Abstract
Bovine tuberculosis (bTB) is a major disease of cattle in the UK, placing a significant economic burden on the taxpayer. The causative agent, Mycobacterium bovis, has a wide host range, including the European badger (Meles meles). While badgers have been implicated in the transmission and maintenance of infection in cattle in areas of endemic disease, their role at the edge of the endemic area is poorly understood. Here we present data on the prevalence of infection in badgers collected along the southern edge of England’s bTB epidemic. Stakeholders across five counties (Oxfordshire, Berkshire, Buckinghamshire, Hampshire, and East Sussex) submitted found-dead badgers for post-mortem examination and testing by bacterial culture. The overall prevalence, as confirmed by whole genome sequencing, was 6.5% (28/428), ranging between 1.1% (1/88) in Hampshire and 13.0% (14/108) in Oxfordshire. The commonest M. bovis clade in badgers was B6-62, which was predominant in 4/5 counties. B6-62 was also the commonest clade found in cattle and was detected in all counties except East Sussex where, although absent from the cattle population, it was detected in local badgers. This study highlights the co-incidence of infection in badgers and cattle in parts of the southern edge area consistent with localised clustering of infection in both species.
Similar content being viewed by others
Introduction
Mycobacterium bovis (M. bovis) is a member of the M. tuberculosis complex (MTC), a group of host-associated, tuberculous disease-causing bacteria of high genetic similarity1,2. Despite this high level of clonality, the group varies significantly in host tropism with M. bovis having the widest known host range3. Indeed, in the United Kingdom (UK) alone, M. bovis has been detected in nine species of farmed or companion mammals, and at least 20 species of wild mammals4,5,6,7,8,9. In cattle, M. bovis causes bovine tuberculosis (bTB), one of the most significant challenges affecting cattle health in England, which is estimated to cost the UK taxpayer over £150 million per annum, with further costs to the cattle industry10. In addition to the financial burden is the cost of livestock life, with over 31,000 cattle slaughtered in 2023, of which 20,200 were from England11.
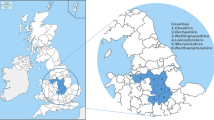
English counties are categorised by their historic prevalence and epidemiology of bTB infection in cattle (see Fig. 1). In the high-risk area (HRA), which encompasses the southwest of the country, the disease is endemic in cattle and extensive disease surveillance is in place, including standard six-monthly herd testing and pre-movement testing. Surrounding the HRA is the edge area (EA), where bTB prevalence has been historically lower than the HRA but into which the epidemic is expanding, resulting in highly variable disease prevalence and either six-monthly or annual cattle testing, in addition to pre-movement testing. Though not contiguous with the rest of the EA, East Sussex is included within this zone due to the prevalence of bTB in the county. Finally, in the low-risk area (LRA) the incidence of bTB in cattle has been historically low and stable, with most cattle herds routinely tested for TB every four years, supplemented by targeted enhanced testing of certain herds and mandatory post-movement testing of cattle introduced from the rest of England and Wales12.
Map showing the counties of England coloured by perceived bTB risk; red indicates the high-risk area, orange the edge area, and blue the low-risk area. The studied counties are outlined in bold and non-faded. The insert shows the counties of the southern Edge Area which were the focus of the present study. Mapping software: Mapping software: QGIS version 3.36.1, QGIS Association ( http://www.qgis.org).
While cattle movements undoubtedly make a substantial contribution to the spread and persistence of bTB in herds across the three risk areas, the presence of infection in the primary wildlife reservoir, the European badger (Meles meles) adds further complexity to disease control13,14,15. Infectious badgers can excrete M. bovis in their saliva, urine, faeces or from ruptured abscesses, presenting a hazard to cattle through indirect transmission, and potentially via direct transmission in cattle housing16,17,18,19. During the Randomised Badger Culling Trial (RBCT, 1998–2005), 15.9% of culled badgers tested positive by culture or by having lesions containing acid-fast organisms20. There was a demonstrable association between the proportion of positive badgers and cattle TB incidents at a distance of 1–2 km20,21. In addition, spoligotyping and multi-locus variable number tandem repeat analyses (MLVA) have demonstrated the co-localisation of related M. bovis isolates from badgers and cattle21,22,23,24. Whole genome sequencing (WGS) has further evidenced the link between infections in cattle and badgers, with highly related isolates being cultured from sympatric populations indicating recent and bi-directional transmission25,26,27. In English areas of high cattle incidence, efforts to reduce transmission risks from wildlife have included licenced culls of badgers, with over 53,000 animals culled during the years 2022 to 202320,22,28,29,30,31,32.
