Abstract
Macaques are important reservoirs of zoonotic malaria in Southeast Asia. Although cross-sectional malaria surveys have been conducted in macaques, little is known about intra-host infection dynamics and host variation in susceptibility to infection in these infectious reservoirs. We performed a longitudinal monitoring of Plasmodium and Hepatocystis infections by microscopy, species-specific polymerase chain reaction (PCR) and targeted amplicon deep sequencing (TADS) in three long-tailed macaques and 20 pig-tailed macaques in two districts of Narathiwat Province, southern Thailand. In total, 104 macaques’ blood samples were obtained during 5 visits with sequential time intervals of 9, 4, 7 and 12 months. Transiently patent Plasmodium infections with low parasite density ( ≤ 1,050 parasites/µL) occurred in 7 pig-tailed macaques, while PCR and TADS diagnosed infections in 45 (43.27%) blood samples with one or more species of parasites, including Plasmodium knowlesi, P. cynomolgi, P. inui, P. fieldi, P. coatneyi, P. aff. coatneyi and Hepatocystis sp. in one long-tailed and 12 pig-tailed macaques. Compared with PCR, TADS additionally detected co-infecting species in 22 of 45 ( 48.89%) samples. Although living in close proximity to other infected macaques, seven macaques were free from infection during the 32-month period. Infections for 4 to 32 months with malaria parasites carrying identical complete mitochondrial genome sequences were reaffirmed in 10 macaques. Potentially new infections were detected transiently or over a long period during the course of the infections while competitive exclusion seemed to occur between Hepatocystis sp. and Plasmodium taxa. Macaques’ Duffy phenotypes did not influence differential susceptibility to Plasmodium infections. These results suggest the ecological complexity of hemoparasite infections in natural reservoirs of zoonotic malaria. The long period of Plasmodium infections in macaques could affect the transmission and control of the disease.
Similar content being viewed by others
Introduction
In Southeast Asia, zoonotic malaria is an important emerging infection amid a decline in malaria prevalence in this region. Unlike Malaysian Borneo, where the nonhuman primate malaria parasite Plasmodium knowlesi is predominantly identified in humans, zoonotic malaria in Thailand has a relatively low prevalence1,2,3,4. It was not until the turn of the decade that P. knowlesi was increasingly identified among malaria patients in Thailand, while P. vivax remarkably outnumbered P. falciparum5. More importantly, other nonhuman primate malaria parasites known to circulate among long-tailed macaques (Macaca fascicularis) and pig-tailed macaques (M. nemestrina) including P. cynomolgi, P. inui, and P. fieldi have recently been diagnosed as the causative agents of human malaria in Thailand6,7. Furthermore, P. coatneyi and probably other species have been detected among inhabitants residing in the vicinity of natural macaque hosts on the Malaysian Peninsula8. The importance of zoonotic malaria in Southeast Asia in terms of ecological, molecular, and epidemiological perspectives has been recently reviewed9,10,11.
Although the zoonotic transmission of nonhuman primate Plasmodium species from naturally infected macaques to humans via mosquito vectors is undisputed, it has been suggested that simian malaria in humans may not be strictly zoonotic12,13. The occurrence of morphologically mature microgametocytes and macrogametocytes of P. knowlesi in the circulations of infected patients2,14 and the transmission of infections from humans to mosquitoes to humans under experimental conditions have suggested biologically possible anthroponotic transmission15,16,17. However, the lack of indigenous cases of zoonotic malaria in humans outside the geographic ranges of naturally infected macaques has underscored the importance of the acquisition of disease from zoonotic transmission13,18.
During the invasion of erythrocytes by malarial merozoites, a number of parasite ligands and host cell receptors are involved in this complex process. In P. vivax, transferrin receptor 1 (CD71), a reticulocyte-specific receptor, and Duffy antigen receptor for chemokines (DARC) act as key invasion molecules in human erythrocytes19,20. Like P. vivax, P. knowlesi merozoites use DARC to trigger internalization into red blood cells20,21. Analysis of the Plasmodium interaction domain located in the N-terminal region of DARC has shown sequence conservation in the human population, while high levels of sequence divergence occur between DARC orthologs in nonhuman primate species22. Therefore, it is likely that the polymorphism at the DARC locus could play a role in the differential susceptibility to simian malaria parasites among primates.
To date, little is known about the temporal changes of natural Plasmodium infections in long-tailed and pig-tailed macaques that serve as the main reservoirs for zoonotic malaria in Southeast Asia, including Thailand. All macaques in Thailand are arboreal and live in troops of varying size. However, they are equally at home on the ground and commonly reside in close proximity to human settlements due to forest destruction. In certain communities, such as local villages in southern Thailand, long-tailed and pig-tailed macaques are kept as pets and are also deployed for coconut picking. These domesticated macaques remain susceptible to simian malaria due to the coexistence of infected wild macaques and susceptible mosquito vectors in the regions23,24,25. To investigate whether there was a chronological variation in Plasmodium infections among domesticated macaques, we performed a longitudinal surveillance of macaques’ blood samples from two nearby districts in Narathiwat Province in southern Thailand using microscopy detection, species-specific polymerase chain reaction (PCR) diagnosis, and targeted amplicon deep sequencing (TADS). The phenotype and genotype of the DARC genes of these macaques were also analyzed. The results revealed that Plasmodium infections in macaques exhibited ecological complexity, while persistent infections throughout the study period highlight the importance of these reservoir hosts as a long-term storage source for the transmission of zoonotic malaria.
