Abstract
Gastric cancer (GC) is a huge threat to global health, there is no effective treatment or just delay the progression of advanced GC until now. Micro-RNAs were reported to participate in the progression of GC. Clonal formation, MTT, caspase-3 activity, sperm DNA fragmentation, flow cytometry assay, cell adhesion, transwell assays were performed to detect the functions of miR-32-5p or anti-miR-32-5p on the growth and metastasis of GC cells. Western blot, qRT-PCR, Co-immunoprecipitation, and luciferase reporter analysis were performed to explore the associated mechanisms. We established mouse tumor xenografts and mouse metastasis models to explore the role of miR-32-5p and anti-miR-32-5p in vivo. We found that miR-32-5p significantly promoting the proliferation and metastasis of GC cells at both in vitro and in vivo levels. The underlying mechanism maybe that miR-32-5p complementary paired with the 3′-UTR of DSC2, and inhibited the expression of DSC2. Furthermore, we found that DSC2 suppressed the transcription of Cyclin B1, and induced G2/M phase arrest through inhibiting the complex of β-catenin/TCF4 in nucleus. We concluded that miR-32-5p negatively regulated the DSC2 expression, might be a potential therapeutic targeting of cancers, most especially in GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a huge threat to global health and the third leading cause of cancer-related death worldwide1. The progression of GC involves multiple steps and is considerably complicated involving numerous factors, the median survival time of GC patients remains short2. So, there is an urgent need to further decipher the molecular mechanisms, explore new therapeutic strategies to inhibit the progression and improve the clinical outcomes of advanced GC3.
Micro-RNAs (miRNAs) are a family of small, non-coding RNAs. MiRNAs regulate tumor cell viability, cycle, differentiation, invasion and metastasis through suppressing their targeted mRNA translation via targeting 3’ untranslated region of mRNA. Till now, the role and regulation mechanisms of miRNAs in the progression of GC remain unclear4. Yu L, et al. reported that miR-200 family might be a new tumor suppressor of GC5. While, Eto K, et al. found that overexpression of miR-223 significantly suppressed trastuzumab-induced apoptosis of GC6.
As everyone knows, GC is of epithelial origin7. Desmosome junctions are prominent complexes to harden epithelial cell-cell adhesion, which is necessary to maintain epithelial cell stability8,9. Desmosome junctions formed vital barrier for inhibiting the development of cancers. Desmocollin 2 (DSC2) is the key component of functional desmosomal junction and also the only isoform of DSCs expressed in normal stomach tissue10. In the previous studies, we found that DSC2 inhibited the viability, migration and invasion of GC both in vitro and in vivo11. In general, DSC2 is in the state of dynamic equilibrium10, the expression of DSC2 might be regulated by certain factors, while the detailed regulators and mechanisms need further study. MiRNAs are highly conserved among different species and regulate various biological functions in an epigenetic manner12,13. We performed a bioinformatic analysis using eight databases (miRWalk, Mircot4, miRanda, miRDB, miRMap, PITA, RNAhybrid and Targetscan), selected the cancer-related genes that have not been reported in GC, and speculated that DSC2 is a candidate target of miR-32-5p in GC cells.
In this work, we assessed miR-32-5p as a specific modulator of DSC2 expression in GC cells, investigated the effect of miR-32-5p and anti-miR-32-5p on the viability and metastasis on GC cells in vitro and in vivo, as well as the mechanisms of action underlying.
Materials and methods
Materials
TRIzol Reagent was the products of Invitrogen (USA). qPCR RT Kit was purchase from Toyobo (Japan). LF3 was purchase from InvivoChem (USA).
Cell culture
Human GES-1 cells, human GC cell lines BGC-823 cells were purchased from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human GC cell lines MGC-803 and SGC-7901 cells were kindly provided by School of Basic Medical Sciences, Shandong University. GES-1 cells, BGC-823 cells and SGC-7901 cells were cultured in RPMI-1640 medium, MGC-803 cells were cultured in DMEM medium, supplemented with 10% fetal bovine serum (Gibico, CA, USA) and 100 μg/μL streptomycin/penicillin, in a humidified 5% CO2 incubator at 37 °C.
Stably overexpression of human DSC2 gene
Cells (2 × 105 cells per well) were seeded into 6-well plates. After transfecting with LV-EFS promoter > hDSC2[NM_024422.6]-CMV > EGFP/T2A/Puro plasmid for 48 h, EGFP positive cells (High-DSC2 group) were sorted. Negative control cells (Lenti-NC group) were transfected with the vector of LV-CMV > EGFP/T2A/Puro.
CRISPR/Cas9-induced knockout of DSC2 gene
The small guide RNA (sgRNA) against human DSC2 was inserted into the CRISPR EGFP plasmid. MGC-803 cells (2 × 105 cells per well) were seeded into 6-well plates. After transfecting with CRISPR DSC2-EGFP plasmid for 48 h, EGFP positive cells were sorted. After 6 weeks, the clones were collected, positive clone MGC-803 cells were photoed. The knockdown of DSC2 expression (sgRNA-DSC2) was detected by photoed and qRT-PCR assay. Negative control cells (sgRNA-NC group) were transfected with the empty vector.