For the purposes of surveillance, found-dead surveys are the most viable, practicable and consistent approach for collecting a sufficiently large sample of badgers (typically the results of road traffic accidents, RTAs) for investigating infection prevalence at a broad geographic scale (e.g., county-level). This approach was extensively validated alongside the RBCT and found to be a reliable proxy for M. bovis infection prevalence within the sampled population15. Since then, it has been employed across Wales23,33, Northern Ireland24 and in the county of Cheshire to investigate an emerging cluster of TB in cattle34. In addition, an investigation of bTB in found dead badgers in the northern EA is of relevance to the design of the current study, which this study was designed to complement35. The northern EA study took place in 2016–2017 and identified a headline MTC infection rate in badgers of 8.3% (range: 4–15%), and common areas of high-prevalence in cattle and badgers as also reported in other studies23,24,33,36.
Effective control of bTB in cattle at the edge of the endemic area requires a better understanding of the potential role of badgers in the spread and perpetuation of infection in this transitional zone. Here we present the results of an investigation into the prevalence of M. bovis in found-dead badgers in the southern EA, undertaken between 1st of April 2021 and the 30th of April 2023. Our results are interpreted alongside contemporaneous data on infection in cattle, gathered during routine surveillance and monitoring, and the results of the earlier study undertaken in the northern EA35.
Results
Badger carcasses
In total, 525 badger carcasses were collected from across the five counties of the southern EA. Of these, 428 (81.5%) were deemed suitable for examination and tissue sampling, and four (0.8%) were excluded due to not being collected within the study area. RTAs accounted for 84.3% of submissions (n = 443), sixteen (3.0%) were found dead elsewhere and were typically emaciated and in poor condition, whilst the causes of death could not be determined in the remainder, usually owing to the degree of carcass decomposition. The number of carcasses submitted varied by county (χ2 = 46.0, df = 4, P < 0.0001), with Oxfordshire yielding the highest number (n = 138) and Berkshire providing the fewest (n = 49) (Table 2). The majority of carcasses were collected by farmers and farming groups (n = 231, 44.0%), followed by conservation groups (n = 132, 25.1%), veterinarians and their affiliates (n = 85, 16.2%), government agencies or local authorities (n = 18, 3.4%), and miscellaneous other groups or individuals (n = 59, 11.2%).
Of the carcasses submitted for which the sex was determined (n = 443), 226 (51.0%) were males and 217 (49.0%) were females. No temporal variation was observed in the average number of carcasses submitted by month. Comparisons between the submissions of male and female carcasses with reference to month had to account for variable sample sizes, the low number of repeat data and the unequal number of repeats. On average, there were fewer submissions of males across the calendar year, with the exception of December – March (Fig. 2). However, only in April were significantly more females submitted than males (t = 9.5, df = 2, P ≤ 0.01). The majority of carcasses were of adult badgers (estimated to be over 1 year old), with only 52/525 (9.9%) of those collected and 43/444 (9.7%) of those examined recorded as cubs.
Mycobacterium Bovis infection in surveyed badgers
Across the southern EA, 31/428 (7.2%, 95% CI: 5.0-9.7%) badger carcasses tested positive for MTC bacteria by culture, and subsequently confirmed using IS6110 PCR for MTC, and hsp65 sequencing for presumptive identification of M. bovis (see methods for details). Of these, 28 were confirmed as M. bovis by WGS giving an apparent prevalence of 6.5% (95% CI: 4.4–8.9%). Despite the low number of positive badgers, and the aforementioned study design, efforts were made to examine factors that increased the likelihood of positive carcasses being detected. The prevalence of infected badgers varied significantly by county (χ2 = 12.7, df = 4, P ≤ 0.01), ranging from 1.1% in Hampshire to 13.0% in Oxfordshire (Table 2). Additionally, the detection of positive carcasses varied with the time of year, being most likely in March (OR (95% CI) = 10.58 (1.794–20.14), P < 0.05), and least likely in October (not significant) (Fig. 2).
Conservation/vaccination-affiliated collectors provided a higher percentage of positive submissions than other collectors (Table 1). However, underlying geographic bias skewed this figure as this group submitted significant numbers from counties with the highest TB burden; 43.2% (57/132) of their submissions were from Oxfordshire and 25.8% (34/132) were from Berkshire. In contrast, badgers submitted by farming-affiliated collectors were more evenly distributed across the study area with percentage positivity estimates skewed by the relatively smaller contribution this group made in the counties of highest TB burden. Collector affiliation was not thought to influence the likelihood of a positive submission.
All of the confirmed positives were adults (28/384 confirmed adults, 7.3%, 95% CI: 4.9–10.4%) and the prevalence of infection was more than 3 times higher in male badgers than in females (χ2 = 8.3, df = 1, P < 0.01), with 22/215 male badgers testing positive (10.2%, 95% CI: 6.5–15.1%) and 6/210 females (2.9%, 95% CI: 1.1–6.1%). The distances between any given positive badger and its nearest positive neighbour were significantly smaller than those between any negative badger and their nearest positive neighbour (W = 4193, P < 0.05). The median distance from a positive badger to the nearest other positive badger was 6.1 km (interquartile range, IQR: 2.4–11.6 km), whilst negative badgers were on average 8.4 km (IQR: 5.1–16.5 km) from their nearest positive neighbour.