Results
Macaques and blood sample collection
Twenty pig-tailed macaques (Mn) and 3 long-tailed macaques (Mf) of 20 households located in three local geographic clusters in Sukhirin and Waeng Districts were longitudinally monitored for Plasmodium and Hepatocystis infections (Fig. 1). Of these, 20 were females (87%), the age ranged from 2 years to 6 years (median, 4 years) comprising 5 juveniles and 18 subadults. The weight ranged from 2 kg to 10 kg (mean ± S.D., 5.95 ± 2.36 kg). During a 32-month period, multiple blood samples were taken from each macaque: twice (n = 2), three times (n = 1), four times (n = 3) and five times (n = 17). The intervals between blood sample collections spanned 9, 4, 7 and 12 months, respectively.
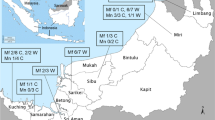
Map of the study area showing 3 clusters of households in (A) Sukhirin and Waeng Districts in Narathiwat Province (yellow area in an inset), Thailand. B, C, and D indicate the locations of the households corresponding to the 3 clusters shown in A. The images were obtained from Google Earth Pro Software version 7.3.6.9345 (https://www.google.com/intl/en_uk/earth/versions/#earth-pro) under the Google Privacy Policy and Terms of Service46. The boundaries of districts and the locations of households were created using Adobe Photoshop 2021.
Microscopy detection
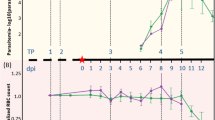
Plasmodium infections were determined by microscopic examination of thin and thick blood smears for 104 blood samples throughout the study period. Six of 23 macaques, five in cluster 3 and one in cluster 2, had patent parasitemia in one or more blood samples during the sampling period (Fig. 2). In total, 11 of 104 (10.58%) blood samples had patent parasitemia. The density of patent infections ranged from 50 to 1,050 parasites/µL of blood (mean ± S.D.: 331.8 ± 345.2 parasites/µL) in which the ring stages were detected in all microscopy-positive samples; two of these had coexisting growing trophozoites (Supplemental Fig. S1). However, species identification by microscopy was not possible. All blood samples showing patent parasitemia belonged to subadult macaques (age > 3 years). Microscopy did not detect Plasmodium infections in 17 of 23 macaques (73.91%).
Distribution of Plasmodium taxa and Hepatocystis sp. diagnosed by PCR and TADS in 23 macaques. Cross marks indicate no amplicon for TADS analysis. Unavailable samples are shown as blank areas. Plasmodium species are displayed in corresponding colors to the key colors on the upper panel. Negative results are unfilled boxes. An oval circle inside the filled box indicates the sample with the complete mitochondrial genome sequence. Kn, Cy, In, Fi, Co, Sp and He represents P. knowlesi, P. cynomolgi, P. inui, P. fieldi, P. coatneyi, P. aff. coatneyi and Hepatocystis sp., respectively. Boxes with red outside border indicate microscopy-positive samples. Asterisks, hashtags and silcrows indicate macaques in the same households.
Plasmodium species-specific PCR
Species-specific PCR targeting Plasmodium and Hepatocystis mitochondrial cytochrome c oxidase subunit I (cox1 nested PCR) revealed that 37 of 104 blood samples were infected with one or more species of Plasmodium and 10 blood samples contained Hepatocystis. During 32 months of follow-up blood samples, cox1 nested PCR detected Plasmodium infections in 12 pig-tailed macaques and one long-tailed macaque, while three pig-tailed macaques were coinfected with Plasmodium and Hepatocystis. In total, 45 of 104 (43.27%) blood samples yielded positive results from cox1 nested PCR analysis. Of these, P. cynomolgi was most frequently detected (n = 22, 21.15%), followed by P. inui (n = 19, 18.27%). P. knowlesi (n = 16, 15.38%), P. feldi (n = 13, 12.50%), P. coatneyi (n = 11, 10.58%) and Hepatocystis (n = 10, 9.62%), respectively (Fig. 2).
Targeted amplicon deep sequencing
The cox1 gene fragments and the apicoplast caseinolytic protease C gene (clpc) fragments from primary PCR products were used as templates for targeted amplicon deep sequencing (TADS) with the Illumina HiSeq2500-PE150 platform. Therefore, no result from TADS was available for all cox1-nested PCR-negative samples. TADS could detect all the corresponding Plasmodium species and Hepatocystis identified by cox1 nested PCR. However, TADS diagnosed additional coinfecting species in 22 blood samples, including the identification of a previously reported Plasmodium species closely related to P. coatneyi26 which is here referred to as ‘P. aff. coatneyi’. Therefore, the prevalence of hemoparasites diagnosed with TADS increased accordingly: P. cynomolgi (n = 27, 25.96%), P. coatneyi (n = 22, 21.15%), P. fieldi (n = 21, 20.19%), P. inui (n = 21, 20.19%), P. knowlesi (n = 17, 16.35%), Hepatocystis (n = 11, 10.58%) and P. aff. coatneyi (n = 4, 3.85%) (Figs. 2, 3). In total, 7 of 23 macaques were not infected with Plasmodium and/or Hepatocystis according to the PCR and TADS analyses.