Quantitative real-time PCR
Total RNA was extracted from cells pellets or tumor specimen using Trizol reagent, and reverse transcribed with the PrimeScript RT-PCR kit. The A260/A280 ratio was calculated to assess RNA quality and purity. qRT-PCR analysis was performed on LightCycler 480II RT-PCR system (Roche, Basel, Switerland) at the recommended thermal cycling settings: one initial cycle at 95 °C for 30 s followed by 60 cycles of 15 s at 95 °C, 20 s at 60 °C and 15 s at 72 °C. Gene primers are represented in (Table 1).
Western blot analysis
Cells or tissues were lysed in RIPA buffer and the protein concentration was determined by BCA protein assay. Then proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrophoresis, the proteins were electrotransferred to PVDF membrane (Millipore, Billerica, USA) and then blocked with 5% non-fat milk for 4 h. The primary antibodies against DSC2 (#53485, Santa Cruz Biotechnology), MMP9 (#AF5228, Affbiotech), CD44 (#15675, proteintech), N-cadherin (#22018, proteintech), TCF4 (#DF6275, Affbiotech), Cyclin B1 (#AF6168, Affbiotech), β-catenin (#G0709, Santa Cruz Biotechnology) and β-actin (#ZF-0313, ZS Bio).
MTT assay
Cells from each group (3 ~ 4 × 103 cells per well) were inoculated into 96-well plates and incubated for 24 h. After washing with PBS, cells were incubated in culture medium containing 20 μL of MTT solution (5 mg/mL in phosphate buffered saline; PBS) for a further 4 h at 37 °C. The medium was removed and replaced with 150 μL of DMSO to dissolve the formazan crystal. The absorbance was measured at 570 nm using Thermo Multiskan GO microplate reader (Thermo-1510, CA, USA).
Sperm DNA fragmentation assay
Cells of each groups were labeled with 5-bromodeoxyuridine (BrdU), then lysed in incubation buffer. The supernatant was collected after centrifuging and treated with anti-DNA antibody coated Quantikine ELISA kits (R&D Systems, MN, USA) at 4 °C overnight. In brief, 100 µL of supernatant per well were added into the Quantikine ELISA kits, and cultured at room temperature for 90 min. The DNA was denatured by heating with medium fire for 5 min to expose BrdU for easy detection. Then, the samples were incubated with 100 µL of anti-BrdU-POD antibody per well at room temperature for 90 min. We added 100 µL tetramethylbenzidine (TMB) per well, and the absorbance was detected at 450 nm with a Thermo Multiskan GO microplate reader (Thermo-1510, CA, USA).
Caspase-3 activity assay
Cells from each group were lysed in lysis buffer. Then, the protein concentration was determined by Bradford protein assay. 150 μg protein, 10 µL caspase-3 substrate (5 μM Ac-DEVD-AMC), and 10 µL caspase-3 inhibitor (1 μM Ac-DEVD-CHO) were mixed and seeded into 96-well plates at 37 °C for 1 h. The absorbance was determined at 355 nm using the Microplate Luminometer (LB960, Berthold, German).
Flow cytometry assay
Cells were cultured in 6-well plates. Adherent cells were detached with 0.25% trypsin without EDTA in 1 × PBS. Cells were harvested in complete culture medium and centrifuged at 1000 rpm for 5 min. Each of the cells were washed with 1 × PBS, fixed with 70% ethanol, and stored at 4 °C overnight. Subsequently, cells were stained with 50 μg/mL propidium iodide (BD Pharmingen, USA) following the manufacturer’s instructions and analyzed using the FACScan flow cytometer (Becton Dickinson, USA). Cell cycle analysis was performed using FlowJo V.10 software.
Transwell assay
Cells (3 ~ 5 × 103 cells per well) of each groups suspended in medium without serum, then delivered into the top side of transwells. For cell invasion assay, the upper chamber was coated with Matrigel, while for cell migration assay, no Matrigel was added. After incubation at 37 °C for 24 h, cells remained in the top chambers (non-invasive or non-migrative cells) were removed together with the medium. The cells on the bottom side of the transwell chamber were photographed and counted under a microscope with 200 magnifications (NIKON Ti-U, Nikon, Japan).
Adhesion assay
The Matrigel gel was diluted eightfold with serum-free medium, and the diluted Matrigel gel was added to a 24-well plates (300 µL per well). After drying and solidification, the unsolidified glue was washed with phosphate buffer saline (PBS). Cells (2 × 104 cells per well) were seeded into the plates, and incubated in a 37 °C sincubator for 90 min. Then washed with PBS and adherent cells were fixed with methanol for 30 min, stained with crystal violet for 10 min, washed with PBS. After drying, the fixed cells were photographed and counted under a microscope with 200 magnifications (NIKON Ti-U, Nikon, Japan).
