Abstract
Brain tumors present a significant global health challenge, and their early detection and accurate classification are crucial for effective treatment strategies. This study presents a novel approach combining a lightweight parallel depthwise separable convolutional neural network (PDSCNN) and a hybrid ridge regression extreme learning machine (RRELM) for accurately classifying four types of brain tumors (glioma, meningioma, no tumor, and pituitary) based on MRI images. The proposed approach enhances the visibility and clarity of tumor features in MRI images by employing contrast-limited adaptive histogram equalization (CLAHE). A lightweight PDSCNN is then employed to extract relevant tumor-specific patterns while minimizing computational complexity. A hybrid RRELM model is proposed, enhancing the traditional ELM for improved classification performance. The proposed framework is compared with various state-of-the-art models in terms of classification accuracy, model parameters, and layer sizes. The proposed framework achieved remarkable average precision, recall, and accuracy values of 99.35%, 99.30%, and 99.22%, respectively, through five-fold cross-validation. The PDSCNN-RRELM outperformed the extreme learning machine model with pseudoinverse (PELM) and exhibited superior performance. The introduction of ridge regression in the ELM framework led to significant enhancements in classification performance model parameters and layer sizes compared to those of the state-of-the-art models. Additionally, the interpretability of the framework was demonstrated using Shapley Additive Explanations (SHAP), providing insights into the decision-making process and increasing confidence in real-world diagnosis.
Similar content being viewed by others
Introduction
The human brain, the seat of consciousness and cognition, is the pinnacle of our intricate biological machinery. This extraordinary organ orchestrates our actions, memories, thoughts, and even the most delicate movements of our limbs under its commanding presence1,2. However, amidst its enigmatic complexity, the brain is susceptible to formidable adversaries known as brain tumors, which arise from the unusual growth of its own cells. This malignant growth ravaged the delicate neural architecture, manifesting as the challenging spectrum of brain cancer—a silent epidemic that leaves no corner of the globe untouched.
Unsettling statistics from the World Health Organization (WHO) show that cancer claimed a staggering 9.6 million lives worldwide in 2018. Owing to this challenge, brain tumors have emerged as a significant contributor3. These tumors are broadly classified into primary and secondary metastatic tumors, and these afflictions present distinct diagnostic and treatment challenges4. Primary brain tumors originate within the confines of the brain, whereas secondary metastatic tumors stealthily infiltrate the cerebral terrain via the bloodstream from other regions of the body. The WHO further divides brain tumors into four categories, which are classified according to their malignant or benign nature, covering the spectrum from insidious growth to aggressive malignancies5,6,7,8.
To confront this formidable focus, early detection and accurate classification of brain tumors are paramount, serving as the cornerstone for devising effective treatment strategies. In this quest, magnetic resonance imaging (MRI) and computer tomography (CT) have emerged as indispensable tools, providing critical insights into the anatomical landscape of these enigmatic masses9. The difficulty, however, lies in precisely separating normal brain tissue from abnormal growth, a task made more difficult by the inherent variability in size, shape, and location of these tumors.
Harnessing the transformative power of machine learning, the realm of medical image processing has experienced a remarkable surge in modernistic computer-aided diagnosis systems aimed at augmenting the expertise of radiologists10,11,12,13,14. By synergizing the advantages of advanced machine and deep learning algorithms and the wealth of information in biomedical images, these innovative systems hold immense promise in enhancing patient care and optimizing brain tumor classification. Accurate early-stage classification of tumors can guide clinicians in formulating personalized treatment plans, ushering in the era of precision medicine8,15,16,17,18.
Among the myriad types of primary brain tumors, three prominent adversaries are in the central stage: gliomas, pituitary tumors, and meningiomas. Gliomas, stemming from glial cells that provide crucial support to neurons, pose a significant challenge due to their infiltrative nature. Pituitary tumors, which are benign in nature, germinate within the pituitary glands and disrupt the delicate hormonal balance vital to bodily homeostasis. Meningiomas, on the other hand, thrive on the protective membranes enveloping the brain and spinal cord, demanding careful scrutiny.
In this context, the formidable task of accurate brain tumor classification necessitates a comprehensive exploration of cutting-edge machine and deep learning techniques and their integration into the domain of biomedical image processing. In addition to merely assisting radiologists in their diagnostic endeavors, these innovative approaches have the potential to revolutionize patient outcomes, mitigating the grim consequences associated with high-grade tumors and bestowing renewed hope and extended lives. Thus, the pursuit of a precise brain tumor diagnosis represents a pivotal milestone in the arduous journey toward effective treatment. To achieve this objective, significant attention has been focused on identifying and classifying brain tumor MRI images. This study introduces a novel approach that combines convolutional neural networks (CNNs) and extreme learning machines (ELMs) to diagnose brain tumors. The extraction of discriminant features from MRI images using a CNN enables the ELM to classify brain tumors precisely. This synergistic fusion of advanced ML techniques holds immense promise for transforming the landscape of brain tumor diagnosis, revolutionizing disease classification, and ultimately reducing patient mortality rates. This paper presents a set of significant contributions that advance the field of brain tumor classification and enhance the interpretability of deep learning models. The key contributions of this study can be summarized as follows:
-
Employing CLAHE improved the visibility and clarity of important tumor features in the MRI images, thereby enhancing the subsequent analysis and classification tasks.
-
A lightweight parallel depthwise separable CNN (PDSCNN) is proposed to extract relevant features from enhanced MRI images. This model efficiently captures tumor-specific patterns while minimizing computational complexity.
-
A hybrid ridge regression ELM (RRELM) model is proposed, which enhances the traditional ELM by replacing the pseudoinverse with ridge regression for improved classification performance.
-
This study compared the proposed framework with various state-of-the-art (SOTA) models based on classification performance, model parameters, and layer sizes.
-
The interpretability of the framework is showcased by employing SHAP, which allows a deeper understanding of the model’s decision making, increasing confidence in its real-world diagnostic application.
Literature review
The precise segmentation of medical images plays a pivotal role in detecting and classifying brain tumors, aiding timely and informed therapeutic decisions. Magnetic resonance imaging (MRI) has emerged as a cornerstone in this endeavor, providing detailed anatomical information for accurate analysis19,20,21,22,23. Over the years, numerous techniques have been proposed to address the challenging task of brain tumor classification in MRI. Ahmad et al.24 proposed a three-step preprocessing technique to enhance MRI image quality, accompanied by a custom CNN architecture. The architecture utilizes batch normalization for rapid training, higher learning rates, and simplified weight initialization. With a compact design comprising few convolutional and max pooling layers, the proposed model achieved exceptional accuracy rates of 98.22% overall, 99% for glioma, 99.13% for meningioma, 97.3% for the pituitary, and 97.14% for normal images. The experimental results demonstrate the robustness and improved detection accuracy of the proposed architecture compared to those of different transfer learning models, such as VGG16 and VGG1925, across various brain diseases within a short timeframe. Similarly, Nayak et al.26 introduced a dense CNN-based variant of EfficientNet with min–max normalization to classify 3,260 brain MRI images into four categories (glioma, meningioma, pituitary, and no tumor). The model exhibited high accuracy in categorizing the limited database by incorporating dense and drop-out layers. Data augmentation and min–max normalization were combined to enhance tumor cell contrast. The experimental results demonstrated exceptional performance, with a training accuracy of 99.97% and a testing accuracy of 98.78%.