Genetic diversity of malaria parasites
To investigate whether strain variations occurred between malaria parasites and Hepatocystis sp. infecting macaques, complete mitochondrial genome sequences of each parasite species were analyzed using DNA templates obtained from two or more blood samples from different periods of collection. For P. knowlesi, two pig-tailed macaques Mn1-3 and Mn3-11 in clusters 1 and 3, respectively, displayed different sequences. However, identical complete mitochondrial genome sequences were observed between different periods of blood sample collection of each monkey: the second and third blood samples from Mn1-3, and the first and fourth blood samples from Mn3-11. Similarly, complete mitochondrial genomes of P. cynomolgi could be determined from the third and fourth blood samples and the third and fifth blood samples of pig-tailed macaques in cluster 3, Mn3-6, and Mn3-11, respectively, whose sequences showed perfect identity within monkeys but different between monkeys. Prolonged infections of P. inui carrying the same complete mitochondrial genome sequences occurred in 4 monkeys Mn3-3 (the first, second, and fifth samples), Mn3-5 (all five samples), Mn3-10 (the first, second and third samples) and Mn3-11 (the second, fourth, and fifth samples).). Interestingly, the same sequences of P. inui were observed only between isolates from monkeys Mn3-3 and Mn3-5. The same complete mitochondrial sequences of P. aff. coatneyi infecting pig-tailed macaque Mn2-4 in cluster 2 were observed in the third, fourth, and fifth blood samples. However, the fourth blood sample also contained a variant sequence. Longitudinal infections with Hepatocystis carrying the same mitochondrial sequences were detected in three monkeys Mn2-2, Mn3-8 and Mn3-9, while infection in each macaque was due to the parasite harboring different sequences. Furthermore, additional distinct sequences were obtained from single blood samples infected with P. cynomolgi (n = 3), P. fieldi (n = 1), P. inui (n = 1) and Hepatocystis (n = 2). The phylogenetic relationship among these mitochondrial genomes is depicted in Fig. 4.
Maximum likelihood tree inferred from the complete mitochondrial genome sequences of Plasmodium and Hepatocytstis. Sequences from clones/isolates in this study are marked with blue dots. Monkeys in clusters 1, 2 and 3 are shown in green, blue and red alphabets, respectively. Sequences obtained from the first to the fifth samples are indicated by A to E, respectively, after the code names for macaques. Numbers on the branches represent the percentage of 1000 bootstrap pseudoreplicates supporting the branch; only values > 50% are shown. GenBank accession numbers of the reference sequences are in parentheses.
Temporal dynamics of Plasmodium and Hepatocystis infections
The results from TADS were considered to represent the status of malaria and Hepatocystis infections in these macaques. Variations in the prevalence of each Plasmodium species and Hepatocystis sp. were observed throughout the study period (Fig. 5). In cluster 1, infection with P. knowlesi occurred in a pig-tailed macaque, while the others were free from infection. Six out of nine monkeys in cluster 2 and nine out of 11 monkeys in cluster 3 were infected with hemoparasites. Two macaques (Mf2-1 and Mn2-2) kept in the same household in cluster 2 were infected with different hemoparasites. Two households in cluster 3, each of which kept two monkeys, had different profiles of hemoparasite infections (Fig. 2). While the prevalence of each hemoparasite fluctuated during the study period, the occurrence of P. aff, coatneyi remained at approximately 5% of total cross-sectional blood samples. It is noteworthy that seven macaques, 2 in cluster 1, 3 in cluster 2 and 2 in cluster 3, were not infected throughout the 32-month period. The status of infections in macaques from the same household differed. Long-term infections for 4 or more months with the same parasite species or strains occurred in 13 of 16 infected monkeys (81.25%) (Fig. 2). All malaria species identified here and Hepatocystis could be detected with TADS from two or more blood samples from the same monkeys collected 4 or more months apart. Coinfection of P. knowlesi, P. cynomolgi, P. inui, P. fieldi and P. coatneyi that persisted for at least 32 months was observed in a pig-tailed macaque in cluster 3 (Mn3-11). In this monkey, identical sequences of the corresponding complete mitochondrial genomes of P. knolwesi, P. cynomolgi, and P. inui were detected from two or more different blood samples. Meanwhile, a transient occurrence of infection with different malaria species could also be observed. For example, macaque Mn3-7 was infected with P. inui and P. fieldi during the third collection of blood samples, while the previous and subsequent blood samples did not show evidence of infection. A similar short-term infection was observed in macaques Mn2-3 and Mn3-2. While P. knowlesi, P. cynomolgi and P. fieldi infections persisted for at least 23 months in macaque Mn2-4, P. inui and P. coatneyi were detected only once in the fourth blood sample. Interestingly, P. aff. coatneyi, a newly identified malaria parasite in Thailand, could establish infection for at least 19 months in macaque Mn2-4. Based on daily activities, behaviors, and appetite informed by their owners, all monkeys did not show evidence of illness during the blood sample collection period.
Coinfection
Taking into account the TADS results, mixed infections were identified in 12 of 16 (75%) infected macaques. Of the 104 blood samples examined, 31 of 45 (68.9%) parasite-positive blood samples contained mixed species infections. To investigate whether the proportions of coinfections with two given species differed from single infections with each species, a pairwise analysis was performed. Because the prevalence of P. aff. coatneyi infections was consistently low across the sampling periods, it was excluded from analysis (Fig. 5). The proportion of coinfection with both P. cynomolgi and P. fieldi was significantly higher than the random expectation (chi-square test, p = 0.0281) (Table 1). A similar finding was observed for coinfection with P. cynomolgi and P. coatneyi (p = 0.0281). On the other hand, the proportions of coinfections with Hepatocystis and Plasmodium species were significantly lower than the random expectation (p < 0.0001).
DARC phenotypes and hemoparasites
The DARC phenotypes of 20 pig-tailed macaques and 3 long-tailed macaques were determined by gel testing to read the reaction of agglutination of antigens Fya (FY1) and Fyb (FY2) in macaques’ erythrocytes using the ID system for human erythrocyte blood group detection. Reactivity to each antigen was quantified as null, weakly positive, 1+, 1.5+, 2 + and 3+. Results showed that all macaque blood samples were not reactive to anti-Fya while variation of reactivity to anti-Fyb was observed. The null phenotype Fy(a-b-) was detected in 2 long-tailed macaques that were not infected with malaria parasites and Hepatocystis sp. throughout the follow-up period. However, no evidence of these infections was also observed in pig-tailed macaques whose erythrocytes had 1+, 1.5 + and 2 + reactivity to anti-Fyb. Mixed species infections were found in macaques with different levels of anti-Fyb reactivity (Table 2).
