition systems often struggle with interpretability, efficiency, overfitting, and generalization issues, especially on smaller datasets. High computational demands and poor dataset imbalance handling further limit their accuracy and suitability for real-time use, resulting in suboptimal performance on standard datasets like ADNI1Complete 1Yr 1.5T, Kaggle, and OASIS-1. To overcome the problems, we proposed that the SCCAN model distinguishes itself from other recent SOTA methods in two key aspects: achieving an average accuracy improvement of 1.31% across three datasets and including an analysis of the FLOPs of our model to determine its efficiency. Our proposed Stacked Convolutional Neural Network with a Channel Attention Network (SCCAN) aims to enhance feature extraction through a hierarchical multi-level approach, reduce computational complexity by focusing on the most relevant features, and improve generalization and robustness. By addressing these specific drawbacks, our model demonstrates superior performance and suitability for real-time deployment, as evidenced by our experimental results on multiple datasets. As shown in Table 4, 5, 6, and Table 7, SCCAN achieves higher accuracy and reduced FLOPs, making it suitable for real-time applications. This comparison Table included most of the state-of-the-art AD recognition methodology with various parameters. Our research contributes to advancing the state-of-the-art (SOTA) in AD detection through the following key innovations:
-
Unlike traditional CNN and attention-based models, SCCAN employs a stacked CNN structure with five integrated CNN modules for multi-level feature extraction. This hierarchical design enhances feature representation while reducing noise and computational demands, evidenced by lower FLOPs compared to conventional attention mechanisms (see Table 5).
-
Our model introduces a channel attention module that refines feature selection based on channel dimensions. This approach addresses the limitations of earlier models focused solely on spatial features, increasing SCCAN’s accuracy by capturing more nuanced AD characteristics in MRI data.
-
SCCANs performance was evaluated against recent SOTA models, demonstrating an average accuracy improvement of 1.31% across the ADNI1 Complete 1-Year 1.5T, Kaggle, and OASIS datasets. This evaluation, along with detailed FLOP and accuracy metrics (see Tables, 4, 5, 6, 7), highlights SCCAN’s efficiency and suitability for real-time applications in AD detection.
AD Dataset
In this study, we used three datasets: ADNI1 Complete 1-Year 1.5T (https://adni.loni.usc.edu), Kaggle55, and OASIS-156,57 to evaluate the proposed model.
ADNI1 1Yr 1.5T
We used the ADNI1 Complete 1-Year 1.5T dataset to perform the multiclass classification. The primary objective of the ADNI is to identify biomarkers for AD to facilitate early diagnosis, improve clinical treatments, and enhance our comprehension of the pathophysiology of AD. The detailed description of the data set used contained 6400 directories, 2294 files, MRI scans, and a total number of 639 subjects labeled AD, CN, and MCI. The scan consists of subjects ranging from 50 years to 91 years, and it was collected in 57 United States and Canada. A total number of 639 subjects were divided into three categories based on the disease stages, namely AD: 133, CN: 195, and MCI: 311. We divide dataset images according to 60% training, 20% testing, and 20% validation based on the previous study1,53, the dimension of each image ( \(265\times 256\)) for the experiment.
Kaggle Dataset
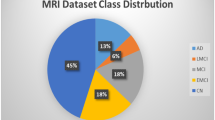
This dataset comprises MRI scan images of unidentified patients and the corresponding class label information. It is worth mentioning that this is a multiclass dataset encompassing diverse perspectives and four distinct classes. These classes consist of an average NOD category and three additional categories that represent various early stages of AD. The AD dataset has a total sample count of 6400, and samples consist of three-channel RGB images with dimensions of \(176 \times 208\) pixels, categorized into four classes. Namely mild demented (MID), moderately demented (MOD), non-demented (NOD), and very mild demented (VMD), as shown in Figure 1. The distribution of images for each class is depicted in Table 1. Notably, the data set is not balanced. To address this issue, neighbors use SMOTETOMEK techniques58,59 to create synthetic data for each underbalanced class to achieve balance with the other classes. The advantage of using the combination of SOMET and TOMEK techniques performance is the ability to mitigate knowledge loss and minimize overfitting60. According to the existing data splitting protocol1,22,23,27, we split it into training, validation, and test sets, with proportions of 60-20-20, respectively.
OASIS-1 dataset
We collected the OASIS-1 dataset from the consisting of four classes: NOD, VMD, MD, and NOD. It contains 80k MRI images. Because of the dataset balancing, the previous study used three classes: NOD, VMD, and MID61. The Table 1 depicted the distribution of images for each class. According to the previous work, we also used three classes in the study where these three classes contained a total of 9600 MRI images, the size of each image \(176 \times 208\) pixels, which are divided into equal amounts for each class. We divided it into training, validation, and test sets, with ratios of 60-20-20 respectively, based on the existing protocol1,62.