Luciferase reporter analysis
We predicted the target of gene binding site through Targetscan (http://www.targetscan.org). Wild-type (wt) and mutant-type (mut) reporter plasmids of DSC2 or Cyclin B1 3’-untranslated region (3’UTR, Fig. 2D) sequences were inserted into pmirGLO vector (Jinan Bo Shang Co., Ltd.). Cells were plated in 24-well plates and were co-transfected with wt or mut plasmids (Table 2) by micropoly (Nanjing Mai Ke Rui, China). The luciferase activity was examined using a dualluciferase reporter assay system (Berthold, German).
Co-immunoprecipitation (Co-IP)
MGC-803 cells that transfected with sgRNA-NC/sgRNA-DSC2/Lenti-NC/ High-DSC2, nuclear extraction from total lysis was performed using Subcellular Structure Cytoplasmic and Nuclear Extraction Kit. Half of the supernatant was taken as input sample, and the left used as IP sample. Specific antibodies conjugated with magnetic protein G beads (Santa Cruz Biotechnology, Beijing, China) were added to the IP samples for overnight incubation at 4 °C. Subsequently, the immunocomplexes of β-catenin/TCF4 were recovered and boiled, respectively. Then the expressions of associated proteins were detected by Western blot assay.
Mouse tumor xenograft model and IVIS imaging
Five-week-old Balb/c nude mice were raised under sterile conditions. All mice were purchased from the Animal Center of China Academy of Medical Science (Beijing, China). All animal experiment procedures confirmed to the Institutional Guidelines of Animal Care and Use committee of Shandong University. The mice were divided randomly into four groups (n = 5 per group): miR-32-5p, miR-NC, anti-miR-32-5p and anti-miR-NC group.
Recombinant adenoviral vectors for stably expression of miR-32-5p and anti-miR-32-5p. PCR product was cloned into shuttle plasmid pAdTrack-CMV-EGFP. pAdTrack-miR linearized with PmeI was used to transform BJ5183-AD-1 cells. Colonies were screened by plasmid miniprep. PacI restriction analysis to obtain clones with recombinant miR-32-5p and anti-miR-32-5p. PacI linearized pAdeasy-miR was used to transfect HEK-293 cells to obtain packaged recombinant miR-32-5p and anti-miR-32-5p adenovirus (designated as miR-32-5p and anti-miR-32-5p, respectively). PacI linearized pAdeasy-miR was used to transfect HEK-293 cells to obtain packaged recombinant miR-NC and anti-miR-NC adenovirus was used as control (designated as miR-NC and anti-miR-NC, respectively). The titers and the multiplicity of infection were determined according to the manufacturer’s protocols. 1 × 107 MGC-803 cells from each group were inoculated to the scapular region of each nude mouse. The tumor sizes were measured every three days. The fluorescence signals of xenografts in all mice were taken with the IVIS Kinetic in vivo imaging system at day 24. Then, all mice were euthanized by CO2 and sacrificed through cervical dislocation, then the xenografts were surgically removed and weighed. The long axes and the short axes of removed tumors were determined by vernier caliper measurements. The formula V = L × W2 / 2 was used to determine the tumor volume (V); W refers to the short axis and L refers to the long axis.
All institutional and national guidelines for the care and use of laboratory animals were followed.
Mouse tumor metastasis model and IVIS imaging
We bought five-week-old Balb/c nude mice and raised under sterile conditions. All mice were purchased from the Animal Center of China Academy of Medical Science (Beijing, China). All animal experiment procedures conformed to the Institutional Guidelines of Animal Care and Use committee of Shandong University. The mice were divided randomly into four groups (n = 5): miR-32-5p, miR-NC, anti-miR-32-5p and anti-miR-NC.
Each mouse was injected with 6 × 106 MGC-803 cells via tail vein (cells construction were consist with 2.15). The body weight of all mice were measured every 3 days. At the end, the fluorescence signals of all mice was taken under the IVIS Kinetic in vivo imaging system at day 30. Subsequently, all mice were sacrificed through cervical dislocation, the lungs were surgically removed and were paraffin-embedded for hematoxylin and eosin (HE) staining.
All institutional and national guidelines for the care and use of laboratory animals were followed. All methods were performed in accordance with the relevant guidelines and regulations.
Statistical analysis
All quantitative data are expressed as mean ± standard error of measurement (SEM). T-test analysis were used to make comparisons between two groups, and paired t-test analysis were used to make comparisons in 41 paired GC samples. A P-value < 0.05 was considered statistically significant. Statistical analysis was performed using the SPSS/Win 16.0 software.
Results
MiR-32-5p promoted the viability of GC cells, anti-miR-32-5p inhibited the viability of GC cells in vitro and in vivo
To determine whether miR-32-5p plays a role in the viability of GC cells, MGC-803 cells were transfecting with miR-32-5p agomir (miR-32-5p mimics), then clonal formation, MTT and sperm DNA fragmentation assay were performed. We found that miR-32-5p mimics significantly increased the colony numbers, survival rate and correspondingly reduced DNA fragmentation level of MGC-803 cells (Fig. 1a,c,d). On the opposite, we found that miR-32-5p antagomir (anti-miR-32-5p mimics) significantly reduced the colony numbers, survival rate and correspondingly increased DNA fragmentation level of MGC-803 and SGC-7901 cells (Fig. 1b,e,f, supplementary Fig. 1a,b). These data suggested that miR-32-5p promoted the growth of GC cells, while anti-miR-32-5p suppressed the viability of GC cells in vitro.