DARC gene sequences
The DARC gene sequences spanning the 5’untranscribed region (5’UTR, 234 bp), exon 1 (21 bp), intron (451–457 bp), and exon 2 (1011–1014 bp) of all long-tailed and pig-tailed macaques were determined. The 5’ UTR was highly conserved, although point mutations were found in monkeys Mf1-1 and Mn2-9 that resulted in heterozygous alleles at positions − 48TG and − 32CT, respectively. The binding site for the transcription factors GATA-1 and GATA-2 was perfectly conserved in all monkeys. A point mutation at -46TC in the GATA-1 sequence that governs transcriptional activity in the erythroid and megakaryocytic lineages27,28 was not detected in the two long-tailed macaques (Mf1-1 and Mf3-1) with the phenotype Fy(a-b-). In the coding region, 26 amino acid positions that were conserved in macaque DARC differed from those in human DARC. Within macaque DARC, amino acid substitutions were identified at five positions: Y28F, I111L, D160G, C224G, and L329P. In total, eight amino acid alleles were identified, characterized by two homozygous and six heterozygous alleles (Table 3). The amino acid substitutions Y28F/S occurred in long-tailed macaques, while the mutations I160L and L329P were found only in pig-tailed macaques. In addition to three long-tailed macaques with null or weak reactivity to anti-Fyb, the levels of antibody reactivity to antigen Fyb seemed to differ in pig-tailed macaques carrying the same DARC alleles (Table 3). Importantly, codon 42 in the N-terminal extracellular domain of all macaque DARC genes encodes aspartic acid, a critical amino acid for reactivity to anti-Fyb. The Plasmodium interaction domain spanning amino acid positions 8–42 (AELSPSTQNSSQLNSEDLWNYSYDGNDSFPDVDYD) of DARC isoform B20 was highly conserved among long-tailed and pig-tailed macaques except for a mutation Y28F/S in two long-tailed macaques that carried the phenotype Fy(a-b-). Despite eight amino acid differences between human and macaque DARC proteins, the Plasmodium interaction domains in the N-terminal domains were predicted to be intrinsically disordered regions (Fig. 6).
Discussion
Natural simian malaria infections in their natural macaque hosts generally run a slow course, causing little or no febrile reaction with very low levels of parasitemia or submicroscopic infection16,29. In patent infection, ring stages are the predominant forms of parasites in peripheral bloods, while symptomatic and severe infections with high parasitemia containing various blood-stage parasites occur in unnatural or accidental hosts including rhesus macaques (M. mullata) and humans2,14,29,30. In this study, most of the infected macaques had submicroscopic parasitemia, while patent infections occurred at very low parasite density. Although we did not systematically assess the health status of these macaques, none had apparent illness as informed by their owners. However, hypoglycemia, anemia, hyperbilirubinemia, and slightly increased levels of alkaline phosphatase and aspartate amino transferase were found in experimentally induced blood-stage infections of P. knowlesi in long-tailed macaques31. It remains unknown whether the acute phase of natural malaria infections in macaques could replicate experimental studies under laboratory conditions. A longitudinal study of P. gonderi and P. mandrilli infections in a cohort of free-ranging mandrills in Gabon has revealed that most infections occurred with low parasite levels, while infections had an impact on skin temperature and neutrophil/lymphocyte ratio. However, the oxidative parameters seemed not to be significantly altered probably due to chronic infections32.
The course of single-species infection of Southeast Asian simian malaria parasites in natural macaque hosts can be inferred from a comprehensive experimental sporozoite inoculation of the long-tailed macaques from Mauritania, a malaria-free area33. These macaques have been shown to be susceptible to seven species of simian malaria parasites, including P. knowlesi, P. cynomolgi, P. inui, P. fieldi, P. coatneyi, P. fragile, and P. gonderi, although they do not serve as a natural host for the latter two species16,29. Once infections were established, persistent fluctuations in parasitemia profiles were observed in infected macaques for more than 3 months unless antimalarial interventions were introduced. Sequential infections with different malaria species after the clearance of previous infections have resulted in patent parasitemia of the newly infecting species with similar prepatent periods for each species33. Although increased levels of parasitemia in these simian malaria parasites occurred after splenectomy, peak levels were significantly lower than those observed in splenectomized macaques, suggesting that factors other than splenic function could also play an important role in maintaining low parasite levels in natural macaque hosts33. These findings seem to represent the course of acute infection, which may differ from the results observed in our study. However, some consistent occurrences were shared between experimentally induced infections and the course of natural infections observed in our study including (i) the fluctuation of parasitemia levels during the course of infections, (ii) the long period of infection without therapeutic intervention, and (iii) the susceptibility to new infection regardless of previous exposure to a different Plasmodium species.