Proposed methodology
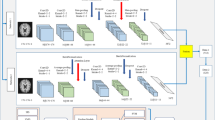
In the study, we first preprocess data and then feed it into the feature extraction and classification module. Figure 2 demonstrated the preprocessing diagram that visualizes the conversion of nii formate to RGB formate based on the axial, sagittal, and coronal axes. The proposed Stacked CNN with Channel Attention Network SCCAN modules are designed for feature extraction and classification module architecture details shown in Figures 3, 4, and 5 respectively. Figure 3 depicted more details of the proposed SCCAN model including convolutional blocks, dens blocks, and channel attention blocks. The architecture of CNN-based methods in the biomedical field is similar to the structure of the human brain. Its predominant use lies in computer vision domains such as the classification of images, segmentation of images, and identification of objects. Newer and previous machine learning and CNN-based methods make it possible to extract meaningful features directly from the dataset45. The CNN-based method shows the best performance as compared to traditional-based methods on predefined features in most computer vision and image processing. We proposed a lightweight Stacked CNN model with a Channel Attention Network SCCAN that performs well in AD classification. The proposed model, a Stacked CNN with channel attention, comprises five CNN blocks (CNNBs), each featuring a channel attention block. Each CNNB is equipped with an activation function ReLU and a 2D average pooling layer. Additionally, the model includes two dropout layers, two dense layers, and a Softmax layer for the classification task. The architecture of the proposed Stacked CNN with Channel Attention SCCAN network, detailing the structural design and flow of data, along with the model summary, which includes specific layer configurations and parameters, is comprehensively examined in Table 2. This architecture consists of four modules: the convolution blocks, channel attention, dense block, and classification module. The M and N represent the width and height of the images, which vary based on the dataset, and M=176, N=208 for the Kaggle dataset. Additional information with details of each module is succinctly provided in the following subsection.
Preprocessing
We downloaded the ADNI-1 complete 1yr 1.5T dataset from the websites (https://adni.loni.usc.edu/.) as a Neuroimaging Informatics Technology Initiative (NIfTI) file (.nii) file, which contains a total of 2294 MRI scans from 639 subjects labeled as AD:133, CN:159, and MCI: 311. In the preprocessing, we follow some of the steps in previous studies25,63,64,65, first, we convert the .nii format into MRI images (.jpg format) of each class, i.e. AD, CN, MCI, for the entire preprocessing step available in this links (https://github.com/najm-h/Alzheimer/). According to previous studies1,53 the dataset is divided in a 60-20-20 ratio for training, testing, and validations. Next, we extracted these MRI images into three views: axial, sagittal, and coronal, using med2image (https://github.com/FNNDSC/med2image). We selected 150 subjects, including those with MCI, CN, and AD (Three classes) with 50 subjects from each class. From these axial slices, only the slices with indices 60 to 120 were used in the study, under the assumption that these images cover the areas that have important features for the classification task as mentioned in the previous study64,65. After checking the quality of the MRI images, we selected 60 MRI images, resulting in a total of \(150\times 60\) = 9000 images. Figure 2 shows the preprocessing stage and three automatic planes extracted from the ADNI dataset. The brain MRI images contain a lot of information that can visualize organs in three planes: axial, sagittal, and coronal.
Feature extraction based on convolutional blocks
To enhance the spatial information in the input data, we utilized a stack of Convolutional Neural Network Blocks (CNNBs), where stack means a series of CNNBs to overcome overfitting and extract hierarchical features. Each CNNB consists of a 2D convolutional layer, an activation function, and a pooling layer, as depicted in Figure 4(a). The convolutional layer captures spatial patterns, while the activation and pooling layers ensure efficient learning and dimensionality reduction. A kernel initializer was employed to initialize the weight matrix in the convolutional layer.
The convolutional operation applied to the input is mathematically described as:
where \(F_{i,j,k}^{(n)}\) is the output feature map at position (i, j) in the n-th convolutional layer for the k-th channel, \(X_{i+p,j+q}^{(n-1)}\) is the input feature map from the previous layer, \(W_{p,q,k}^{(n)}\) is the convolutional kernel, and \(b_k^{(n)}\) is the bias term.
To introduce non-linearity and enable faster learning, we applied the ReLU activation function to the output of the convolutional layer:
Finally, to reduce the dimensionality of the feature maps while retaining the most important information, we employed an average pooling layer, defined as:
where \(P_{i,j,k}^{(n)}\) is the pooled output, and \(m \times m\) is the size of the pooling window. The convolutional layers in the early stages of the model extract low-level patterns, such as lines, edges, and curves, which form the basis for subsequent short-range dependency features. As the data moves through deeper CNNBs, high-level features are extracted, improving the model’s ability to classify images accurately. To capture more detailed and comprehensive features, we employed five CNNBs in sequence. Each block extracts multilevel compelling features that help the model adapt to variations in the data while effectively reducing noise and disregarding irrelevant variations. The hierarchical feature map after N convolutional blocks can be represented as:
where \(F^{(N)}\) is the final hierarchical feature map obtained after the N-th convolutional block which we finally defined as \(F_{StackCN}\) that fed into the channel attention module. This process ensures the extraction of both low-level and high-level spatial patterns, allowing the model to handle complex and variable input data effectively. The integration of multiple CNNBs ensures robustness in feature extraction, reducing overfitting while enhancing generalization and accuracy.
Channel wise Feature Selection
We implemented the channel attention (CA) module on the output of stacked CNN to enhance the representation of features with the channel of the CNN layers. The CA module calculates the channel-wise weight matrix and then sorts the weight matrix among the channels. After that, it selects the top weight matrix and discards the lower weight matrix contained in the channel. Our aim in using the CA module66,67,68 in the study is to select the effective feature that is also known as optimal features and suppress the less optimal features. This effective feature enhances the gen-realizability and discriminative capability of the proposed SCCAN model. In this research, the CA models enhanced the extracted feature maps by subjecting them to global average pooling, producing distinctive outputs for individual channels. Afterwards, every channel underwent a series of fully connected (FC) layers, followed by a batch normalization layer, and was associated with ReLU activation, generating either positive values or 0. The powerful feature vector emerged by multiplying the activation function’s output with the input features, enhancing the channel attention mechanism. To clarify, the CA module allocated a positive value to promising features and 0 to less impactful features. After the multiplication operation, the crucial feature was isolated from the stacked CNN feature, causing unimportant features to be transformed into zeros69. Figure 4(c) depicts the architecture of the CA employed in this study, showcasing how global average pooling received input from N channels. We utilized a dense layer with a size of N/470 and passed it through a batch normalization layer to mitigate internal covariate shift issues and avoid excessively small gradients. Following the application of the ReLU activation layer, we employed another FC layer with a size of N, succeeded by another ReLU activation. The preference for ReLU activation was based on its lower computational complexity compared to the sigmoid function. Mathematically, we can define the channel attention mechanism as below equation 5 where pass \(F_{StackCN}\) through the Channel Attention Module. The global average pooling (GAP) step is applied to aggregate spatial information across the channels, and the resulting vector is passed through two fully connected layers to compute the channel-wise weights. This process is described by:
Where:
-
\(F_{StackCN,i,j,c}\) represents the feature value at location (i, j) for channel c from \(F_{StackCN}\).