MiR-32-5p promoted the viability of MGC-803 cells in vitro. After transfecting with miR-32-5p mimics in MGC-803 cells, the viability of cells was determined by colony formation assay (a), MTT assay (c) and sperm DNA fragmentation assay (d). ***p<0.001 vs. Blank control group (BC). ###p<0.001 vs. miRNA negative control (miR-NC). After transfecting with anti-miR-32-5p mimics in MGC-803 cells, the viability of cells was determined by colony formation assay (b), MTT assay (e) and sperm DNA fragmentation assay (f). ***p<0.001 vs. BC. ###p<0.001 vs. anti-miR-NC. Data above are presented as mean ± SEM (n = 3).
Then, the effects of miR-32-5p or anti-miR-32-5p on the in vivo growth of GC were investigated by establishing mice xenograft models. We measured the xenograft volumes of each group every 3 days (Fig. 2a), and observed the xenograft tumors with the IVIS Kinetic in vivo imaging system at day 24 (Fig. 2b). Subsequently, the xenografts were removed from the mice, the tumor were photographed (Fig. 2c) and the xenografts were weighed (Fig. 2d). We found that the xenograft tumor volumes and weights of the anti-miR-32-5p group were markedly smaller than those in the anti-miR-NC group. On the other hand, xenograft tumor volumes and weights of the miR-32-5p group were significantly larger than those in the miR-NC group. The tumor weights in the miR-NC group, miR-32-5p group, anti-miR-NC group, and anti-miR-32-5p group were (2.80 ± 1.01) g, (4.11 ± 0.37) g, (2.64 ± 1.09) g, and (1.06 ± 0.77) g, respectively. These data suggested that miR-32-5p promoted the growth of GC, while anti-miR-32-5p suppressed the viability of GC in vivo.
MiR-32-5p promoted the growth of GC in vivo. (a) Tumor volume of mice were measured every 3 days. (b) Representative IVIS imaging for scapular bearing tumor. At the day 24, mice were sacrificed and tumors were surgically removed, photoed (c), weighed (d). Data above are presented as mean ± SEM (n = 5).
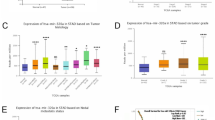
miR-32-5p directly targets DSC2 in GC
Firstly, to analyze the expression of miR-32-5p in GC tissues, the data repositories of the Cancer Genome Atlas (TCGA) were used to obtain 41 paired GC samples. We found that miR-32-5p was upregulated in GC (Fig. 3a). Then, the expression levels of miR-32-5p in GC MGC-823, SGC-7901 and BGC-823 cells, and normal human gastric mucosal cell GES-1 were examined by qRT-PCR assay. The results showed that miR-32-5p expressions increased markedly in all three GC cell lines compared to GES-1 cells (Fig. 3b), which was negatively correlated with the expression of DSC2. Subsequently, we constructed wt-DSC2 3’-UTR and the corresponding mut-DSC2 3’-UTR luciferase reporters (Fig. 3d), and performed luciferase activity assays in MGC-803 cells. The results showed that miR-32-5p reduced the luciferase activity of wt-DSC2 3’-UTR but not that of mut-DSC2 3’-UTR (Fig. 3c). To determine whether DSC2 was regulated by miR-32-5p, MGC-803 cells were transfected with miR-32-5p mimics or anti-miR-32-5p mimics, then the protein and gene expression of DSC2 were measured by Western blot and qRT-PCR assay. The results showed that overexpression of miR-32-5p downregulated DSC2 protein expression in GES-1 cells, whereas silencing of miR-32-5p upregulated DSC2 protein expression in MGC-803 cells (Fig. 3e-f). Neither miR-32-5p nor anti-miR-32-5p affected the mRNA of DSC2 in GC cells (Fig. 3g-h). These data suggested DSC2 might be a direct target gene of miR-32-5p.
MiR-32-5p directly targets DSC2 in GC. (a) Comparison of miR-32-5p expression in 41 paired samples collected from the TCGA database. ∗∗∗P < 0.001. (b) MiR-32-5p expressions in MGC-803, SGC-7901, BGC-823 cells and normal gastric GES-1 cells were detected by qRT-PCR assay. ***p<0.001 vs. GES-1 cells. (c) The binding of DSC2 and miR-32-5p were determined by luciferase reporter assays using co-transfection of luciferase plasmids with wild type (wt)/mutant (mut) DSC2 and miR-32-5p mimics/miR-NC in MGC-803 cells. ***P<0.001 vs. miR-NC. (d) Mut or wt DSC2 fragments with putative binding sites to miR-32-5p were constructed. After transfecting with miR-32-5p mimics in GES-1 cells, the expression of DSC2 were performed by Western blot assay (e) and qRT-PCR assay (f). ***p<0.001 vs. BC. ###p<0.001 vs. miR-NC. After transfecting with anti-miR-32-5p mimics in MGC-803 cells, the expression of DSC2 were performed by Western blot assay (g) and qRT-PCR assay (h). ***p<0.001 vs. BC. ###p<0.001 vs. anti-miR-NC. Data above are presented as mean ± SEM (n = 3).