In addition, Alanazi et al.27 introduced a novel transfer deep learning model for the early diagnosis of brain tumors and their subclasses. Initially, isolated CNN models were built and evaluated for brain MRI images. The 22-layer, binary-classification CNN model was then reutilized with adjusted weights to classify tumor subclasses via transfer learning. The transfer-learned model achieved a high accuracy of 95.75% for MRI images from the same machine and demonstrated adaptability and reliability with 96.89% accuracy on an unseen dataset from another machine. The proposed framework showed potential for assisting doctors and radiologists in early brain tumor diagnosis. Khan et al.28 introduced a hierarchical deep learning-based brain tumor (HDL2BT) classification system using a CNN. The system accurately categorizes glioma, meningioma, pituitary, and nontumor types from MRI images with 92.13% precision and a 7.87% miss rate. The proposed model outperformed previous brain tumor detection and segmentation methods, providing valuable clinical assistance in diagnosis. In contrast, Irmak’s study aimed to enable the early diagnosis of brain tumors from MR images through multiclassification using a custom CNN29. Three distinct CNN models are proposed for different classification tasks. The hyperparameters of the CNN models are automatically optimized via a grid search algorithm. The first model achieved 99.33% accuracy in brain tumor detection. The second model accurately classified brain tumors into five types (normal, glioma, meningioma, pituitary, and metastatic), with 92.66% accuracy. The third model successfully classified brain tumors into three grades (Grade II, Grade III, and Grade IV) with 98.14% accuracy. A comparative analysis with other state-of-the-art (SOTA) CNN models (AlexNet, Inceptionv3, ResNet-50, VGG-16, and GoogleNet) demonstrated the superior performance of the proposed CNN models.
Badza et al.30 proposed a simplified CNN architecture for brain tumor classification that explicitly targets three tumor types (glioma, meningioma, and pituitary) using T1-weighted contrast-enhanced MRI images. The network’s performance was evaluated using various approaches, including cross-validation methods and databases. The augmented image database demonstrated the best accuracy of 96.56% in 10-fold cross-validation (CV). The developed CNN architecture showed excellent generalization capability and efficient execution speed, making it a promising decision-support tool for radiologists in medical diagnostics. Similarly, Mzoughi et al.31 introduced an efficient and fully automatic 3D CNN architecture for classifying glioma brain tumors into low-grade and high-grade gliomas using volumetric T1-Gado MRI sequence images. The architecture effectively integrates local and global contextual information while reducing weights through small kernels. Preprocessing techniques, including intensity normalization and adaptive contrast enhancement, were employed to address data heterogeneity, and augmentation was used for robust training. The proposed approach outperformed the existing methods, achieving an impressive overall accuracy of 96.49% on the validation dataset. This study emphasizes the significance of proper MRI preprocessing and data augmentation in achieving accurate classification using CNN-based approaches. In addition, Gumaei et al. introduced a hybrid feature extraction approach coupled with a regularized extreme learning machine (RELM) for accurate brain tumor classification32. The method involves preprocessing brain images with min–max normalization to enhance contrast, followed by hybrid feature extraction. The classification of tumor types is then performed using the RELM. Experimental evaluations on a new public dataset demonstrated the approach’s superiority over existing methods, achieving an improved classification accuracy from 91.51 to 94.233% using the random holdout technique. Guan et al.3 proposed an efficient method for brain tumor grading using preprocessing, clustering-based location extraction, feature extraction, refinement, alignment, and classification steps. The method demonstrated superior performance to existing approaches on a publicly available brain tumor dataset, achieving an impressive overall classification accuracy of 98.04%. Deepak et al.33 introduced a classification system utilizing deep transfer learning with a pretrained GoogLeNet for feature extraction from brain MRI images. Proven classifier models were integrated to classify the extracted features. The system achieved an impressive mean classification accuracy of 98% using patient-level fivefold cross-validation, outperforming existing SOTA methods. Shaik et al.34 introduced a multilevel attention network (MANet) that incorporates spatial and cross-channel attention for tumor recognition. This approach prioritizes tumor regions while maintaining cross-channel temporal dependencies. Experimental evaluation on the Figshare and BraTS benchmark datasets demonstrated that the proposed model achieved superior performance, with the highest accuracy of 96.51% on the Figshare dataset and 94.91% on the BraTS2018 dataset, outperforming the existing models. Ahuja et al.35 proposed an automated brain tumor classification, localization, and segmentation approach using T1W-CE-MRI datasets. Pretrained DarkNet models (DarkNet-19 and DarkNet-53) were evaluated and achieved high accuracies of 99.60% for training and 98.81% for validation. The best-performing DarkNet-53 model achieved a testing accuracy of 98.54% and an area under the curve (AUC) of 0.99 on 1070 T1W-CE testing images.
An extensive review of the literature revealed that although deep learning (DL) models have demonstrated remarkable success in identifying brain tumors from MRI images, there has been significant oversight in evaluating the computational complexities of these models. Most studies have focused on achieving high classification performance, neglecting crucial factors such as the model’s depth, parameter count, and processing time. Notably, transfer learning (TL) models have proven to be time-consuming due to their training requirements with millions of parameters26,33,34. Furthermore, interpretability, a vital aspect of model analysis, has been largely disregarded, leaving a gap in understanding specific regions in an image that models prioritize for brain tumor classification.
In light of these challenges, this cutting edge study proposes an innovative lightweight framework that ensures accurate brain tumor classification and addresses the limitations of existing models. The authors extracted highly informative features that significantly reduce the number of layers, parameters, and computation time by harnessing the discriminative power of a parallel depthwise separable CNN. Notably, a pioneering approach was introduced by incorporating ridge regression into the proposed hybrid ELM model, replacing the more-pseudoinverse method. This integration enhances the classification performance of the novel hybrid ridge regression ELM (RRELM) model, surpassing the capabilities of previous approaches. In addition, conventional research boundaries were surpassed by achieving an unprecedented level of interpretability within the proposed framework. By developing a hybrid explainable CNN-ELM, the authors gained insights into the inner workings of the proposed model, allowing them to decipher the specific image regions that contribute most prominently to the classification of brain tumors. Transfer learning has emerged as a pivotal approach in image representation, particularly in fields with limited labeled data. The survey on food category recognition Zhang et al.55 highlights transfer learning’s role in leveraging pre-trained models to adapt features for domain-specific tasks, demonstrating its relevance in efficiently handling diverse datasets. Similarly, the study on cerebral microbleed detection (FeatureNet) combines transfer learning with ensemble techniques, freezing early layers to utilize generic features while fine-tuning classifiers like randomized neural networks for enhanced accuracy56. This approach achieved state-of-the-art results, showcasing the potential of transfer learning in complex medical imaging tasks. Furthermore, the CTBViT model for tuberculosis classification demonstrates the integration of transfer learning with vision transformers, employing a Patch Reduction Block and randomized classifiers to optimize performance and mitigate overfitting57. These studies collectively underline the adaptability and efficacy of transfer learning across various image analysis applications, particularly in resource-constrained domains.