-
GAP aggregates the spatial information over the height (H) and width (W) of each channel.
-
\(W_1\) and \(W_2\) are the weights of the fully connected layers, and \(b_1\) and \(b_2\) are their biases.
-
\(\sigma\) is the sigmoid function, producing the attention score for each channel.
-
\(F^{CA}_c\) is the final output feature map after the channel attention is applied.
By multiplying the attention weights with the original feature map \(F_{StackCN,c}\), we obtain the refined feature map \(F^{CA}_c\), where important channels are emphasized, and less relevant channels are suppressed and can be defined as \(F_{CA}\) which fed into the next step classification module. This refined feature map captures the most critical information across channels, enhancing the model’s ability to discriminate between different features for classification.
Classification module
In the classification module, the flattened layer transforms the multi-dimensional feature map into a 1D array, followed by the dropout layer, which serves as a regularization method which is shown in Figure 5. Finally, the softmax layer converts the raw outputs into a probability distribution, selecting the class with the highest probability for precise class predictions. The main components are explained as follows:
Flatten layer
The flattened layer converts the feature map from the channel attention module into a one-dimensional vector. If the input feature map has dimensions \((H \times W \times C)\), the flattening process reshapes it to a vector of size \(H \times W \times C\), denoted as:
This operation ensures that the attention-weighted features are compatible with the fully connected layers, which follow the flattening.
Dense block
The dense block is shown in Figure 4 (b), which consists of a fully connected layer with ReLU. The ReLU activation function is applied to hidden layers for computational efficiency:
where \(W^{(l)}\) and \(b^{(l)}\) are the weight matrix and bias for the dense layer l, and \(\textbf{h}^{(l)}\) is the output after applying ReLU activation.
Dropout Layer
The dropout layer is introduced after the dense block of flattening to reduce the risk of overfitting by randomly turning off some neurons during training. Dropout is controlled by a probability \(p_{dropout}\) that governs the proportion of neurons to deactivate:
where \(h^{(l)}\) represents the activations of layer l, and \(\hat{h}^{(l)}\) is the result after dropout is applied. By deactivating a portion of neurons, the model becomes less sensitive to overfitting, improving its generalization performance the output of the dropout layer fed into the dense layer again.
Probabilistic map with softmax activation
Finally, the output of the dense layer is passed through the SoftMax function for multi-class classification. The SoftMax function converts the logits into probabilities for each class, enabling accurate prediction:
where \(P(y = i \mid X)\) is the probability that the input X belongs to class i, C is the number of classes, and \(W_i\) and \(b_i\) are the weights and bias for class i. The final class prediction is determined by selecting the class with the highest probability:
where \(\hat{y}\) is the predicted class label. Thus, the classification module combines dropout, flattening, and fully connected layers to generate accurate multi-class predictions while preventing overfitting and ensuring generalization.
Experimental evaluation
To evaluate the proposed SCCAN model, we utilized three Alzheimer’s benchmark datasets and included state-of-the-art comparisons. We describe the environmental setup, conduct an ablation study, and present the experimental evaluation results for each dataset in the following sections.
Hyperparameter setting and environmental setup
We implemented the system using the Python environment along with various TensorFlow modules. During hyperparameter tuning, we experimented with different configurations to optimize model performance. The initial learning rate was set to 0.01, which was determined to provide stable convergence through empirical evaluation. We tested several optimization algorithms, including Adam, RMSProp, and Stochastic Gradient Descent (SGD), and after evaluating performance, we selected SGD as the optimizer due to its superior accuracy and convergence across all three datasets. We also tuned the batch size and determined that a size of 12 was optimal for balancing computational efficiency and model accuracy. Additionally, for the classification tasks, we opted for Categorical Cross-Entropy (CCE) as the loss function, as it consistently outperformed Mean Squared Error (MSE) when paired with the SoftMax output layer. The model was trained on an “NVIDIA GeForce RTX 3090 GPU machine” running Ubuntu with NVIDIA version 12.1 and 48 GB RAM. We ensured a comprehensive assessment of model robustness by employing various metrics, including accuracy, loss, AUC, ROC curve, extended ROC curve, F1-Score, precision, and confusion matrix. These metrics provided a detailed analysis of the model’s performance, offering a holistic understanding of its effectiveness in classification tasks.