MiR-32-5p promoted the viability of MGC-803 cells through downregulating the expression of DSC2
To further explore the mechanism of miR-32-5p in promoting the viability of GC cells, MGC-803 cells (Lenti-NC/High-DSC2) were transfected with miR-32-5p mimics, then MTT, caspase-3 activity, sperm DNA fragmentation assays and flow cytometry assay were performed. The results showed that miR-32-5p increased survival rate and correspondingly reduced DNA fragmentation level and caspase-3 activity in both Lenti-NC and High-DSC2 group (Fig. 4a–c). At the same time, miR-32-5p mimics alleviated the G2/M arrest in both Lenti-NC and High-DSC2 group (Fig. 4d). The data above suggested that miR-32-5p promoted the viability of MGC-803 cells at least partly through reducing the expression of DSC2.
MiR-32-5p promoted the viability of MGC-803 cells through downregulating the expression of DSC2. After transfecting with miR-32-5p mimics in MGC-803 cells with over-expressed (High-DSC2) or normal-expressed (Lenti-NC) DSC2, the viability of cells was determined by MTT assay (a), sperm DNA fragmentation assay (b) and caspase-3 activity assay (c). ***p<0.001 vs. Lenti-NC. ###p<0.001 vs. High-DSC2. After transfecting with miR-32-5p mimics in MGC-803 cells (Lenti-NC/High-DSC2), the viability of cells was determined by flow cytometry assay (d). **p<0.01 vs. Lenti-NC + miR-NC. ###p<0.001 vs. miR-NC + High-DSC2. Data above are presented as mean ± SEM (n = 3).
Subsequently, MGC-803 cells (Lenti-NC/High-DSC2) were transfected with anti-miR-32-5p mimics, and MTT, caspase-3 activity, sperm DNA fragmentation assays and flow cytometry assay were performed. The results showed that anti-miR-32-5p mimics reduced survival rate and correspondingly increased DNA fragmentation level and caspase-3 activity in both Lenti-NC and High-DSC2 group, while anti-miR-32-5p mimics neither further reduced the survival rate nor increased caspase-3 activity and DNA fragmentation level in High-DSC2 group (Fig. 5a–c).
Anti-miR-32-5p inhibited the viability of MGC-803 cells through upregulating the expression of DSC2. After transfecting with anti-miR-32-5p mimics in MGC-803 cells (Lenti-NC/High-DSC2), the viability of cells was determined by MTT assay (a), sperm DNA fragmentation assay (b) and caspase-3 activity assay (c). ***p<0.001 vs. Lenti-NC. After transfecting with anti-miR-32-5p mimics in MGC-803 cells (Lenti-NC/High-DSC2), the viability of cells was determined by flow cytometry assay (d). **p<0.01 vs. anti-miR-NC + Lenti-NC. Data above are presented as mean ± SEM (n = 3).
Furthermore, transfected with anti-miR-32-5p mimics and Lenti-DSC2 ( High-DSC2) both increased the G2/M arrest, but anti-miR-32-5p mimics didn’t further increased the G2/M arrest of High-DSC2 group (Fig. 5d). The data above suggested that anti-miR-32-5p inhibited the viability of MGC-803 cells partly through upregulating the expression of DSC2.
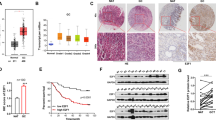
DSC2 downregulated Cyclin B1 through inhibiting the complex of β-catenin/TCF4 in nucleus
We further explore the mechanisms of DSC2 in regulating the cell cycle of GC cells, and Western blot, Co-IP, qRT-PCR and luciferase reporter assays were performed. First, we found that the expression of DSC2 was downregulated, while Cyclin B1 was increased after transfecting with miR-32-5p mimics in both Lenti-NC and High-DSC2 group (Fig. 6a). Anti-miR-32-5p mimics had the opposite effect on the expression of DSC2, Cyclin B1 in Lenti-NC group, but didn’t further affect the expression of DSC2 and Cyclin B1 in High-DSC2 group (Fig. 6b). Then, Co-IP assay results showed that the complex of TCF4/β-catenin increased in the nuclei after transfecting with sgRNA-DSC2 (Fig. 6c), while MGC-803 cells that overexpressed DSC2 gene showed the opposite effects (Fig. 6d).