Taking into account the level of parasitemia, it is likely that most infected macaques in our study appeared to have a chronic phase of infection, as shown by a relatively low prevalence of microscopy positive samples compared to molecular detection (10.58% versus 44.23%) and differential prevalence of infections detected by PCR and a more sensitive TADS, consistent with submicroscopic fluctuation of cryptic parasitemia. Despite the oscillation of parasite density around a detection threshold, the identical complete mitochondrial genome sequences of malaria parasites across blood samples collected from different periods from the same monkeys have suggested that the same parasites could be maintained in the macaques for a long period; for example, up to 32 months for P. inui in the monkey Mn3-5 (Fig. 2). Similarly, chronic infection with P. inui for more than two years has been reported from a captive naturally infected long-tailed macaque whose monthly blood samples showed mainly submicroscopic infection34. However, during preexisting infection with one or more malaria species, new infections with different species could occur transiently or appeared to co-circulate with preexisting species for a certain period (e.g. infections in monkeys Mn2-4, Mn3-3 and Mn3-5). Meanwhile, spontaneous resolution of transient infections with malaria parasites or Hepatocystis sp. was observed based on TADS analysis (e.g. infections in monkeys Mn2-6, Mn2-9, Mn3-2, and Mn3-3). Therefore, the course of natural infections with malaria parasites and Hepatocystis sp. among macaques exhibited an ecological complexity similar to those observed in a longitudinal molecular study of P. gonderi and P. mandrilli infections in wild mandrills32.
Our previous study has shown that Hepatocystis sp. and simian Plasmodium taxa coexisted in the same hosts significantly less frequently than expected by chance on the basis of the overall occurrence in the pig-tailed macaque population. On the other hand, coinfection with dual Plasmodium taxa occurred at a frequency not different from infections with each species24. These results suggest that infection with Hepatocystis sp. reduces the likelihood of infection with Plasmodium species and vice versa. Therefore, some sort of competitive exclusion occurs between these taxa. Importantly, the macaque population in this study differs from that of our previous surveys; therefore, intra-host interaction between Hepatocystis sp and Plasmodium taxa has been reaffirmed24. Although the mechanism of intra-host interaction between these taxa is not immediately obvious, it is likely that macaques infected with Hepatocystis sp. may not readily serve as the reservoir for zoonotic malaria.
In this study, we observed that coinfection with P. cynomolgi and P. fieldi as well as coinfection with P. cynomolgi and P. coatneyi occurred significantly more frequently than random expectation, which was not observed in our previous study24. It is unlikely that infection with a Plasmodium species could benefit from coinfection with another species because within-host competition between phylogenetically related parasites that share similar ecological niche and host resources, along with a possible alteration of host environments favored by one over the other parasite can occur35,36. Alternatively, apart from possible artifacts from a small sample size in this study, these coinfections could be due to the coexistence of simian malaria vectors that coincidentally transmit these Plasmodium species in this area. Indeed, our recent longitudinal surveys have shown an abundant coexistence of Anopheles introlatus and An. latens in these regions, while the former is a potential vector for P. cynomolgi, P. knowlesi, and P. inui and the latter vectors P. inui, P. fieldi, and P. coatneyi25. A significantly more frequent occurrence of these coinfections may suggest multiple infective bites from different species of mosquitoes rather than having a cooperative advantage. In this study, seven monkeys consistently gave negative results by microscopy, species-specific PCR and TADS for Plasmodium and Hepatocystis infections in all blood samples collected over a 32-month period (Fig. 2). In addition to the identification of P. aff. coatneyi infections by TADS that were not detected by species-specific PCR assays, additional detection of malaria parasites by TADS may suggest parasitemia below the PCR detection limit.
It should be noted that monkeys kept in the same households were not uniformly infected with the same hemoparasites, although they had a similar chance to be exposed to the same groups of vectors. Although factors other than host preferences by mosquitoes and biting midges could contribute to differential infections with hemoparasites, variation of within-host environments, including erythrocyte receptors for merozoite invasion, could determine host susceptibility to infection. In this study, we determined the DARC genotypes of macaques by DNA sequencing and phenotypes using antibodies to human Fya and Fyb antigens. All pig-tailed macaque erythrocytes in the study population were reactive to anti-Fyb while the levels of reactivity ranged from weak to 2+. In humans, the C265T mutation that encodes the amino acid R89C confers weak reactivity to the Fyb antigen (Fybweak phenotype). However, the changes in amino acids in macaque DARC did not correspond to known residues conferring the phenotypic variation of DARC in human erythrocytes in addition to the conserved aspartic acid at position 42 that defines the Fyb antigen. Meanwhile, a range of anti-Fyb reactivity of pig-tailed macaque erythrocytes occurred in the same DARC alleles (alleles YIGGL/YIGGL, YIGGP/YIGGL and YIDGL/YIGGL) (Table 3). Although the expression levels of the phenotype Fy(a-b+) varied in these macaques, they remained susceptible to Plasmodium infections, particularly P. knowlesi that requires DARC for invasion of erythrocytes21 (Table 3). Although anti-Fya recognizes the epitope containing glycine at position 42, all macaque erythrocytes were not reactive to this antibody due to the conserved aspartic acid at this position. Therefore, the phenotype Fy (a-b-) in two long-tailed macaques does not directly indicate the absence of DARC expression in macaque erythrocytes. The perfectly conserved GATA-1 sequences in all macaques in this study may suggest the expression of DARC in macaque erythrocytes. Despite a small sample size, it is likely that the DARC polymorphism did not confer differential susceptibility to Plasmodium infections in the study population. Although 8 of 35 (22.9%) amino acid differences occurred between the Plasmodium interaction domain of human and macaque DARC, the shared intrinsically disordered region of these proteins could suggest the phenotypic and functional plasticity of receptor-ligand interaction during the process of erythrocyte invasion, particularly during host-switching of malaria parasites22.