This research offers a comprehensive solution that surpasses existing methodologies. By combining cutting-edge techniques in feature extraction, classification, and interpretability, an impressive framework has been presented that achieves superior accuracy and addresses the crucial concerns of computational complexity and model transparency. This research represents a significant advancement toward revolutionizing the field of brain tumor identification, setting a new standard for achieving performance and interpretability in medical image analysis.
Methodology
Proposed framework
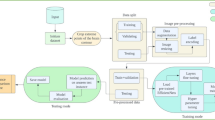
Figure 1 illustrates an innovative framework devised for the classification of three distinct types of brain tumors (BTs) from MRI images, alongside the classification of images depicting the absence of tumors. During training, the RRELM model uses fivefold cross-validation to evaluate and save the best-performing model, which is then used for final testing. When tested, the optimized model provides robust four-class classification, effectively distinguishing between different types of brain tumors and non-tumor cases. The entire framework showcases the power of combining a parallel CNN for feature extraction with ridge regression regularization in the classifier, achieving an efficient and accurate medical image classification solution.
An explanation generated using the Shapley Additive Explanation (SHAP) method was employed to gain insights into the black-box nature of the PDSCNN-RRELM model. This innovative visualization technique provided a deeper understanding of the model’s decision-making process and highlighted the most influential features contributing to the classification outcomes. In the subsequent sections, each step of the proposed framework is presented in greater detail, offering a comprehensive explanation of the methodology employed and the remarkable contributions made toward the accurate identification and classification of BT in MRI scan images.
Brain tumor dataset
The brain tumor dataset utilized in this study was acquired from the esteemed Kaggle repository. The dataset comprises four distinct classes: glioma, meningioma, no tumor, and pituitary tumor. It encompasses a total of 7,023 MRI images distributed across the following classes: glioma (1,621 images), meningioma (1,645 images), no tumor (2,000 images), and pituitary (1,757 images). A rigorous evaluation strategy was employed to ensure the reliability and generalizability of the model. Considering the critical role of brain tumor data in medical and clinical applications, ensuring patient privacy and adhering to ethical standards are fundamental to the integrity of this study. This dataset36 is sourced from publicly available repositories (the figshare, SARTAJ, and Br35H datasets), all of which are intended for research purposes. These repositories are typically structured to anonymize personal data, removing any patient-identifying information, thus aligning with the HIPAA and GDPR guidelines for data privacy. Additionally, dataset collection and processing followed the ethical standards and guidelines established for publicly available medical imaging data. No identifying features are linked to individual patients in the dataset, ensuring that the privacy of individuals is maintained throughout the study. The dataset was divided such that 80% of the images were allocated for training purposes, whereas the remaining 20% were reserved for testing the novel RRELM model with a fivefold cross-validation methodology. Figure 2 shows a selection of MRI samples to provide a visual representation of the dataset.
While Fig. 2 visually represents tumor shapes, it is imperative to highlight the inherent variability of the dataset, encompassing background characteristics, imaging artifacts, and tumor appearances. The presence of surrounding tissues such as the brain parenchyma, blood vessels, or ventricles introduces considerable background variations across MRI scans. Additionally, factors such as imaging parameters, scanner settings, and patient-specific traits further contribute to image diversity. Moreover, brain tumors manifest diverse shapes, sizes, and locations, leading to a spectrum of appearances on MRI images. The differentiation of gliomas, which are known for having irregular margins and infiltrative patterns, from healthy tissues is challenging. Conversely, meningiomas typically present as well-defined extra-axial masses, often adjacent to the dura mater.
The complexity of classifying these images extends beyond distinguishing tumors from backgrounds. Accurate identification and categorization of tumor types based on morphological characteristics and spatial relationships within the brain are paramount. Hence, while Fig. 2 provides insight into tumor shape diversity, it is essential to underscore the complexity of the classification task. The proposed method aims to address these challenges by leveraging advanced machine learning techniques to extract discriminative features effectively, facilitating accurate classification across tumor types. By recognizing the nuances in the dataset’s composition and the intricacies of brain tumor characterization, our study endeavors to provide a comprehensive understanding of the complexities involved in MRI-based tumor classification.
Data preprocessing
In the domain of image preprocessing, researchers have recognized the paramount importance of achieving optimal results. To this end, various methodologies have emerged, each aiming to enhance medical images for improved outcomes. In this study, the researchers employed the Contrast Limited Adaptive Histogram Equalization (CLAHE) technique to address the challenge of enhancing image contrast and model accuracy. Originally designed to address the inherent limitations of low-contrast medical images, CLAHE operates by constraining the amplification process within a user-defined threshold referred to as the clip limit37. This threshold plays a crucial role in smoothing out the noise in the histogram, consequently dictating the extent of contrast improvement achieved. For this study, the researchers judiciously set the clip limit at 2.0 while also adopting an (8 × 8) tile grid size to optimize the transformative potential of CLAHE in their pursuit of superior image enhancement. Figure 3 shows a selection of original MRI images alongside their corresponding counterparts, which have undergone enhancement using the CLAHE method. The transformative impact of CLAHE can be observed as it effectively enhances the visual quality and details of the MRI images, enabling improved clarity and contrast for accurate analysis and interpretation. The database encompasses a diverse range of images, each presenting varying sizes, necessitating a harmonization process to facilitate seamless integration into the CNN model. In pursuit of this objective, the images underwent a conversion procedure, aligning them to a standardized size of 124 × 124 pixels. By employing the powerful CLAHE technique, the images were then enhanced, ensuring optimal visual quality and facilitating detailed analysis.
Recognizing the significance of managing complexity and promoting uniformity, the researchers implemented a normalization procedure. Given the abundance of intensity values employed to represent an image, normalization serves as a crucial step to mitigate the intricacies arising from the vast number of pixels. For this purpose, the scale of the images was meticulously adjusted from the original range of 0–255 to a normalized range of 0–1. This strategic adjustment involved dividing the pixel values of each image by 255, effectively streamlining the subsequent analysis by reducing complexity and enhancing computational efficiency.
Parallel depthwise separable convolutional neural network
The primary objective was to develop a CNN model capable of effectively identifying essential features while maintaining a compact structure suitable for diverse real-world applications. To balance the trade-off between parameter limitations and layer complexity, a streamlined PDS-CNN model was crafted and specifically designed to extract meaningful features with minimal resource utilization. The architecture depicted in Fig. 4 exemplifies the elegance of this lightweight approach for capturing distinctive features within the MRI images.