Six M. bovis clades were identified amongst the isolates obtained from confirmed positive badger carcasses. Of these, B6-62 was the most common, representing 21/28 (75.0%) of M. bovis confirmed isolates. The remaining clades (B6-61, B6-71, B6-85, B6-91, and B1-11) were each detected only once while two isolates (7.1%) were not identified to clade level (Fig. 3). The distance to the nearest badger with the same strain (where available) had a mean of 7.9 km (95% CI: 5.4–10.7 km) while the distance to the nearest badger with a different strain was 17.4 km (95% CI: 12.7–22.0 km). Statistical analysis identified a significant difference between the two distributions (t = -4.57, df = 20, P < 0.001).
The spatial distribution of the sampled badger carcasses, and herd level TB test results during the study period (1st April 2021–30th April 2023). Stars or black crosses indicate badger carcass test results; filled circles or red crosses indicate cattle herds where Officially bTB Free status was withdrawn (OTF-W) due to a TB incident (breakdown) with at least one lesion and/or M. bovis culture-positive animal removed, while unfilled circles indicate suspended herd status (OTF-S). Mapping software: QGIS version 3.36.1, QGIS Association (http://www.qgis.org).
Mycobacterium bovis in cattle
During the study period, there was an average of 2639 (range: 2606–2711) active herds in the southern EA per annum. Of these, 277 herds (10.5%) experienced at least one TB breakdown (i.e. detection of at least one TB test positive animal and/or laboratory-positive case at routine slaughter). During the study period, a total of 308 breakdowns occurred in the study area (including 31 repeat breakdowns) of which 132 (42.9%) had their ‘Officially bTB Free’ (OTF) status withdrawn (OTF-W), 174 (56.5%) had their OTF status suspended (OTF-S) and the remainder were unclassified. Of the breakdowns involving at least one animal with visible tuberculous lesions or positive culture (OTF-W), WGS clades were identified in 84.1% (n = 111) of incidents, meaning that amongst all incidents of OTF status being suspended or withdrawn a clade was identified in 36.0% of cases.
The proportion of herds with reactor cattle during the period of study varied by county (χ2 = 13.7, df = 4, P < 0.01); Oxfordshire herds were the most likely to have a TB test reactor detected with 20.9% of herds experiencing a breakdown during the study, compared to only 5.0% of herds in Hampshire (Table 2). Of those herds where a clade associated reactor was identified (n = 111), the most common M. bovis clade was B6-62 which was found in 83 incidents (74.8%), followed by B6-11 in twelve incidents (10.8%).
Comparison between badgers and cattle
A statistically significant, positive relationship was identified between the percentage of positive badger carcasses and the herd level prevalence (defined as the percentage of herds that experienced at least one breakdown during the study period) at the county level (Fig. 4; Pearson’s r(3) = 0.97 (95% CI: 0.82-1.0), t(3) = 10.9, P < 0.01).
County-level comparison of the percentage of M. bovis infected badgers (detected by culture and WGS) and infected cattle herds (as determined by the single intradermal comparative cervical tuberculin skin test (SICCT)) in the Southern Edge Area Study (April 2021–April 2023). Repeat breakdowns are excluded from these summary data.
The distances between badger carcasses and cattle infected with the same and different clades were analysed. The two East Sussex B6-62 infected badgers were removed from the analyses as outliers, due to the non-detection of the clade in local herds and because this county was not contiguous with any other EA county. Overall, the distance from a positive badger to a herd with the same clade was 3.6 km (IQR: 2.9–5.6 km) while the mean distance to the nearest herd infected with a different clade was 9.9 km (95% CI: 7.6–12.2 km). Badgers were significantly more likely to be found near herds infected with the same clade, than to be found near herds infected with different clades (V = 41, P < 0.01).
Non-tuberculous mycobacteria (NTM)
Fifteen putative species of non-tuberculous mycobacteria (NTM) were detected in 30 badgers (apparent prevalence 7.0%, 95% CI: 5.1–9.4%). M. avium was the most prevalent NTM with an apparent prevalence of 2.6% (95% CI: 1.3–4.5%, 11/429), followed by M. vaccae with an apparent prevalence of 1.2% (95% CI: 0.4–2.7%, 5/429); some isolates were not fully identified by this method. More males tested positive for M. bovis (22/215, 10.2%) than for NTM (16/215, 7.4%) and more females tested positive for NTM (13/210, 6.2%) than for M. bovis (6/210, 2.9%), although neither difference was significant. One unsexed badger was also positive for NTM.