Ablation study
We performed an ablation study of the proposed model to prove the system’s superiority, as demonstrated in Table 3. We analyzed the impact of different configurations on the model’s accuracy, providing insights into the effectiveness of specific components and techniques in the proposed SCCAN model. Our objective in using a stacked CNN is to extract long-range dependency features, which reveal complex relationships among the pixels in the spatial domain, thereby enhancing the feature maps. We then apply a channel attention module to select the most significant features from these long-range dependency features. We enhanced spatial feature maps in the ablation study using 3, 4, and 5 convolutional blocks. We found that two blocks or more than five blocks decreased performance accuracy. Additionally, we highlighted the necessity of channel attention for feature selection and the impact of data balancing techniques. While the ablation study may seem trivial, our goal was to illustrate the power of stacked CNN features with and without channel attention. Our findings indicate that fewer than three convolutional blocks produce low-performance accuracy, suggesting that less complex relationships fail to accurately capture the essential features needed to represent the AD class accurately. Conversely, overly complex relationships with more than five blocks do not effectively represent the actual AD class either. In the first ablation study, we stacked 4 CNN modules, including Channel attention and balancing, which shows 98.29%. We also experimented with 3 CNN blocks, focusing on the balancing technique that generated 98.21% accuracy. Using five blocks of CNN without a channel attention module, including the data balancing technique, it produced 97.24% accuracy. Employing the number of CNN block five beside the channel attention module and data balancing, it reported 99.22% accuracy.
Experimental evaluations with kaggle dataset
We evaluated the performance of the proposed SCCAN model utilizing the overall metrics as depicted in Table 4. We compared the result of a stacked CNN with channel attention network SCCAN to the SOTA algorithms regarding the model accuracy, loss, ROC curve, extension of the ROC curve, and confusion matrix. These comparative metrics are briefly explained as follows.
Performance metrics : Accuracy, Precision, F1-Score and Confusion matrix
The experimental performance matrics and the SOTA comparison are shown in Table 4 including the existing methods, dataset name, number of images, and data splitting ratio. We observe that the proposed model achieves remarkable accuracy, precision, and F1-Score compared to all other models. Figure 8 visualizes the accuracy curves of the proposed model and SOTA models after each epoch, while Figure 10 presents the confusion matrix. Besides the proposed SCCAN model, we also experimented with recent models to make the SOTA comparison, including InceptionResNet V2, ResNet50, AlexNet28, VGG1671, Deep-ensemble72, Demnet23, CNN-SVC22, DenseNet201-SVM27, ADD-Net1, FDCT-WR26, and CNN24, in terms of accuracy as depicted in Table 4. Among all existing systems, we observed the most comparable performance accuracy in Demnet23, CNN-SVC22, DenseNet201-SVM27, ADD-Net1, FDCT-WR26, Conv-BLSTM with SMOTE73 and CNN24. Kamal et al.22 proposed an AD detection system using SpinalNet with a CNN module employing the Kaggle MRI AD dataset. They used microarray gene expression data to recognize the disease, employing several machine learning algorithms, and achieved 97.60% accuracy. Finally, they employed several machine learning algorithms for classification, reporting 91.75% accuracy and a 96.50% F1 score. Chabib et al.26 utilized a cruvlet transformer-based CNN module, namely DeepCurvMRI, achieving 98.62% accuracy. Kim et al.24 employed an ensemble CNN, specifically 1D CNN, to classify AD. They combined ten 1D CNNs and reported a 98.60% accuracy and a 0.0386% loss for their model. Murugan et al.23 proposed a CNN network to build a framework for AD detection using MRI images. They utilized the Kaggle dataset to evaluate their model and reported 95.23% accuracy and a 97% AUC. Sharma et al.27 introduced a hybrid AI-based model that combines permutation-based ML and transfer learning-based voting classifier in two phases of feature extraction. Fareed et al.1 employed image augmentation techniques to balance class labels among AD disease classes. They utilized a CNN-based feature extraction and classification module, reporting a 97.05% accuracy. Besides the existing model, our proposed model achieved 99.22% accuracy, which is high-performance accuracy and surpasses all existing models in this domain.
Loss and required model parameters
Table 5 details the number of parameters, model loss, and FLOPs of the proposed SCCAN model in comparison to SOTA models. The SCCAN model achieved a loss value of 0.0284, requiring fewer parameters (1,241,140) and FLOPs, demonstrating superior performance.. Figure 9 illustrates the loss curves of the proposed SCCAN model and the AddNet model after each epoch.
ROC, extensive ROC curves
The AUC information of the proposed model is shown in Table 4, where we observed that the proposed SCCAN model achieved the best value of 99.92% of AUC. The ROC and extensive ROC curves of the proposed model are demonstrated in Figure 6 and 7, respectively, including the SOTA Add-net1 model.