MiR-32-5p regualtes DSC2/Cyclin B1 signaling pathway. After transfecting with miR-32-5p mimics or anti-miR-32-5p mimics in MGC-803 cells (Lenti-NC/High-DSC2), the expression of DSC2 and Cyclin B1 were performed by Western blot assay . ***p<0.001 vs. Lenti-NC + miR-NC, ###p<0.001 vs. High-DSC2 (a). ***p<0.001 vs. anti-miR-NC + Lenti-NC (b). After knocking down (sgRNA-NC/sgRNA-DSC2) or stably overexpressing DSC2 gene (Lenti-NC/High-DSC2) in MGC-803 cells, the β-catenin/TCF4 complex in the nuclues was detected by Co-IP assay. ***p<0.001 vs. sgRNA-NC (c). ***p<0.001 vs. Lenti-NC (d). After treating with LF3, the expression of Cyclin B1 in MGC-803 cells (sgRNA-NC/sgRNA-DSC2) were performed by Western blot assay (e) and qRT-PCR assay (f). ***p<0.001 vs. sgRNA-NC. ###p<0.001 vs. sgRNA-DSC2. After knocking down DSC2 gene in MGC-803 cells, the binding of TCF4 and the promoter of Cyclin B1 were determined by luciferase reporter assays using co-transfection of luciferase plasmids with wt or mut Cyclin B1 promoter (g). ***P<0.001 vs. sgRNA-NC. (h) The expressions of DSC2 and Cyclin B1 in tumor xenograft tissues were detected by Western blot assay. ***p<0.001 vs. miR-NC. ###p<0.001 vs. anti-miR-NC. Data above are presented as mean ± SEM (n = 3).
Following, MGC-803 cells (sgRNA-NC/sgRNA-DSC2) were treated with the TCF4/β-catenin inhibitor LF3 (30 μM), and the expression of Cyclin B1 were performed with Western blot and qRT-PCR assay. The results showed that the protein level of Cyclin B1 in both sgRNA-NC and sgRNA-DSC2 group were suppressed after treating with LF3, without regulating the level of Cyclin B1 mRNA (Fig. 6e,f). Subsequently, we constructed wt-Cyclin B1 promoter and the corresponding mut-Cyclin B1 promoter luciferase reporters, and performed luciferase activity assays in MGC-803 cells (sgRNA-NC/sgRNA-DSC2). The results showed that downregulated the expression of DSC2 reduced the luciferase activity of wt-Cyclin B1 promoter, but not that of mut-Cyclin B1 promoter (Fig. 6g). The results suggested that TCF4 directly combined with Cyclin B1 promoter.
At last, along with the expression of DSC2 increased, while Cyclin B1 was reduced in the xenograft tumor tissues of anti-miR-32-5p group. We observed the opposite effect in the tissues of miR-32-5p group (Fig. 6h). These data suggested that miR-32-5p promoted the growth of GC through regulating DSC2/Cyclin B1 signaling pathway. While anti-miR-32-5p increasd G2/M arrest of GC cells through upregulating the expression of DSC2, reducing the complex of TCF4/β-catenin in the nuclei, and suppressing the transcription of Cyclin B1, mainly through inhibiting the combination of the TCF4/β-catenin complex with Cyclin B1 promoter.
MiR-32-5p promoted the migration, invasion and adhesion of GC cells through downregulating the expression of DSC2 in vitro and in vivo
Next, we aimed to detect whether miR-32-5p or anti-miR-32-5p influence the migratory, invasive and adhesion behavior of GC cells, and transwell migration, transwell invasion and adhesion assay were performed. We found that the amount of cells that passed the chamber with or without Matrigel, and the adherent cells significantly reduced after transfecting with anti-miR-32-5p in both MGC-803 and SGC-7901 cells (Supplementary Figs. 1c–e, 2a–c). The data above suggested that miR-32-5p promoted the migration, invasion and adhesion of GC cells.
Subsequently, the results showed that the amount of cells that passed the chamber with or without Matrigel, and the adherent cells didn’t markly reduced after transfecting with anti-miR-32-5p in High-DSC2 group (Supplementary Fig. 3a–c). The Western blot assay results showed that the expression of EMT associated proteins, such as N-cadherin, MMP9 and CD44 were downregulated after transfecting with anti-miR-32-5p mimics in Lenti-NC group, but didn’t affect these proteins expression of High-DSC2 group (Supplementary Fig. 3d). Moreover, the amount of cells that passed the chamber with or without Matrigel, and the adherent cells significantly increased after transfecting with miR-32-5p mimics in both Lenti-NC and High-DSC2 group (Fig. 7a-c). In addition, the expression of N-cadherin, MMP9 and CD44 were upregulated after transfecting with miR-32-5p mimics in both Lenti-NC and High-DSC2 group (Fig. 7d). The data above suggested that miR-32-5p promoted the migration, invasion and adhesion of GC cells partly through downregulating the expression of DSC2.
MiR-32-5p promoted the migration, invasion and adhesion of GC through downregulating the expression of DSC2. After transfecting with miR-32-5p mimics in MGC-803 cells (Lenti-NC/High-DSC2), the migration and invasion were detected by transwell migration assay (a, 200×), transwell invasion assay (b, 200×) and adhesion assay (c, 200×). The expression of DSC2, N-cadherin, MMP9 and CD44 were performed by Western blot assay (d). ***p<0.001 vs. miR-NC. ###p<0.001 vs. High-DSC2. Data are presented as mean ± SEM (n = 3).