A refined strategy was adopted to simplify the proposed CNN model compared with the TL models. The model comprised nine convolutional layers (CLs) and two fully connected layers (FCs), which were carefully calibrated to strike a balance. Through iterative experimentation, it was observed that a solitary CL would fail to capture crucial features, while a sequence of five CLs would introduce unnecessary depth and complexity. Consequently, the first five CLs were thoughtfully executed in parallel, leveraging the insights gained from meticulous trial and error. Each CL was equipped with 256 kernels of varying sizes, ranging from 11 × 11 to 3 × 3. The choice of employing different filter sizes ranging from 11 × 11 to 3 × 3 is strategic. If identical filter sizes were utilized, they would extract redundant features, ultimately leading to a lack of diversity in the feature space. However, by incorporating varying filter sizes, each filter can capture distinct types of features. Upon concatenating the feature maps obtained from these filters, a rich and diverse set of features is derived. This diversity aids in capturing intricate patterns and nuanced details present in the data, thus enhancing the model’s ability to discern and classify objects accurately. The rationale behind selecting a range from 3 × 3 to 11 × 11 is rooted in the characteristics of the filter sizes. A 3 × 3 filter is considered ideal for detecting local features such as edges, corners, and texture details due to its smaller receptive field. Conversely, an 11 × 11 filter possesses a larger receptive field, enabling it to capture more global features or overarching patterns present in the input data. By encompassing filter sizes across this spectrum, the model can effectively leverage both local and global information, striking a balance between detailed feature extraction and holistic pattern recognition.
A constant padding size was employed across the initial five CLs, enabling comprehensive information extraction even from the border elements. The resulting feature maps generated by the parallel CLs were harmoniously merged and subsequently fed into a sequential CL.
To further refine the model’s simplicity without compromising its classification performance, a novel approach involving depthwise separable convolution (DSC) was adopted, supplanting conventional convolution methods. The DSC expertly dissects the convolution process into depthwise convolution and pointwise convolution, dramatically reducing the parameter count from 2.2 million to a mere 0.53 million. This judicious reduction in computational complexity not only streamlined the model but also enhanced its classification performance. Following the last four CLs, a batch normalization (BN) layer and a max pooling layer were integrated, and their sizes were optimized to 128, 64, 32, and 16, respectively, with 3 × 3 kernels and VALID padding. The integration of the BN expedited execution while bolstering the model’s stability. To introduce nonlinearity and facilitate training, a rectified linear unit (ReLU) activation function was employed across all the CLs. Furthermore, to combat overfitting and expedite training, dropout layers were strategically inserted, randomly disregarding 50% of the nodes. Specifically, two dropout layers were placed after the final two CLs, and an additional layer was positioned after the first FC layer. Considering that the learning rate was set at 0.001 using the ADAM optimizer, the model loss was adeptly managed through the application of the sparse categorical cross-entropy loss function, enabling efficient feature extraction. Employing a systematic trial-and-error approach, 256 distinctive features were thoughtfully selected from the final FC layer, which encapsulates the essence of the model’s capabilities. Algorithm 1 demonstrates the kernel concatenation algorithm used in Separable Conv2d.
Algorithm 1

In a concatenated layer, the number of filters returned after merging depends on the specific configuration of the layer. When concatenating two or more layers, each layer may have a different number of filters, and the resulting concatenated layer will have a total number of filters equal to the sum of the filters in each individual layer.
The total parameters in the sequential model after concatenation depend on the type of layers and their configurations. Each layer in the model contributes a certain number of parameters based on its architecture and the number of filters or units it contains. When concatenating layers, the total number of parameters in the sequential model will be the sum of the parameters from all the layers involved in the concatenation, including the concatenated layer itself.
In this study, the true power of the proposed model was determined by extracting the most discriminative features from intricate MRI images. To convey the remarkable results with the utmost clarity and visual appeal, the exceptional ability of the t-Distributed Stochastic Neighbor Embedding (t-SNE) technique was harnessed, as shown in Fig. 5. With unparalleled finesse, t-SNE seamlessly unraveled the inherent structure and intricate relationships within high-dimensional MRI data. This remarkable algorithm skillfully transforms the complexity of the data into a lower-dimensional representation while preserving the subtle nuances and intricate similarities among the data points.
To ensure the optimal performance of the Extreme Learning Machine (ELM) algorithm, the extracted features underwent a crucial step of standardization using the standard scaler, which transformed the feature values to a standardized scale, thereby enhancing the ELM model’s effectiveness in handling the data. By standardizing all the features, the subsequent feeding of these standardized features into the ELM model facilitated improved performance and more accurate analysis10,38,39,40.
Interaction between PDSCNN and RRELM
The feature extraction process uses a parallel depthwise separable convolutional neural network (PDSCNN) to capture spatial features efficiently. Through its structure, the PDSCNN extracts 256 prominent features from each MRI image, providing a rich representation of tumor-specific patterns essential for accurate classification. After feature extraction by the PDSCNN, the network produces 256 feature maps (as shown in Fig. 6), each capturing distinct spatial features from the MRI images. These feature maps are then processed to form a single 1D feature vector, which serves as the basis for classification. Mathematically, if each feature map has dimensions of h×w, then flattening these maps results in a vector of size 256×h×w. However, for simplicity and computational efficiency, the mean or another statistical representation across each feature map is often taken, compressing the information into a 256-dimensional feature vector f, where:
Here, \({f}_{i}\) represents the summary value (such as the mean) of the i-th feature map, providing a compact, descriptive vector of the most prominent features detected by the PDSCNN.
Next, normalization is applied to this feature vector to scale the values within a consistent range. Let \({f}_{\text{norm}}\) denotes the normalized vector, where each component \({f}_{\text{i}}\) is transformed as:
where \({\mu}_{i}\) and \({\sigma}_{i}\) represent the mean and standard deviation of the i-th feature across the training set, respectively. This normalization ensures that the features are standardized, making them compatible for input into the next stage without being affected by scale differences.
In this pipeline, the PDSCNN and RRELM work as tightly integrated systems. The PDSCNN acts as the feature extraction module, producing \({f}_{\text{norm}}\) as its output, which becomes the direct input to the RRELM classifier.
The normalized feature vector \({f}_{\text{norm}}\) is then fed into the ridge regression extreme learning machine (RRELM), which is chosen for its robustness in handling high-dimensional and potentially correlated features. The RRELM model is an enhanced version of the classic extreme learning machine (ELM) that incorporates ridge regression regularization to control for overfitting, which is especially useful when dealing with intercorrelated features such as those derived from the PDSCNN.
In RRELM, the classification output y is calculated as:
where W is a matrix of weights learned during training, which maps the 256-dimensional feature vector to the four-class output (three tumor types and one non-tumor class), and b is a bias term added to adjust the final output.
The regularization term in RRELM adds a penalty proportional to the square of the weight matrix W:
where λ is a hyperparameter controlling the regularization strength. This regularization ensures that the model does not overfit, particularly when features are highly intercorrelated or redundant, as it encourages smaller, more stable weights.