Experimental evaluations with ADNI1 Complete 1Yr 1.5T dataset
We evaluate the proposed SCCAN model using the “ADNI1 Complete 1Yr 1.5T” dataset and compare its performance with existing models, as shown in Table 6. The Table includes performance metrics along with details such as reference number, type of input, number of MRIs, number of images, dataset splitting, and methods, where reported. The proposed model is more comparable with recent models such as VGGNet1664, DCNN75, VGGNet1676, CNN-VGG1665, Deep ensemble72 TriAd25 and ViT53. Gunawardena et al.76 developed two methods: SVM and CNN base utilized the slices-based MRI image involving the preprocessing steps. First, they converted the 3D MRI to 2D Images and selected 1615 coronal plane images. Further they randomly 1292 images for training and 323 for testing. Billones et al.64 used different tools for the reconstruction process and obtained coronal image slices with size \(256\times 256\) in the PNG format. They selected the slice indices 111 to 130 due to the assumption that these slices cover more informative areas for feature extraction. They used the CNN with VGGnet16 for the classification of AD, CN, and MCI and reported an accuracy of 91.85%. Jain et al.65 selected the ADNI dataset of 150 subjects, including 50 AD, 50 CN, and 50 MCI, for the classification task. Each brain MRI is in NIfTI format, and the NIfTI images are volumetric (3D) images of size \(256\times 256\times 256\) after preprocessing. These images contain 2D images called MRI slices. They selected 4800 slices (150 subjects \(\times\) 32 slices, which consist of 1600 AD, 1600 NC, and 1600 MCI slices). They used a transfer learning-based approach, including a CNN architecture and the VGG16-trained model for feature extraction, and reported an accuracy of 95.18%. Kundram et al. proposed a DL-based method using the ADNI-1 dataset for classification subjects with AD, MCI, and NC; they reported a good accuracy of 98%75. Mercaldo et al.25 extracted 6768 MRI images from the same ADNI dataset and developed a DL-based model, TriAD, consisting of two CNN blocks called Tblocks for AD detection. They reported an accuracy of 95.18%. Alp et al.53 proposed a vision transformer (ViT) based model to extract features from the ADNI1 complete 1Yr 1.5T dataset and utilize a time series transformer to classify features. They achieve an accuracy of 91.24%, though at the cost of computational efficiency. Similarly, Liu et al.74 proposed an attention-based CNN mechanism to improper the feature presentation capability. They performed segmentation tasks using ADNI MRI data to obtain gray matter and white matter. They performed binary classification tasks and reported an accuracy of 95.37% AD vs MCI. Abdulazeem et al.77 proposed a CNN-based model for AD classification using 211,655 images extracted from the ADNI dataset and reported an accuracy of 97.50% for multi-class classification. Similarly, Savas et al.63 used the same analysis of the preprocess steps of our study. First, they converted the .nii format to .png with Python code and exported 166 frames from each nii image, which were selected as the middle two images. They select 4364 slices of images and split them in the 80:10:10 training testing and validation ratio, respectively. Next, they suggest 29 pre-trained models as a comparison and determine that EfficientNetB3 achieves a high accuracy of 97.28%. The main difference between their method of exploring the pre-trained model and our proposed model is a novel approach. In summary, we follow the same analysis of preprocessing steps in previous studies such as25,63,64,65 the proposed SCCAN model achieved the best performance accuracy of 99.58%, a loss of 0.0174, a precision of 99.58%, a recall of 99.58%, and an F1 score of 99.66%, which demonstrates that outperforming than the existing methods. Figures 11 and 12(a) depict the proposed model’s accuracy, loss, and ROC curves, respectively, demonstrating that our model performs well during training and validation. Figure 12(b) shows the confusion matrix of the SCCAN model.
Experimental evaluations with OASIS-1 dataset
In Table 7, we present the numerical results regarding performance accuracy for AD classification on the OASIS-1 dataset with existing models. The existing models Mohammad et al. proposed a hybrid model that consists of (AlexNet+SVM and ResNet-50+SVM) for AD classification and reported an accuracy of 94.80%78. Kabir et al.79 presented the DL-based approach with 18-layer architecture using the same dataset for multi-classification and reported an accuracy of 92.00% through the cost computational. Most of the comparable accuracy results in the existing methods we investigated are related to our study62,72. Loddo et al.72 presented a deep ensemble strategy using the OASIS dataset with slices based on AD diagnosis and reported an accuracy of 98.51%. Almufareh et al.62 employed attention-based techniques with the Vision Transformer (ViT) approach for AD detection using the same OASIS dataset and reported an accuracy of 99.06%. Besides this, our proposed SCCAN model achieved an accuracy of 99.70%, demonstrating a notable improvement over existing methods. Figure 13 shows the accuracy and loss curves for the proposed model. The curves indicate that our model performs well on this dataset, with consistent accuracy and loss metrics for both the training and testing phases.
Discussion
Our contribution lies in the novel combination and adaptation of these methodologies to address the specific challenges of AD classification using MRI images. Our approach, SCCAN, integrates multiple CNN modules in a stacked architecture, enhancing feature extraction and reducing noise in MRI images. Our stacked CNN approach provides comprehensive multi-level feature extraction, surpassing traditional single CNN architectures in capturing complex patterns in medical images. The channel attention mechanism in our model prioritizes important features, achieving higher accuracy, especially with limited training data. Our model achieves superior performance with accuracies of 99.58%, 99.22%, and 99.70% on the ADNI-1, Kaggle, and OASIS-1 datasets, respectively, demonstrating robustness and generalizability across different datasets. This performance accuracy proved the effectiveness of our approach in advancing the state-of-the-art in this domain. We compared the previous model with our proposed model, including DL-based and channel attention-based in terms of the performance accuracy demonstrated in Table 4, 6 and 7. We recognize the need for a more detailed error analysis and have provided insights based on confusion matrices for multi-class classification tasks as depicted in Figures 10 and 12(a). Most errors occurred between closely related stages, such as VMD and NOD in the four-class problem and NC and MCI in the three-class problem. Despite these minimal error rates (less than 1%), further feature refinement and data augmentation could enhance the model’s performance. Overall, the model’s strong handling of false positives and negatives confirms its reliability for real-world diagnostic applications. The computational complexity of the proposed SCCAN model is addressed through a detailed comparison with SOTA as shown 5. Besides the reduced parameter count, we evaluated the model regarding FLOPs and model loss, which are critical indicators of computational efficiency. This significant reduction in parameters and FLOPs indicates that our model is computationally more efficient while maintaining competitive accuracy. These reductions are essential for clinical deployment, where real-time performance and resource efficiency are necessary. Lower-value FLOPs ensure faster inference times and reduced computational resource needs, making our model suitable for deployment in environments with limited hardware, such as portable medical devices or systems with limited computational power. Our proposed model’s architecture, combining stacked CNN modules with a channel attention mechanism, is highly versatile and can be applied to various fields beyond Alzheimer’s Disease classification. Cancer Detection, Cardiovascular Disease Diagnosis, Anomaly Detection, Facial Recognition.