Next, we detected the effect of miR-32-5p and anti-miR-32-5p on the lung metastasis of GC cells in nude mice by tail vein injection of MGC-803 cells (miR-32-5p/miR-NC/anti-miR-32-5p/anti-miR-NC). Results showed that compared to miR-NC group, more micro-/macro- metastatic pulmonary lesions were generated in the mice of miR-32-5p group through the IVIS Kinetic in vivo imaging system and hematoxylin–eosin (HE) staining, while anti-miR-32-5p had the opposite effect (Supplementary Fig. 4a,b). At the same time, the body weight of nude mice was negatively associated with the degree of lung metastasis (Supplementary Fig. 4c). These suggested that miR-32-5p promoted the lung metastasis of MGC-803 cells in vivo.
Discussion
GC is the most frequent and the third most leading cause of cancer-linked death worldwide1. It is urgent to identify the gene expression patterns and biomarkers in GC to advanced GC research and improve patient survival14. Emerging evidences showed that miRNAs could regulate the stemness characteristics of GC cells, and anti-microRNAs agents has been confirmed to be the new strategy for clinical anti-tumor therapy15,16. Therefore, investigation of the biological functions of dysregulated miRNAs in GC may contribute to the identification of novel biomarkers and therapeutic strategies for patients with advanced GC.
DSC2 is the key component of functional desmosomal junction and also the only isoform of DSCs expressed in normal gastric tissue17,18, being essential for the gastric epithelial morphogenesis19. In our previous research, we found that DSC2 was significantly downregulated in GC tissues, DSC2 deletion played a key role in the development of GC, and up-regulating the expression of DSC2 may be a potential strategy for the treatment of advanced GC.
In general, miRNAs exert gene regulatory expression, and have great role in the progression of many cancers through complete or incomplete complementary pairing with the 3′-UTR of target genes20,21. The role of miR-32-5p in the pathogenesis of cancers is still controversial. In the previous work, we screened out miR-205-5p, miR-25-3p and miR-32-5p from databases, which may regulate the expression of DSC2 in GC cells. Q-PCR results indicated that miR-32-5p downregulated the expression of DSC2 gene, while neither miR-205-5p nor miR-25-3p significantly regulated the expression of DSC2 gene (supplement Fig. 2d-e). MiR-32-5p was upregulated in colorectal cancer, pancreatic cancer tissues22,23, but downregulated in lung adenocarcinoma tissues24. To the best of our knowledge, this work first demonstrated miR-32-5p-mediated cancer-promoting activity in GC through downregulating the expression of DSC2, the mechanism was mainly through complementary pairing with the 3′-UTR of DSC2.
In this work, we found miR-32-5p not only promoted the viability and metastasis of GC through suppressing the expression of DSC2, but also alleviated the G2/M arrest base on overexpression of DSC2 through inhibiting the expression of Cyclin B1.
Be universally known, Cyclin B1 is the key regulatory subunit for gastric cells that leading G2/M phase transition. Unfortuantely, the mechanism that regulating the expression of Cyclin B1 is still unclear. In the previous study, we found that DSC2 suppressed the nucleus translocation of β-catenin. β-Catenin also has been shown to translocate to the nucleus, binded to the TCF/LEF families in the nuclei of GC cells, and regulated the transcription of target genes25. As far as we know, this work first demonstrated that the expression of Cyclin B1 was upregulated in GC cells after downregulating the expression of DSC2, mainly through increasing the complex of β-catenin/TCF4 in the nuclei.
Although there are important discoveries revealed by this study, some limitations of its are miRNAs have multiple mRNA targets. The effect of miR-32-5p in promoting the viability and migration of GC cells cannot be blamed on DSC2 downregulation alone. Target protectors which would mask the miRNA binding site of miR-32-5p in the 3’ UTR of DSC2, might be attributed to targeting of DSC2 alone and not the other targets of miR-32-5p.
In conclusion, anti-miR-32-5p suppressed the tumorigenicity of GC cells at least partly by targeting DSC2. Based on our findings, we first identified miR-32-5p- mediated cancer-promoting activity in GC through downregulating the expression of DSC2 mainly through complementary pairing with the 3′-UTR of DSC2. At the same time, we first demonstrated that the expression of Cyclin B1 was upregulated in GC cells after downregulating the expression of DSC2 through activating the transcriptional activity of β-catenin/TCF4. These point out the vulnerable parts of anti-miR-32-5p for the therapeutic targeting of GC.
Data availability
The data that support the findings of this study are available from the corresponding author, [ttsky313@163.com], upon reasonable request.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Wei, L. et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol. Cancer 19, 62 (2020).
Joshi, S. S. & Badgwell, B. D. Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 71, 264–279 (2021).