Mature gametocytes are indispensable for the transmission of malaria parasites from vertebrate hosts to mosquito vectors. Gametocyte levels in the peripheral circulation seem to oscillate temporally during the course of malaria infection in monkeys16,33. For P. falciparum, gametocytemia exhibits variation in host age, clinical symptom, season, therapeutic intervention, and complexity of infection37. The peak gametocytemia of human and simian malaria parasites occurs periodically around midnight when the feeding period of the mosquito vectors is maximum; thus, the parasites and their vectors behave synchronously to ensure successful transmission31,38,39. However, several lines of evidence have supported that the transmission of submicroscopic gametocytemia during the course of infections with P. falciparum and P. vivax, including asymptomatic carriages, remains infective to their mosquito vectors37,40. Therefore, the occurrence of zoonotic simian malaria parasites in their natural hosts, regardless of the patency of gametocytes in their peripheral circulations, can potentially pose a risk to human infections.
The macaque populations in this study were chosen on the basis of the relatively higher prevalence of zoonotic malaria in humans in Narathiwat Province than in other regions of the country3,4,7 and the presence of known anopheline vectors for simian malaria in the areas where the macaques were domesticated25. Macaques in Sukhirin and Waeng Districts were chosen based on the cooperativeness of their owners and accessibility to the study areas. Macaque blood samples were collected longitudinally with an interval of at least 4 months to ensure the possibility of covering the period of complete vector-host transmission cycles16,29. Due to occasional political unrest at the study sites, a regular interval of blood sample collection was not possible. Meanwhile, these macaques were reared as pets and at times used to assist in coconut-picking; therefore, some of them were not available during the blood sample collection period. Although the small sample size, the non-probability sampling strategy, and the uneven interval of blood sample collections could potentially bias the results in this study, the temporal dynamics of natural infections with hemoparasites have been observed among macaques.
In conclusion, longitudinal monitoring of sequential blood samples collected over a 32-month period from 3 long-tailed macaques and 20 pig-tailed macaques in Narathiwat Province has revealed the ecological complexity of Plasmodium and Hepatocystis infections. Based on microscopy, PCR and TADS, seven monkeys were free from infections amid the transmission of simian hemoparasites in communities. Although long-term infections with the same malaria parasite strains were reaffirmed in 10 macaques, superimposed infections with other species could occur transiently or within an extended period. Within-host competitive interactions were observed in coinfection with Hepatocystis sp. and Plasmodium species. Phenotypic and genotypic analysis of DARC in these macaques did not provide supporting evidence for its role in susceptibility to infections. Although patent gametocytemia was not observed in infected macaques, it is likely that they remain important reservoirs for simian malaria in the study population. These findings could be useful for the surveillance and potential control of emerging zoonotic malaria.
Materials and methods
The study population
From May 2017 to February 2020, three long-tailed macaques and 20 pig-tailed macaques were recruited from the Sukhirin and Waeng Districts of Narathiwat Province in southern Thailand. Our previous surveys have shown that macaque populations in this region were infected with malaria and Hepatocystis parasites7,24, while Anopheles mosquitoes in these areas were previously reported to be potential vectors of zoonotic simian malaria parasites25. These macaques were domesticated in 20 households (Fig. 1). The distances between the approximate centers of clusters 1 and 2, 1 and 3, and 2 and 3 were 6.2, 11.7 and 11 km, respectively. The households in each cluster were located in a close vicinity (Fig. 1). Each macaque was identified as Mf or Mn, representing M. fascicularis and M. nemestrina, followed, respectively, by the cluster number and numerical order of the monkeys in each cluster. Two to five blood samples (< 1 ml each) were sequentially obtained from the superficial veins of the legs of each monkey without prior sedation. The study was conducted in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Microscopy
Microscopy diagnosis of Plasmodium and Hepatocystis infections was performed by experienced microscopists who were blinded to the results of PCR detection. For each slide, the Giemsa-stained thick blood film was examined for at least 200 leucocytes, and the thin blood film for at least 200 microscopic fields with the 100X objective as described4.
Preparation of DNA
Genomic DNA was prepared from 200 µL of each blood sample using QIAamp DNA mini kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendation and kept at −40 º C until use.
PCR diagnosis of Plasmodium and Hepatocystis
Identification of Plasmodium species was performed by species-specific nested PCR targeting the mitochondrial cytochrome c oxidase subunit I gene (cox1) from genomic DNA of each isolate. The primary PCR primers targeted genera Plasmodium and Hepatocystis while the nested PCR primers are specific to P. knowlesi, P. cynomolgi, P. inui, P. fieldi, P. coatneyi and Hepatocystis sp. (Supplemental Table S1).
Amplification and sequencing of the mitochondrial genome
The complete mitochondrial genomes of Plasmodium taxa and Hepatocystis sp. were amplified by nested PCR using forward and reverse sequences (Supplemental Table S2). The amplified amplicons from primary PCR were purified using QIAquick PCR purification kit (Qiagen, Hilden, Germany). The concentration of DNA was determined using NanoDrop ND-1000 Microvolume Spectrophotometer (Thermo Fischer Scientific). The purified amplicons from primary PCR were serially diluted to obtain approximately 1–3 copies of the Plasmodium / Hepatocystis genomes and used as DNA templates for secondary PCR. At least five separate nested PCR assays were performed for each diluted genomic DNA template. The resulting positive PCR products were sequenced until two distinct nonsuperimposed signals of the electropherograms were obtained.
Illummina PCR amplicon sequencing
The cox1 gene fragments from primary PCR of each sample were purified using QIAquick PCR purification kit (Qiagen). In addition, the apicoplast caseinolytic protease C gene (clpc) fragments of genera Plasmodium and Hepatocystis spanning 750 bp were amplified by PCR using primers CLPCF and CLPCR (Supplemental Table S3). Both cox1 and clpc PCR-amplified products were used as templates for amplicon deep sequencing with Illumina HiSeq2500-PE150 platform41. Therefore, Plasmodium species that was not included in the species-specific PCR assays could be diagnosed by TADS, while the higher sensitivity of the latter has enabled additional detection of co-circulating hemoparasites in the samples.