Conclusion
In the study, we proposed the SCCAN model as a groundbreaking solution for AD classification through Magnetic Resonance Imaging (MRI). We introduce several advancements aligning the deep learning technique with the contemporary needs of medical image diagnosis. Its distinctive feature extraction mechanism, leveraging a hierarchical approach through multi-level extraction, contributes to a reduced parameter model tailored for efficient training on smaller datasets. Moreover, the SCCAN model enhances interpretability by leveraging channel attention mechanisms for effective feature selection.. The high-performance accuracy with extensive experiments proved that our proposed SCCAN model achieved the goal by achieving better performance compared to the existing SOTA models. For future work, we plan to collect more data, explore the integration of advanced transfer learning techniques, and investigate the SCCAN model’s performance across diverse datasets, which could provide valuable insights and further enhance its applicability in broader medical imaging contexts.
Data availability
Datasets are available online for academic research that can be available in the below link. ADNI1 Complete 1Yr 1.5T dataset: (https://adni.loni.usc.edu) Kaggle Dataset55 OASIS-1 dataset56,57.
References
Fareed, M. M. S. et al. Add-net: an effective deep learning model for early detection of alzheimer’s disease in mri scans. IEEE Access 10, 96930–96951 (2022).
Wang, H. et al. Mst1 mediates neuronal loss and cognitive deficits: A novel therapeutic target for alzheimer’s disease. Progress in Neurobiology 214, 102280 (2022).
Li, H. et al. Untargeted metabolomics analysis of the hippocampus and cerebral cortex identified the neuroprotective mechanisms of bushen tiansui formula in an a\(\beta\)25-35-induced rat model of alzheimer’s disease. Frontiers in Pharmacology 13, 990307 (2022).
Thaver, A. & Ahmad, A. Economic perspective of dementia care in pakistan. Neurology 90, e993–e994 (2018).
Przedborski, S. & Vila, M. et v. jackson-lewis,“neurodegeneration: What is it and where are we?”. J. Clin. Invest 111, 3–10 (2003).
Tufail, A. B., Ma, Y.-K. & Zhang, Q.-N. Binary classification of alzheimer’s disease using smri imaging modality and deep learning. Journal of digital imaging 33, 1073–1090 (2020).
Nichols, E. et al. Global, regional, and national burden of alzheimer’s disease and other dementias, 1990–2016: a systematic analysis for the global burden of disease study 2016. The Lancet Neurology 18, 88–106 (2019).
Doi, K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Computerized medical imaging and graphics 31, 198–211 (2007).
Tiwari, S., Atluri, V., Kaushik, A., Yndart, A. & Nair, M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. International journal of nanomedicine 5541–5554 (2019).
Duyckaerts, C., Delatour, B. & Potier, M.-C. Classification and basic pathology of alzheimer’s disease. Acta neuropathologica 118, 5–36 (2009).
de la Torre, J. C. Alzheimer’s disease is incurable but preventable. J. Alzheimer’s Disease 20, 861–870 (2010).
Casey, D. A., Antimisiaris, D. & O’Brien, J. Drugs for alzheimer’s disease: are they effective?. Pharmacy and Therapeutics 35, 208 (2010).
Prince, M. Dementia u.k.: Overview. Tech. Rep. (2014).
Nichols, E. & Vos, T. The estimation of the global prevalence of dementia from 1990–2019 and forecasted prevalence through 2050: an analysis for the global burden of disease (gbd) study 2019. Alzheimer’s & Dementia 17, e051496 (2021).
Tanveer, M. et al. Machine learning techniques for the diagnosis of alzheimer’s disease: A review. ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM) 16, 1–35 (2020).
Ebrahimighahnavieh, M. A., Luo, S. & Chiong, R. Deep learning to detect alzheimer’s disease from neuroimaging: A systematic literature review. Computer methods and programs in biomedicine 187, 105242 (2020).
Zhuang, J., Cai, J., Wang, R., Zhang, J. & Zheng, W.-S. Deep knn for medical image classification. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part I 23, 127–136 (Springer, 2020).
Suganthe, R. et al. Multiclass classification of alzheimer’s disease using hybrid deep convolutional neural network. NVEO-NATURAL VOLATILES & ESSENTIAL OILS Journal| NVEO 145–153 (2021).
Jiang, X., Chang, L. & Zhang, Y.-D. Classification of alzheimer’s disease via eight-layer convolutional neural network with batch normalization and dropout techniques. Journal of Medical Imaging and Health Informatics 10, 1040–1048 (2020).
Kalavathi, P. & Prasath, V. S. Methods on skull stripping of mri head scan images-a review. Journal of digital imaging 29, 365–379 (2016).
Basheera, S. & Ram, M. S. S. A novel cnn based alzheimer’s disease classification using hybrid enhanced ica segmented gray matter of mri. Computerized Medical Imaging and Graphics 81, 101713 (2020).
Kamal, M. S. et al. Alzheimer’s patient analysis using image and gene expression data and explainable-ai to present associated genes. IEEE Transactions on Instrumentation and Measurement 70, 1–7 (2021).
Murugan, S. et al. Demnet: a deep learning model for early diagnosis of alzheimer diseases and dementia from mr images. Ieee Access 9, 90319–90329 (2021).
Kim, C.-M. & Lee, W. Classification of alzheimer’s disease using ensemble convolutional neural network with lfa algorithm. IEEE Access (2023).
Mercaldo, F. et al. Triad: A deep ensemble network for alzheimer classification and localisation. IEEE Access (2023).
Chabib, C., Hadjileontiadis, L. J. & Al Shehhi, A. Deepcurvmri: Deep convolutional curvelet transform-based mri approach for early detection of alzheimer’s disease. IEEE Access (2023).
Sharma, S. et al. Htlml: Hybrid ai based model for detection of alzheimer’s disease. Diagnostics 12, 1833 (2022).