Ridge regression extreme learning machine
Huang et al.41 introduced the ELM, a remarkable forward feed network based on supervised learning, to harness the power of neural networks (NNs) for feature classification. By employing a single hidden layer, the ELM revolutionized the training process by eliminating the need for backpropagation, enabling training times that were a thousandfold faster than those of conventional NNs. This groundbreaking approach endowed the model with exceptional generalization capabilities and unparalleled classification performance. Notably, in the realm of large-scale multiclass classification tasks, the ELM exhibited remarkable progress, surpassing traditional methods10,42,43,44. While the parameters linking the input and hidden layers were initialized randomly, the parameters connecting the hidden and output layers were determined using the pseudoinverse technique. However, in this study, the pseudoinverse was replaced by ridge regression, which injected an additional layer of sophistication. The input layer boasted a grand ensemble of 256 nodes, the hidden layer consisted of 1,500 nodes, and the output layer was composed of four nodes for the classification of four types of BT from MRI images. The following steps outline the implementation details:
Initialization: The input weight matrix (input_weights) of size (input_size × hidden_size) and biases were randomly initialized using a normal distribution.
Activation function: The rectified linear unit (ReLU) function was used as the activation function for the hidden layer. It can be defined as relu(x) = max (0, x), where x represents the input.
Hidden layer computation: The hidden_nodes() function computes the hidden layer output (H_train) using the following equations.
G = X_train * input_weights + biases, where X_train is the input feature matrix.
H_train = relu(G), where relu(x) is the ReLU activation function applied elementwise to G.
Ridge Regression: Ridge regression was utilized to optimize the output weight matrix (output_weights). The ridge regression formulation can be expressed as follows.
output_weights = inv (H_train. T @ H_train + alpha * I) @ H_train. T @ y_train, where alpha is the regularization parameter, I represents the identity matrix of size hidden_size, and y_train denotes the target labels.
Prediction : The predict() function was introduced to make predictions using the trained model. It computes the output of the hidden layer for the given input features (X) using the following equations.
G = X * input_weights + biases,
H = relu(G),
predictions = H @ output_weights, where predictions represent the final predicted output.
By incorporating Ridge Regression within the ELM (RRELM) framework, this methodology aimed to strike a balance between effective feature learning and regularization, enhancing the model’s ability to generalize and produce accurate predictions.
Assessment matrices and implementation
The effectiveness of the lightweight PDSCNN-RRELM model was evaluated using a confusion matrix (CM), which is a valuable tool for assessing classification performance. The following equations were employed to derive key performance metrics, including accuracy, precision, recall, F1 score, and area under the curve (AUC), from the CM45,46.
where true positives, true negatives, false positives, and false negatives are represented by \({T}_{P}\), \({T}_{N}\), \({F}_{P}\) and \({F}_{N}\), respectively.
The implementation of all the deep learning algorithms and Explainable Artificial Intelligence (XAI) techniques was accomplished using Keras, a powerful framework, in conjunction with TensorFlow as the backend. The entire system was orchestrated within PyCharm Community Edition (2021.2.3), a reliable software environment. Model training and testing were conducted on a high-performance computing platform comprising an 11th generation Intel(R) Core (TM) i9–11,900 CPU operating at 2.50 GHz, an impressive 128 GB RAM configuration, and an NVIDIA GeForce RTX 3090 graphics processing unit with a substantial 24 GB memory capacity. This robust hardware infrastructure, complemented by the 64-bit Windows 10 Pro operating system, facilitated efficient and expedient execution of the training and testing processes, enabling the exploration of complex deep learning architectures and XAI methodologies.
Results and discussion
An ELM with pseudoinverse
Following the extraction of the most discriminant 256 features using the PDSCNN, the ELM model with pseudoinverse (PELM) was trained on 80% of the available data, whereas the remaining 20% was reserved for evaluating the model’s performance. Fivefold cross-validation was employed to assess the model’s precision, recall, F1 score, and accuracy comprehensively. Figure 7 shows the confusion matrices for each fold, which were instrumental in computing the class-specific precision, recall, and F1 score.
The average accuracy, precision, and recall achieved remarkable values of 99%, 99.1%, and 99.3%, respectively, for the classification of four types of brain tumors (glioma: 0, meningioma: 1, no tumor: 2, and pituitary: 3) based on MRI images, as shown in Table 1. Fold 2 had exceptional average accuracy and recall values of 99.34% and 99.5%, respectively. Furthermore, Fig. 8 clearly illustrates a classwise receiver operating characteristic (ROC) curve, demonstrating the ability of the PDSCNN-PELM model to effectively differentiate between the four classes of brain tumors. Notably, the highest Area Under the Curve (AUC) values for all classes exceeded 99.70%, exemplifying the robustness of the proposed framework. The best AUC of 99.94% further attests to the model’s capacity for achieving outstanding classification accuracy.
ELM with ridge regression
The experimental procedures were replicated, with the exception of replacing the pseudoinverse with ridge regression (RRELM) to enhance the classification performance. The resulting confusion matrices for each fold are presented in Fig. 9, highlighting the model’s ability to classify brain tumor types accurately. Notably, the fourth-fold method demonstrated exceptional performance, achieving average accuracy, precision, and recall values of 99.50%, 100%, and 100%, respectively, as summarized in Table 2.
Comparison between PELM and RRELM
Table 3 provides a comprehensive comparison between the PELM and RRELM. It is evident that the PDSCNN-RRELM exhibited superior performance metrics compared to the PDSCNN-PELM. Specifically, the average precision of the PDSCNN-RRELM reached an impressive value of 99.35% ± 0.379, surpassing the precision of 99.10% ± 0.137 achieved by the PDSCNN-PELM. In terms of recall, RRELM outperformed PDSCNN-PELM, achieving an average recall of 99.30% ± 0.411, which is approximately 0.50% higher. Moreover, the novel PDSCNN-RRELM model achieved a remarkable accuracy of 99.50%, surpassing the 99.21% accuracy of the PDSCNN-PELM model by a margin of 0.30%. The class discriminant capability was assessed through ROC analysis, where the PDSCNN-RRELM achieved the best AUC of 99.99%, outperforming the AUC of 99.94%, achieved by the PDSCNN-PELM as shown in Fig. 10. These results underscore the superior performance of the novel ELM with ridge regression in accurately classifying the three types of brain tumors from MRI images.
In summary, the introduction of ridge regression in the ELM framework led to significant enhancements in classification performance. The PDSCNN-RRELM demonstrated superior accuracy, precision, recall, and AUC values, indicating its efficacy in accurately classifying brain tumor types.
Performances comparison of PDSCNN-RRELM with SOTA models
The proposed PDSCNN-RRELM model was evaluated via a comprehensive comparison of its classification performance and model characteristics with those of state-of-the-art (SOTA) models, as shown in Table 4. Among the SOTA models, those of Khan et al.28 achieved an exceptional accuracy of 99.13%. However, the proposed model surpassed this benchmark by achieving an even higher accuracy of 99.22%, showing an improvement of 0.10%. Furthermore, precision, a crucial metric in medical diagnosis, revealed the superiority of the proposed model. With a precision of 99.35%, the PDSCNN-RRELM outperformed the model of Khan et al. by 0.20%. Similarly, the proposed model exhibited remarkable performance in terms of recall, achieving a recall rate of 99.30%, surpassing all other SOTA models. The discriminative capability of the proposed model was assessed through ROC analysis. Impressively, the PDSCNN-RRELM model achieved the best AUC of 99.99%, highlighting its superior discriminative power in comparison to that of the SOTA models, which achieved a maximum AUC of 99.90%. These compelling findings support the conclusion that the proposed PDSCNN-RRELM framework outperforms the SOTA models in terms of classification performance. Notably, it achieved higher accuracy, precision, and recall while exhibiting exceptional discriminative capability, as evidenced by the outstanding AUC of 99.99%.