DARC phenotyping
DARC phenotypes of macaques were determined by the gel test to read agglutination reaction of the Fya (FY1) and Fyb (FY2) antigens on macaques’ erythrocytes using the ID system for human erythrocyte blood group detection according to the manufacturer’s protocol (DiaMed GmbH, Switzerland). Each blood sample was tested twice. Results were evaluated based on the aspects of agglutination reactions with a five degree scale (0–4) according to the manufacturer’s recommendation.
Data analysis
Chi-square test was deployed to compare differences between the proportions of coinfection with two different species. Sequences were aligned using the CLUSTAL_X program42. The phylogenetic tree was reconstructed by the maximum likelihood method using GTR + G + I as the best substitution model. The confidence levels of the clustering patterns in the phylogenetic tree were evaluated by 1000 bootstrap pseudoreplicates. For bioinformatics, the filtered reads were classified using Kraken2 v2.0.6 based on the k-mer classification algorithm43. We used a custom database of mitochondrial genomes and apicoplast caseinolytic protease C genes of Plasmodium and Hepatocystis species from the NCBI database (Supplemental Table S4). A cutoff level with a threshold of 2% of the total mapped reads is used to determine the presence of Plasmodium species. The Kraken2 outputs were visualized with Pavian metagenomis data explorer44. To verify hemoparasite detection, interpretations were performed using the consensus from 3 approaches by (i) mapping all reads to Plasmodium apicoplast caseinolytic protease C gene and then the unmapped reads were aligned to the Plasmodium mitochondrial cytochrome c oxidase subunit I gene, (ii) mapping all reads to Plasmodium mitochondrial cytochrome c oxidase subunit I gene and then the unmapped reads were aligned to the Plasmodium apicoplast caseinolytic protease C gene and (iii) mapping all reads simultaneously to the Plasmodium mitochondrial cytochrome c oxidase subunit I gene and Plasmodium apicoplast caseinolytic protease C gene. Prediction of intrinsically disordered domains of the protein was performed using the DisoFLAG Web server45.
Data availability
The datasets generated and/or analysed during the current study are available in the NCBI GenBank repository under accession numbers PQ362384-PQ362418.
References
Singh, B. et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363, 1017–1024 (2004).
Jongwutiwes, S., Putaporntip, C., Iwasaki, T., Sata, T. & Kanbara, H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg. Infect. Dis. 10, 2211–2213 (2004).
Jongwutiwes, S. et al. Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg. Infect. Dis. 17, 1799–1806 (2011).
Putaporntip, C. et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J. Infect. Dis. 199, 1143–1150 (2009).
Putaporntip, C., Cheng, C. W., Rojrung, R., Kuamsab, N. & Jongwutiwes, S. Non-human primate malaria in travellers. J. Travel Med. 30, taad135 (2023).
Putaporntip, C. et al. Plasmodium cynomolgi co-infections among symptomatic malaria patients, Thailand. Emerg. Infect. Dis. 27, 590–593 (2021).
Putaporntip, C. et al. Cryptic Plasmodium inui and Plasmodium fieldi infections among symptomatic malaria patients in Thailand. Clin. Infect. Dis. 75, 805–812 (2022).
Yap, N. J. et al. Natural human infections with Plasmodium cynomolgi, P. inui, and 4 other simian malaria parasites, Malaysia. Emerg. Infect. Dis. 27, 2187–2191 (2021).
Jeyaprakasam, N.K., Liew, J.W.K., Low, V.L., Wan-Sulaiman, W.-Y. & Vythilingam, I. Plasmodium knowlesi infecting humans in Southeast Asia: What’s next? PLoS Negl. Trop. Dis. 14, e0008900 (2020).
Johnson, E. et al. Landscape drives zoonotic malaria prevalence in non-human primates. Elife 12, RP88616 (2024).
Vythilingam, I. & Jeyaprakasam, N. K. Deforestation and non-human primate malarias will be a threat to malaria elimination in the future: insights from Southeast Asia. Acta Trop. 257, 107280 (2024).
Putaporntip, C., Thongaree, S. & Jongwutiwes, S. Differential sequence diversity at merozoite surface protein-1 locus of Plasmodium knowlesi from humans and macaques in Thailand. Infect. Genet. Evol. 18, 213-219 (2013).
Ruiz Cuenca, P. et al. Is there evidence of sustained human-mosquito-human transmission of the zoonotic malaria Plasmodium knowlesi? A systematic literature review. Malar. J. 21, 89 (2022).
Nakaviroj, S. et al. An autochthonous case of severe Plasmodium knowlesi malaria in Thailand. Am. J. Trop. Med. Hyg. 92, 569–572 (2015).
Chin, W., Contacos, P. G., Collins, W. E., Jeter, M. H. & Alpert, E. Experimental mosquito-transmission of Plasmodium knowlesi to man and monkey. Am. J. Trop. Med. Hyg. 17, 355–358 (1968).
Coatney, G. R., Collins, W. E., Warren, M. & Contacos, P. G. The Primate Malarias (Original Book Published 1971) [CD-ROM]. Version 1.0 (Centers for Disease Control and Prevention, 2003).
Collins, W. E. Plasmodium knowlesi: a malaria parasite of monkeys and humans. Annu. Rev. Entomol. 57, 107–121 (2012).
Moyes, C. L. et al. Predicting the geographical distributions of the macaque hosts and mosquito vectors of Plasmodium knowlesi malaria in forested and non-forested areas. Parasit. Vectors 9, 242 (2016).