Al-Adhaileh, M. H. Diagnosis and classification of alzheimer’s disease by using a convolution neural network algorithm. Soft Computing 26, 7751–7762 (2022).
Shah, S. M. A. H. et al. Computer-aided diagnosis of alzheimer’s disease and neurocognitive disorders with multimodal bi-vision transformer (bivit). Pattern Analysis and Applications 27, 76 (2024).
Alhassan, A. M., Initiative, A. D. N., Biomarkers, A. I. & of Ageing, L. F. S. Enhanced fuzzy elephant herding optimization-based otsu segmentation and deep learning for alzheimer’s disease diagnosis. Mathematics 10, 1259 (2022).
Weiner, M. W. et al. The alzheimer’s disease neuroimaging initiative: a review of papers published since its inception. Alzheimer’s & Dementia 9, e111–e194 (2013).
Marcus, D. & Wang, T. Oasis: Cross-sectional, mri data in young, middle-aged, nondemented, and demented, older adults. J. Cogn. Neurosci. To be published.
Islam, J. & Zhang, Y. Early diagnosis of alzheimer’s disease: A neuroimaging study with deep learning architectures. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops, 1881–1883 (2018).
Szegedy, C., Ioffe, S., Vanhoucke, V. & Alemi, A. A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proc. 31st AAAI Conf. Artif. Intell., 1–7 (2017).
Khan, S. H., Hayat, M., Bennamoun, M., Sohel, F. A. & Togneri, R. Cost-sensitive learning of deep feature representations from imbalanced data. IEEE Trans. Neural Netw. Learn. Syst. 29, 3573–3587 (2017).
Xu, B., Wang, N., Chen, T. & Li, M. Empirical evaluation of rectified activations in convolutional network. arXiv preprint [SPACE]arXiv:1505.00853 (2015).
Agarap, A. F. Deep learning using rectified linear units (relu). arXiv preprint[SPACE]arXiv:1803.08375 (2018).
Lau, M. M. & Lim, K. H. Review of adaptive activation function in deep neural network. In 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), 686–690 (IEEE, 2018).
Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1251–1258 (2017).
Howard, A. G. et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint[SPACE]arXiv:1704.04861 (2017).
Pradhan, A., Gige, J. & Eliazer, M. Detection of alzheimer’s disease (ad) in mri images using deep learning. Int. J. Eng. Res. Technol. (IJERT) 10, 580–585 (2021).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 4700–4708 (2017).
Battineni, G., Chintalapudi, N., Amenta, F. & Traini, E. Deep learning type convolution neural network architecture for multiclass classification of alzheimer’s disease. In Bioimaging, 209–215 (2021).
Shereen A El-Aal, N. I. G. A proposed recognition system for alzheimer’s disease based on deep learning and optimization algorithms. Journal of Southwest Jiaotong University 56 (2021).
Wen, F. & David, A. K. A genetic algorithm-based method for bidding strategy coordination in energy and spinning reserve markets. Artif. Intell. Eng. 15, 71–79 (2001).
Wang, L., Wang, X., Fu, J. & Zhen, L. A novel probability binary particle swarm optimization algorithm and its application. J. Softw. 3, 28–35 (2008).
Illakiya, T., Ramamurthy, K., Siddharth, M., Mishra, R. & Udainiya, A. Ahanet: Adaptive hybrid attention network for alzheimer’s disease classification using brain magnetic resonance imaging. Bioengineering 10, 714 (2023).
Dutta, T. K., Nayak, D. R. & Zhang, Y.-D. Arm-net: Attention-guided residual multiscale cnn for multiclass brain tumor classification using mr images. Biomedical Signal Processing and Control 87, 105421 (2024).
Guo, M.-H. et al. Attention mechanisms in computer vision: A survey. Computational visual media 8, 331–368 (2022).
Dutta, T. K. & Nayak, D. R. Cdanet: Channel split dual attention based cnn for brain tumor classification in mr images. In 2022 IEEE international conference on image processing (ICIP), 4208–4212 (IEEE, 2022).
Woo, S., Park, J., Lee, J.-Y. & Kweon, I. S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on computer vision (ECCV), 3–19 (2018).
Chen, Y. An alzheimer’s disease identification and classification model based on the convolutional neural network with attention mechanisms. Traitement du Signal 38 (2021).
Alp, S. et al. Joint transformer architecture in brain 3d mri classification: its application in alzheimer’s disease classification. Scientific Reports 14, 8996 (2024).
Dhinagar, N. J., Thomopoulos, S. I., Laltoo, E. & Thompson, P. M. Efficiently training vision transformers on structural mri scans for alzheimer’s disease detection. In 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 1–6 (IEEE, 2023).
Dubey, S. Alzheimer’s dataset: A dataset of 4 classes of images. https://www.kaggle.com/datasets/tourist55/alzheimers-dataset-4-class-of-images (2019). Kaggle.
Marcus, D. S. et al. Open access series of imaging studies (oasis): cross-sectional mri data in young, middle aged, nondemented, and demented older adults. Journal of cognitive neuroscience 19, 1498–1507 (2007).
D. Marcus, J. C. e. a., R. Buckner. Oasis-1: Cross-sectional: Principal investigators, morris; p50 ag05681, p01 ag03991, p01 ag026276, r01 ag021910, p20 mh071616, u24 rr021382. https://www.kaggle.com/datasets/ninadaithal/imagesoasis/data. Kaggle.
Mansourifar, H. & Shi, W. Deep synthetic minority over-sampling technique. arXiv preprint[SPACE]arXiv:2003.09788 (2020).
Pan, T., Zhao, J., Wu, W. & Yang, J. Learning imbalanced datasets based on smote and gaussian distribution. Information Sciences 512, 1214–1233 (2020).