Considering computational resources, a comparison was made between the lightweight PDSCNN-RRELM model and Shaikh et al.‘s34 modification of DarkNet53. Shaikh et al.‘s model employed 2.2 million parameters and 53 layers to classify brain tumors with an accuracy of 96.51%. In contrast, the proposed PDSCNN-RRELM model outperformed it significantly. With only 0.53 million parameters (approximately five times fewer than the best SOTA model) and ten layers (approximately five times fewer than the best SOTA model), it achieved an outstanding accuracy of 99.22%. In conclusion, the proposed lightweight PDSCNN-RRELM model not only demonstrated superior classification performance compared to that of the SOTA models but also showed a decrease in the number of parameters and layers. This achievement holds great potential to support medical practitioners in accurately diagnosing brain tumors, instilling confidence, and ultimately contributing to saving patients’ lives.
The PDSCNN-RRELM model also performs better than SOTA transformer-based architectures such as LCDEiT47, FTVT-b3248, and RanMerFormer49 in several important ways. The findings demonstrate the model’s outstanding classification ability, effectively balancing true positive detection and false positive reduction. In addition to its accuracy, the PDSCNN-RRELM demonstrates remarkable efficiency, including 0.53 million parameters and a streamlined 10-layer architecture, in contrast to the more intricate transformer models, such as LCDEiT, which has 8 million parameters. Transformer models excel in capturing long-range relationships and managing sequential data, making them particularly adept at addressing complicated data patterns and activities necessitating contextual comprehension. Nonetheless, their complex and parameter-intensive architecture sometimes requires substantial computing resources and prolonged training durations, thus restricting their practicality for real-time or resource-limited applications. Consequently, while the PDSCNN-RRELM demonstrates superior efficiency and efficacy in certain domains, contexts that prioritize the interpretability of intricate connections over computational efficiency may favor transformer-assisted models.
Interpretability of PDSCNN-RRELM using SHAP
Explainable artificial intelligence (XAI) has emerged as a groundbreaking concept within the realm of deep learning. Its significance lies in its ability to unravel the inner workings of complex deep neural networks, shedding light on their decision-making processes. This is particularly crucial for deep learning models, which are known for their enigmatic and intricate nature. To address this challenge, a remarkable study harnessed the power of SHAP, an advanced technique that effectively removes the “black box” cloak enveloping deep learning models39,40,50. By employing SHAP in conjunction with the PDSCNN-RRELM model, this research enabled medical professionals to comprehensively evaluate and elucidate the results, facilitating real-world application in the domain of brain tumor categorization.
The impact of SHAP is profound, as it not only ranks the importance of individual features within the model but also quantifies their contributions through meticulous calculations. In this study, the scores assigned to each pixel in a predicted image vividly portrayed the intricate functionalities of those pixels, thus enabling precise categorizations51. The meticulously derived Shapley values, obtained by considering all possible combinations of BT disease features, were combined to form pixelated representations. The remarkable findings revealed a fascinating pattern: red pixels significantly enhanced the likelihood of accurate tumor identification, while blue pixels indicated the potential for misclassification.
The visualization of the SHAP results, elegantly depicted in Fig. 11, bestowed upon medical professionals a treasure trove of explanation images for the distinct tumor classes: glioma, meningioma, no tumor, and pituitary tumors. An intriguing aspect was the near-invisibility of the relevant gray explanation backgrounds when juxtaposed with the input images on the left. In the first row of the image, the dominance of red pixels within the SHAP explanation image effortlessly conveyed that the MRI image pertained to a glioma tumor, while the absence of blue pixels unequivocally signaled the exclusion of the other tumor types: meningioma, no tumor, and pituitary. Astoundingly, the second row revealed a contrasting pattern: an absence of red pixels in the SHAP explanation images for glioma, no tumor, and pituitary, accompanied by a profusion of red pixels in the SHAP explanation image for meningioma. This remarkable observation accurately indicated the classification of the image as belonging to the meningioma class. The third row continued to dazzle, showcasing a concentration of red pixels within the SHAP explanation image for the no tumor class, while a substantial presence of blue pixels in the SHAP explanation images for glioma, meningioma, and pituitary precisely identified the original image as containing no tumor. The fourth row followed suit, with the SHAP explanation image predominantly illuminated by a vibrant array of red pixels, suggesting the presence of a pituitary tumor. This visual elucidation through SHAP images not only validated the model’s findings but also bestowed upon medical specialists an unmistakable indication of the diverse types of brain tumors under consideration.
The integration of SHAP in this study not only dismantled the impenetrable black box of deep learning but also provided a captivating narrative of the inner workings of the PDSCNN-RRELM model. This breakthrough empowered medical professionals with an awe-inspiring level of confidence in their tumor categorization endeavors, forever transforming the landscape of brain tumor analysis and enriching the field of medical specialization.
Strengths, limitations and future work
One of the major contributions of this paper lies in the introduction of a novel hybrid approach that surpasses the performance of the traditional pseudoinverse of the ELM by leveraging ridge regression. This innovative methodology leads to remarkably accurate classification of brain tumors using MRI images, demonstrating its efficacy in solving complex classification tasks. Moreover, the proposed model achieves these impressive results while maintaining a smaller number of parameters and layers, further highlighting its efficiency and practicality.
However, it is essential to acknowledge and address a significant limitation of the current study, which is related to the validation and generalizability of the proposed model. While the framework demonstrates high performance during cross-validation, validating its effectiveness via independent real-world datasets is crucial. Given the structure of the dataset used in this study, data leakage could occur if images from the same patient appeared in both the training and testing sets, as this would enable the model to leverage prior information instead of learning to generalize. Since MRI images are inherently 3D but 2D slices are used, this could further complicate potential leakage. The dataset does not provide explicit patient IDs, making strict patient-level separation difficult. Although data leakage could not be fully avoided, an effort was made to employ strategies such as cross-validation to minimize the impact of any potential leakage and accurately assess model performance. Involving domain experts and collaborating with clinical institutions to obtain such datasets will not only enhance the credibility of the research but also ensure the applicability of the proposed model in real-world clinical settings.
Deploying this model in real clinical settings may pose challenges, such as high computational needs, data privacy concerns, and the requirement for standardized preprocessing to ensure consistent inputs. Additionally, adapting to variations in MRI settings and patient demographics is essential for broader use. Solutions include developing a lighter model for clinical hardware, using anonymization to protect privacy, and thoroughly testing diverse data sources. Collaborating with medical experts to address these aspects will improve the model’s readiness for practical application, supporting brain tumor diagnosis and treatment planning.