Gruszczyk, J. et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 359, 48-55 (2018).
Chitnis, C. E., Chaudhuri, A., Horuk, R., Pogo, A. O. & Miller, L. H. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J. Exp. Med. 184, 1531–1536 (1996).
Miller, L. H., Mason, S. J., Dvorak, J. A., McGinniss, M. H. & Rothman, I. K. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189, 561–563 (1975).
Demogines, A., Truong, K. A. & Sawyer, S. L. Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi. Mol. Biol. Evol. 29, 445–449 (2012).
Seethamchai, S., Putaporntip, C., Malaivijitnond, S., Cui, L. & Jongwutiwes, S. Malaria and Hepatocystis species in wild macaques, southern Thailand. Am. J. Trop. Med. Hyg. 78, 646–653 (2008).
Putaporntip, C. et al. Ecology of malaria parasites infecting southeast Asian macaques: evidence from cytochrome b sequences. Mol. Ecol. 19, 3466–3476 (2010).
Yanmanee, S. et al. Natural vectors of Plasmodium knowlesi and other primate, avian and ungulate malaria parasites in Narathiwat Province, Southern Thailand. Sci. Rep. 13, 8875 (2023).
Muehlenbein, M. P. et al. Accelerated diversification of nonhuman primate malarias in Southeast Asia: adaptive radiation or geographic speciation? Mol. Biol. Evol. 32, 422–439 (2015).
Orkin, S. H. GATA-binding transcription factors in hematopoietic cells. Blood 80, 575–581 (1992).
Tournamille, C., Colin, Y., Cartron, J. P. & Le Van Kim, C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 10, 224–228 (1995).
Garnham, P. C. C. Simian quartan malaria parasites Plasmodium inui, Plasmodium shortti and Plasmodium brasilianum. In: Malaria Parasites and Other Haemosporidia. 1st ed. 285–322. (Oxford: Blackwell Scientific, 1966).
Cox-Singh, J. et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 46, 165–171 (2008).
Anderios, F., Noorrain, A. & Vythilingam, I. In vivo study of human Plasmodium knowlesi in Macaca fascicularis. Exp. Parasitol. 124, 181–189 (2010).
Charpentier, M. J. E. et al. A longitudinal molecular study of the ecology of malaria infections in free-ranging mandrills. Int. J. Parasitol. Parasites Wildl. 10, 241–251 (2019).
Collins, W. E. et al. Susceptibility of Macaca fascicularis monkeys from Mauritius to different species of Plasmodium. J. Parasitol. 78, 505–511 (1992).
Huang, Y. et al. Isolation and identification of a South China strain of Plasmodium inui from Macaca fascicularis. Vet. Parasitol. 176, 9–15 (2011).
Bashey, F. Within-host competitive interactions as a mechanism for the maintenance of parasite diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140301 (2015).
Ramesh, A. & Hall, S. R. Niche theory for within-host parasite dynamics: analogies to food web modules via feedback loops. Ecol. Lett. 26, 351–368 (2023).
Bousema, T. & Drakeley, C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 24, 377–410 (2011).
Hawking, F. et al. The biological purpose of the blood-cycle of the malaria parasite Plasmodium cynomolgi. Lancet 288, P422–424 (1966).
Gautret, P. & Motard, A. Periodic infectivity of Plasmodium gametocytes to the vector. Parasite 6, 103–111 (1999).
Ouédraogo, A. L. et al. Substantial contribution of submicroscopical Plasmodium falciparum gametocyte carriage to the infectious reservoir in an area of seasonal transmission. PLoS ONE 4, e8410 (2009).
Illumina HiSeq 2500 system. https://www.illumina.com/systems/sequencing-platforms/hiseq-2500.html. (Accessed 5 February 2021) (2021).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 1–3 (2019).
Breitwieser, F. P. & Salzberg, S. L. Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 36, 1303–1304 (2020).
Pang, Y. & Liu, B. DisoFLAG: accurate prediction of protein intrinsic disorder and its functions using graph-based interaction protein language model. BMC Biol. 22, 3 (2024).
Google Earth Pro. Map data of Thailand. https://www.google.com/intl/en_uk/earth/versions/#earth-pro (2022).
Acknowledgements
The authors are grateful to Siriporn Thongaree for valuable advice and support. The authors wish to thank all staff at Hala-Bala Research Station, Department of National Parks, Wildlife and Plant Conservation for assistance in field studies. We thank all macaques’ owners for excellent cooperation during the blood sample collections. C.W.C. is supported by BHF Mautner Career Development Fellowship.
Funding
This research is funded by Thailand Science Research and Innovation Fund Chulalongkorn University (HEA663000037) to C.P. and S.J.
Author information
Authors and Affiliations
Contributions
“C.P. and S.J. designed the study. C.P., S.Y. and S.K. performed field studies. C.P., J.S., N.K., R.R. and U.P. performed the experiments. C.W.C., C.P. and S.J. performed data analysis. C.P. and S.J. drafted the manuscript. C.P. finalized the manuscript. All authors approved the manuscript.”
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was reviewed and approved by the Institutional Animal Care and Use Committee in Animal Research of Chulalongkorn University Laboratory Animal Center, Thailand. Prior to blood sample collection, informed consent from macaques’ owners was obtained for handling the animals. All procedures for blood sample collection were carried out in accordance with the relevant guidelines and regulations. The animal study was conducted in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Putaporntip, C., Yanmanee, S., Somkuna, J. et al. Ecological complexity of zoonotic malaria in macaque natural hosts. Sci Rep 15, 4893 (2025). https://doi.org/10.1038/s41598-025-86415-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-86415-y