Liu, Q. et al. Construction of a circular RNA-microRNA-messengerRNA regulatory network in stomach adenocarcinoma. J. Cell Biochem. 121 (2), 1317–1331 (2020).
Yu, L. et al. Complete loss of miR-200 family induces EMT associated cellular senescence in gastric cancer. Oncogene 41 (1), 26–36 (2022).
Eto, K. et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int. J. Cancer 136 (7), 1537–1545 (2015).
Khan, K. et al. Desmocollin switching in colorectal cancer. Br. J. Cancer 95 (10), 1367–1370 (2006).
Kurinna, S. et al. A novel Nrf2-miR-29-desmocollin-2 axis regulates desmosome function in keratinocytes. Nat. commun. 5, 5099–5102 (2014).
Lowndes, M. et al. Different roles of cadherins in the assembly and structural integrity of the desmosome complex. J. Cell Sci. 127 (10), 2339–2350 (2014).
Sun, C., Wang, L., Yang, X. X., Jiang, Y. H. & Guo, X. L. The aberrant expression or disruption of desmocollin2 in human diseases. Int. J. Biol. Macromol. 131, 378–386 (2019).
Sun, C. et al. DSC2 suppresses the growth of gastric cancer through the inhibition of nuclear translocation of γ-catenin and PTEN/PI3K/AKT signaling pathway. Aging (Albany NY) 15, 1–20 (2023).
Demircan, T., Sibai, M., Avşaroğlu, M. E., Altuntaş, E. & Ovezmyradov, G. The first report on circulating microRNAs at pre- and post-metamorphic stages of axolotls. Gene 768, 145258 (2021).
Jin, Y. & Wang, H. Circ_0078607 inhibits the progression of ovarian cancer via regulating the miR-32-5p/SIK1 network. J. Ovarian Res. 15 (1), 3 (2022).
Jiang, T. et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 20 (1), 66 (2021).
Dogra, P. et al. Translational modeling-based evidence for enhanced efficacy of standard-of-care drugs in combination with anti-microRNA-155 in non-small-cell lung cancer. Mol. Cancer 23 (1), 156 (2024).
Ruggieri, V. et al. The role of microRNAs in the regulation of gastric cancer stem cells: a meta-analysis of the current status. J. Clin. Med. 8 (5), 639 (2019).
Holthöfer, B., Windoffer, R., Troyanovsky, S. & Leube, R. E. Structure and function of desmosomes. Int. Rev. Cytol. 264, 65–163 (2007).
Pozo, F. M., Hunter, T. & Zhang, Y. The ‘New(Nu)-clear’ evidence for the tumor-driving role of PI3K. Acta Mater. Med. 1 (2), 193–196 (2022).
Nekrasova, O. E., Amargo, E. V., Smith, W. O., Chen, J. & Kreitzer, G. E. Green KJ desmosomal cadherins utilize distinct kinesins for assembly into desmosomes. J. Cell Biol. 195 (7), 1185–1203 (2011).
Chen, D. L. et al. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol. Cancer 20 (1), 166 (2021).
Gao, Z., Jiang, J., Hou, L. & Zhang, B. Dysregulation of MiR-144-5p/RNF187 axis contributes to the progression of colorectal cancer. J. Transl. Intern. Med. 10 (1), 65–75 (2022).
Liang, H., Tang, Y., Zhang, H. & Zhang, C. MiR-32-5p regulates radiosensitization, migration and invasion of colorectal cancer cells by targeting TOB1 gene. Onco Targ. Ther. 12, 9651–9661 (2019).
Gao, Z. Q. et al. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 7, 66 (2017).
Zhang, J. X. et al. MicroRNA-32-5p inhibits epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting SMAD family 3. J. Cancer 12 (8), 2258–2267 (2021).
Wang, Y. et al. Wnt/beta-catenin signaling confers ferroptosis resistance by targeting GPX4 in gastric cancer. Cell Death Differ. 29 (11), 2190–2202 (2022).
Funding
This work was supported by Qilu Special Clinical Research Fund Of Shandong Provincial Medical Association (No. YXH202202177), Cultivation Fund of the Second Hospital of Shandong University (No. 2022YP69).
Author information
Authors and Affiliations
Contributions
Conceptualization: D.Q.S., and C.S.; Data curation: L.G.H., C.C., and T.T.G.; Formal analysis: L.G.H., G.Y.T, and Y.D.; Funding acquisition: C.S.; Methodology: L.G.H., Y.D., R.J.L., and T.T.G.; Writing-original draft: C.S., B.L., and L.G.H.; Writing-review and editing: D.Q.S. and T.T.G.; Supervision: D.Q.S. and T.T.G.; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Written informed consent was obtained from all participants. All the experiments were approved by the Ethics Committee of the Second Hospital of Shandong University. All methods were carried out in accordance with relevant guidelines and regulations, and all methods are reported in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, C., Huang, Lg., Leng, B. et al. MicroRNA-32-5p promotes the proliferation and metastasis of gastric cancer cells. Sci Rep 15, 2282 (2025). https://doi.org/10.1038/s41598-025-86367-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-86367-3