To improve the credibility and impact of future work, expanding the dataset collection process by obtaining patientwise data is recommended since the present dataset does not share any patientwise information. The proposed framework can be extended by incorporating specific patient characteristics and individual data to enable patientwise classification. This advancement would contribute to a more personalized and accurate classification of brain tumors, addressing different patients’ unique characteristics and complexities. Consequently, incorporating patientwise classification will undoubtedly enhance the framework’s accuracy, practicality, and overall utility in real-world clinical scenarios.
To further increase the accuracy of brain tumor classification and extend the model’s capabilities, future research may explore the integration of multi-modal data, combining MRI imaging with other patient data (e.g., genetic or clinical data). This approach could provide a more comprehensive representation of patient characteristics, facilitating better diagnosis. Additionally, advanced architectures, such as multi-layer transformers, could be investigated to capture more complex relationships within the data and improve classification accuracy. Relevant studies have shown the effectiveness of such methods in medical imaging and may provide important insights for future studies52,53,54.
Conclusion
In conclusion, accurate classification of brain tumors is crucial for early detection and effective treatment strategies. This study presents a novel approach that combines the PDSCNN and ELM to diagnose brain tumors based on MRI images. The subsequent analysis and classification tasks have been improved by enhancing the visibility and clarity of essential tumor features using CLAHE. The proposed PDSCNN efficiently captured tumor-specific patterns while minimizing computational complexity (only − 5.3 million parameters with 10 layers). Additionally, a hybrid ridge regression ELM model was introduced, which enhanced the classification performance by replacing the pseudoinverse with ridge regression. The results obtained from fivefold cross-validation demonstrated remarkable accuracy, precision, and recall values of 99.22%, 99.35%, and 99.30%, respectively, for the classification of glioma, meningioma, no tumor, and pituitary based on MRI images. Compared with the PDSCNN-PELM, the PDSCNN-RRELM model exhibited superior performance (almost 0.20% higher than that for each metric) metrics, establishing its efficacy in accurately classifying brain tumor types. SHAP allowed confident interpretation of the model’s outcomes, helping doctors and radiologists correctly diagnose brain tumors.
Data availability
Code and data are available here: https://github.com/hafsa-kibria/Brain_Tumor_Classification.
Change history
11 March 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-92325-w
References
Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131, 803–820 (2016).
Chahal, P. K., Pandey, S. & Goel, S. A survey on brain tumor detection techniques for MR images. Multimed. Tools Appl. 79, 21771–21814 (2020).
Guan, Y. et al. A framework for efficient brain tumor classification using MRI images (2021).
Komninos, J. et al. Tumors metastatic to the pituitary gland: Case report and literature review. J. Clin. Endocrinol. Metab. 89, 574–580 (2004).
Ahamed, M. F. et al. A review on brain tumor segmentation based on deep learning methods with federated learning techniques. Comput. Med. Imaging Graph. 110, 102313 (2023).
Faysal Ahamed, M., Robiul Islam, M., Hossain, T., Syfullah, K. & Sarkar, O. Classification and segmentation on multi-regional brain tumors using volumetric images of MRI with customized 3D U-Net framework. In Proceedings of International Conference on Information and Communication Technology for Development: ICICTD 2022 223–234 (Springer, 2023).
Titu, M. M. T., Mary, M. M., Ahamed, M. F., Oishee, T. M. & Hasan, M. M. Utilizing customized 3D U-Net framework for the classification and segmentation of multi-regional brain tumors in volumetric MRI images. In 2024 3rd International Conference on Advancement in Electrical and Electronic Engineering (ICAEEE) 1–6 (IEEE, 2024).
Ahamed, M. F. et al. Automated detection of colorectal polyp utilizing deep learning methods with explainable AI. IEEE Access (2024).
Varuna Shree, N. & Kumar, T. N. R. Identification and classification of brain tumor MRI images with feature extraction using DWT and probabilistic neural network. Brain Inf. 5, 23–30 (2018).
Nahiduzzaman, M., Islam, M. R. & Hassan, R. ChestX-Ray6: Prediction of multiple diseases including COVID-19 from chest X-ray images using convolutional neural network. Expert Syst. Appl. 211, 118576 (2023).
Islam, M. R. & Nahiduzzaman, M. Complex features extraction with deep learning model for the detection of COVID19 from CT scan images using ensemble based machine learning approach. Expert Syst. Appl. 195, 116554 (2022).
Nahiduzzaman, M. et al. Diabetic retinopathy identification using parallel convolutional neural network based feature extractor and ELM classifier. Expert Syst. Appl. 217, 119557 (2023).
Nahiduzzaman, M. et al. Hybrid CNN-SVD based prominent feature extraction and selection for grading diabetic retinopathy using extreme learning machine algorithm. IEEE Access 9, 152261–152274 (2021).
Nahiduzzaman, M. et al. A novel method for multivariant pneumonia classification based on hybrid CNN-PCA based feature extraction using extreme learning machine with CXR images. IEEE Access 9, 147512–147526 (2021).
Hossain, M. M., Islam, M. R., Ahamed, M. F., Ahsan, M. & Haider J. A collaborative federated learning framework for lung and colon cancer classifications. Technologies 12, 151 (2024).
Hossain, M. M., Ahamed, M. F., Islam, M. R. & Imam, M. D. R. Privacy preserving federated learning for lung cancer classification. In 2023 26th International Conference on Computer and Information Technology (ICCIT) 1–6 (IEEE, 2023).
Ahamed, M. F., Nahiduzzaman, M., Ayari, M. A., Khandakar, A. & Islam, S. M. R. Malaria parasite classification from RBC smears using lightweight parallel depthwise separable CNN and ridge regression ELM by integrating SHAP techniques (2023).
Sarkar, O. et al. Multi-scale CNN: An explainable AI-integrated unique deep learning framework for lung-affected disease classification. Technologies 11, 134 (2023).
Ullah, F., Nadeem, M. & Abrar, M. Revolutionizing brain tumor segmentation in MRI with dynamic fusion of handcrafted features and global pathway-based deep learning. KSII Trans. Internet Inform. Syst. 18 (2024).
Anwar, R. W., Abrar, M. & Ullah, F. Transfer learning in brain tumor classification: Challenges, opportunities, and future prospects. In 2023 14th International Conference on Information and Communication Technology Convergence (ICTC) 24–29 (IEEE, 2023).
Akbar, M. et al. An effective deep learning approach for the classification of Bacteriosis in peach leave. Front. Plant Sci. 13. https://doi.org/10.3389/fpls.2022.1064854 (2022).
Nazar, U. et al. Review of automated computerized methods for brain tumor segmentation and classification. Curr. Med. Imaging 16, 823–834 (2020).
Ahamed, M. F., Sarkar, O. & Matin, A. Instance segmentation of visible cloud images based on mask R-CNN applying transfer learning approach. In 2020 2nd International Conference on Advanced Information and Communication Technology (ICAICT) 257–262 (IEEE, 2020). https://doi.org/10.1109/ICAICT51780.2020.9333531.