El-Assy, A., Amer, H. M., Ibrahim, H. & Mohamed, M. A novel cnn architecture for accurate early detection and classification of alzheimer’s disease using mri data. Scientific Reports 14, 3463 (2024).
Paleczny, A., Parab, S. & Zhang, M. Enhancing automated and early detection of alzheimer’s disease using out-of-distribution detection. arXiv preprint[SPACE]arXiv:2309.01312 (2023).
Almufareh, M. F., Tehsin, S., Humayun, M. & Kausar, S. Artificial cognition for detection of mental disability: a vision transformer approach for alzheimer’s disease. In Healthcare, vol. 11, 2763 (MDPI, 2023).
Savaş, S. Detecting the stages of alzheimer’s disease with pre-trained deep learning architectures. Arabian Journal for Science and Engineering 47, 2201–2218 (2022).
Billones, C. D., Demetria, O. J. L. D., Hostallero, D. E. D. & Naval, P. C. Demnet: a convolutional neural network for the detection of alzheimer’s disease and mild cognitive impairment. In 2016 IEEE region 10 conference (TENCON), 3724–3727 (IEEE, 2016).
Jain, R., Jain, N., Aggarwal, A. & Hemanth, D. J. Convolutional neural network based alzheimer’s disease classification from magnetic resonance brain images. Cognitive Systems Research 57, 147–159 (2019).
M., A. S. M., H., M. A. M., N., S. & S., J. Sign language recognition using graph and general deep neural network based on large scale dataset. IEEE Access 12, 34553–34569 (2024).
Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks. In 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 7132–7141, https://doi.org/10.1109/CVPR.2018.00745 (2018).
Zhang, Y. e. a. Image super-resolution using very deep residual channel attention networks. In Ferrari, V. e. a. (ed.) Computer Vision – ECCV 2018, 294–310 (Springer International Publishing, Cham, 2018).
Li, H. et al. Scattnet: Semantic segmentation network with spatial and channel attention mechanism for high-resolution remote sensing images. IEEE Geoscience and Remote Sensing Letters 18, 905–909. https://doi.org/10.1109/LGRS.2020.2988294 (2021).
Egawa, R., Miah, A. S. M., Hirooka, K., Tomioka, Y. & Shin, J. Dynamic fall detection using graph-based spatial temporal convolution and attention network. Electronics 12 (2023).
Mggdadi, E., Al-Aiad, A., Al-Ayyad, M. S. & Darabseh, A. Prediction alzheimer’s disease from mri images using deep learning. In 2021 12th International Conference on Information and Communication Systems (ICICS), 120–125, https://doi.org/10.1109/ICICS52457.2021.9464543 (2021).
Loddo, A., Buttau, S. & Di Ruberto, C. Deep learning based pipelines for alzheimer’s disease diagnosis: a comparative study and a novel deep-ensemble method. Computers in biology and medicine 141, 105032 (2022).
Stamate, D., Smith, R., Tsygancov, R. & Vorobev. Applying deep learning to predicting dementia and mild cognitive impairment. In Artificial Intelligence Applications and Innovations: 16th IFIP WG 12.5 International Conference, AIAI 2020, Neos Marmaras, Greece, June 5–7, 2020, Proceedings, Part II 16, 308–319 (Springer, 2020).
Liu, Z. et al. Diagnosis of alzheimer’s disease via an attention-based multi-scale convolutional neural network. Knowledge-Based Systems 238, 107942 (2022).
Kundaram, S. S. & Pathak, K. C. Deep learning-based alzheimer disease detection. In Proceedings of the Fourth International Conference on Microelectronics, Computing and Communication Systems: MCCS 2019, 587–597 (Springer, 2021).
Gunawardena, K., Rajapakse, R. & Kodikara, N. D. Applying convolutional neural networks for pre-detection of alzheimer’s disease from structural mri data. In 2017 24th international conference on mechatronics and machine vision in practice (M2VIP), 1–7 (IEEE, 2017).
AbdulAzeem, Y., Bahgat, W. M. & Badawy, M. A cnn based framework for classification of alzheimer’s disease. Neural Computing and Applications 33, 10415–10428 (2021).
Mohammed, B. A. et al. Multi-method analysis of medical records and mri images for early diagnosis of dementia and alzheimer’s disease based on deep learning and hybrid methods. Electronics 10, 2860 (2021).
Kabir, A. et al. Multi-classification based alzheimer’s disease detection with comparative analysis from brain mri scans using deep learning. In TENCON 2021-2021 IEEE Region 10 Conference (TENCON), 905–910 (IEEE, 2021).
Funding
This work was supported by the Competitive Research Fund of The University of Aizu, Japan.
Author information
Authors and Affiliations
Contributions
All the authors have contributed equally to this article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hassan, N., Miah, A.S.M., Suzuki, K. et al. Stacked CNN-based multichannel attention networks for Alzheimer disease detection. Sci Rep 15, 5815 (2025). https://doi.org/10.1038/s41598-025-85703-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-85703-x
Keywords
This article is cited by
-
A token efficient vision framework using patch residual transformer for Alzheimer’s disease diagnosis
Scientific Reports (2026)
-
Enhancing medical image classification via ConvKC-ViT: a hybrid vision transformer model integrating convolutional insights and K-class mean clustering
The Visual Computer (2026)
-
Hybrid stacking of Squeeze Net features and ML models for accurate Alzheimer’s diagnosis
Discover Artificial Intelligence (2026)
-
Deep ensemble learning with transformer models for enhanced Alzheimer’s disease detection
Scientific Reports (2025)
-
Developing a QSPR model for Alzheimer’s drugs using topological indices and M-polynomial: A computational study
Scientific Reports (2025)