Musallam, A. S., Sherif, A. S. & Hussein, M. K. A new convolutional neural network architecture for automatic detection of brain tumors in magnetic resonance imaging images. IEEE Access 10, 2775–2782 (2022).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014).
Nayak, D. R., Padhy, N., Mallick, P. K., Zymbler, M. & Kumar, S. Brain tumor classification using dense efficient-net. Axioms 11, 34 (2022).
Alanazi, M. F. et al. Brain tumor/mass classification framework using magnetic-resonance-imaging-based isolated and developed transfer deep-learning model. Sensors 22, 372 (2022).
Khan, A. H. et al. Intelligent model for brain tumor identification using deep learning. Appl. Computat. Intell. Soft Comput. 2022, 8104054 (2022).
Irmak, E. Multi-classification of brain tumor MRI images using deep convolutional neural network with fully optimized framework. Iran. J. Sci. Technol. Trans. Electr. Eng. 45, 1015–1036 (2021).
Badža, M. M. & Barjaktarović, M. Č. Classification of brain tumors from MRI images using a convolutional neural network. Appl. Sci. 10, 1999 (2020).
Mzoughi, H. et al. Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J. Digit. Imaging 33, 903–915 (2020).
Gumaei, A., Hassan, M. M., Hassan, M. R., Alelaiwi, A. & Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 7, 36266–36273 (2019).
Deepak, S. & Ameer, P. M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 111, 103345 (2019).
Shaik, N. S. & Cherukuri, T. K. Multi-level attention network: Application to brain tumor classification. Signal. Image Video Process. 16, 817–824 (2022).
Ahuja, S., Panigrahi, B. K. & Gandhi, T. K. Enhanced performance of Dark-nets for brain tumor classification and segmentation using colormap-based superpixel techniques. Mach. Learn. Appl. 7, 100212 (2022).
Brain Tumor MRI Dataset. https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset/data.
Pisano, E. D. et al. Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. J. Digit. Imaging 11, 193–200 (1998).
Ahamed, M. F., Salam, A., Nahiduzzaman, M., Abdullah-Al-Wadud, M. & Islam, S. M. R. streamlining plant disease diagnosis with convolutional neural networks and edge devices. Neural Comput. Appl. 36, 18445–18477 (2024).
Ahamed, M. F. et al. Detection of various gastrointestinal tract diseases through a deep learning method with ensemble ELM and explainable AI. Expert Syst. Appl. 256, 124908 (2024).
Ahamed, M. F. et al. Interpretable deep learning model for tuberculosis detection using X-ray images. In Surveillance, Prevention, and Control of Infectious Diseases: An AI Perspective (eds. Chowdhury, M. E. H. & Kiranyaz, S.) 169–192 (Springer, 2024).
Huang, G. B., Zhu, Q. Y. & Siew, C. K. Extreme learning machine: Theory and applications. Neurocomputing 70, 489–501 (2006).
Nahiduzzaman, M. et al. Parallel CNN-ELM: A multiclass classification of chest X-ray images to identify seventeen lung diseases including COVID-19. Expert Syst. Appl. 229, 120528 (2023).
Nahiduzzaman, M., Nayeem, M. J., Ahmed, M. T. & Zaman, M. S. U. Prediction of heart disease using multi-layer perceptron neural network and support vector machine. In 2019 4th International Conference on Electrical Information and Communication Technology (EICT) 1–6 (IEEE, 2019).
Kibria, H. B., Nahiduzzaman, M., Goni, M. O. F., Ahsan, M. & Haider, J. An ensemble approach for the prediction of diabetes mellitus using a soft voting classifier with an explainable AI. Sensors 22, 7268 (2022).
Powers, D. M. W. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061 (2020).
Swets, J. A. Measuring the accuracy of diagnostic systems. Science (1979) 240, 1285–1293 (1988).
Ferdous, G. J., Sathi, K. A., Hossain, M. A., Hoque, M. M. & Dewan, M. A. A. LCDEiT: A linear complexity data-efficient image transformer for MRI brain tumor classification. IEEE Access 11, 20337–20350 (2023).
Reddy, C. K. K. et al. A fine-tuned vision transformer based enhanced multi-class brain tumor classification using MRI scan imagery. Front. Oncol. 14 1–23 (2024).
Wang, J., Lu, S. Y., Wang, S. H. & Zhang, Y. D. RanMerFormer: Randomized vision transformer with token merging for brain tumor classification. Neurocomputing 573, 127216 (2024).
Lundberg, S. A unified approach to interpreting model predictions. arXiv Preprint arXiv:1705.07874 (2017).
Bhandari, M., Shahi, T. B., Siku, B. & Neupane, A. Explanatory classification of CXR images into COVID-19, Pneumonia and Tuberculosis using deep learning and XAI. Comput. Biol. Med. 150, 106156 (2022).
Ullah, F. et al. Evolutionary model for brain cancer-grading and classification. IEEE Access 11, 126182–126194 (2023).
Ullah, F. et al. Enhancing brain tumor segmentation accuracy through scalable federated learning with advanced data privacy and security measures. Mathematics 11, 4189 (2023).
Ullah, F. et al. Brain tumor segmentation from MRI images using handcrafted convolutional neural network. Diagnostics 13, 2650 (2023).
Zhang, Y. et al. Deep learning in food category recognition. Inf. Fusion 98, 101859 (2023).
Lu, S. Y., Nayak, D. R., Wang, S. H. & Zhang Y.-D. A cerebral microbleed diagnosis method via FeatureNet and ensembled randomized neural networks. Appl. Soft Comput. 109, 107567 (2021).
Lu, S. Y., Zhu, Z., Tang, Y., Zhang, X. & Liu, X. CTBViT: A novel ViT for tuberculosis classification with efficient block and randomized classifier. Biomed. Signal. Process. Control 100, 106981 (2025).
Author information
Authors and Affiliations
Contributions
All authors have contributed equally during the research work and preparing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article omitted an affiliation for author Lway Faisal Abdulrazak. The correct affiliations are listed in the correction notice.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nahiduzzaman, M., Abdulrazak, L.F., Kibria, H.B. et al. A hybrid explainable model based on advanced machine learning and deep learning models for classifying brain tumors using MRI images. Sci Rep 15, 1649 (2025). https://doi.org/10.1038/s41598-025-85874-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-85874-7
Keywords
This article is cited by
-
TriGWONet a lightweight multibranch convolutional neural network using gray wolf optimization for accurate oral cancer image classification
Discover Artificial Intelligence (2026)
-
Advancements in Machine Learning for Brain Tumor Classification and Diagnosis: A Comprehensive Review of Challenges and Future Directions
Archives of Computational Methods in Engineering (2026)
-
AI-based neoadjuvant immunotherapy response prediction across pan-cancer: a comprehensive review
Cancer Cell International (2025)
-
XcepFusion for brain tumor detection using a hybrid transfer learning framework with layer pruning and freezing
Scientific Reports (2025)
-
Transparent brain tumor detection using DenseNet169 and LIME
Scientific Reports (2025)