Discussion
Mycobacterium bovis infection in badgers collected from the southern EA was spatially heterogeneous, with the county level apparent prevalence ranging from 1.1% in Hampshire to 13.0% in Oxfordshire. This concurs with findings from a similar study of found dead badgers in the northern EA where county-level prevalence ranged from 4–5 to 15%35. This marked spatial heterogeneity of the disease has also been demonstrated in Wales23 and the Republic of Ireland36. Our study wide prevalence estimate of 6.5% (95% CI: 4.4–8.9%) is also similar to that of other studies in the northern EA (8.3%, 95% CI: 6.4–11.0%) and Wales (7.3%, 95% CI: 5.6–9.5%) which are contiguous with England’s HRA23,35. Despite nearly two decades of separation, the overall apparent prevalence remains lower than that of the HRA (2002–2005), in which 16.6% of culled badgers and 15% of RTA badgers were positive for M. bovis15 demonstrating the slow expansion of the disease. However, direct comparisons between studies can be problematic owing to minor methodological differences. For example, the current study used WGS to confirm M. bovis isolation whilst Swift et al. (2021) used IS6110 to confirm only to the MTC level, and Schroeder et al.. (2020) used spoligotyping. WGS excluded three samples that were identified as MTC members by IS6110 PCR and hsp65 sequencing (putative M. bovis due to the host species), reducing the overall apparent prevalence from 7.2% to the reported 6.5%. This highlights that differing methodologies can produce slight differences in prevalence estimates.
The prevalence of M. bovis in badgers was positively correlated with the number of incident cases in cattle at the county level, consistent with findings in the northern Edge Area, and in endemic areas of England, Wales and the Republic of Ireland (RoI)22,23,33,35,36. At the clade level, infected badgers were geographically closer to herds infected with the same clade than those with different clades, as previously shown in the HRA using spoligotyping21 and in Northern Ireland using MLVA24. Clustering of positive badgers was also evident as has been observed in the RoI36 and England’s HRA37.
The ratio of male to female badgers in the present study was circa 1:1, similar to that observed in other found-dead studies33,35,38 and as identified in capture-mark-recapture studies of badger populations39,40,41. The prevalence of confirmed M. bovis infection in male badgers in the present study was higher than in females, consistent with previous work showing that males are more likely to become infected and experience more rapid disease progression, thus increasing the likelihood of yielding a positive culture at post-mortem examination PME42,43. It is also hypothesised that immunological differences may enhance male susceptibility to disease43.
The present study resulted in a lower percentage of sampled juvenile badgers (9.9%) compared to other found dead surveys using similar collection methods (c. 30% 34,35 and population studies (c. 20–30%)41,44,45. As TB infection is more often observed in adult badgers than in juveniles, the high proportion of adults in the present study is likely to have inflated overall population-level prevalence20,46,47. The reasons for the low percentage of juveniles in our sample of carcasses are not clear, but could potentially reflect poor survival related to unfavourable conditions48. For example, April 2021 and 2022 and the summer of 2021 were unusually dry49,50 which is likely to have reduced food availability51,52 and impacted on cub survival in particular48.
Routine WGS of M. bovis requires the culture of bacterial isolates from infected individuals. However, the success of culture can be limited by early-stage infection53,54, the metabolic state of the bacteria55,56, the slow growth rate of M. bovis57 and the potential for overgrowth by competing microorganisms58; it is therefore highly probable that the apparent prevalence given here is an underestimation of the true prevalence. Furthermore, the current routine approach of sequencing a single isolate per badger or entire cattle herd means that if multiple infections with different strains or clades were present then they would not be detected. Despite these limitations, six clades were cultured from badger carcasses during the present study, of which B6-62 was the most common, being identified in 4/5 counties. Two badgers collected from East Sussex were infected with B6-62, outside the putative home range of the clade, and at that time the clade was undetected in the county’s cattle59. The probability that these two samples were similarly mislabelled seems low given that both the examination of carcasses and subsequent sequencing of isolates were carried out several weeks apart. Furthermore, B6-62 has recently been detected in the county’s cattle, within two herds which were OTF-S during the present study, and therefore may have been present for some time60. Two further clades which were identified during the study (B1-11, B6-85) were also isolated from badgers in areas which were well beyond their typical home range, and one nationally rare isolate with no designated home range was detected in Oxfordshire (B6-61)59. While B6-85 was identified in a badger found in close proximity to cattle herd breakdowns associated with that clade, the same was not true for B1-11 and B6-61. The identification of M. bovis clades in badgers that are distinct from those detected over the same period in cattle could arise as a result of historic spillover from cattle into badgers, or the limited sensitivity of M. bovis surveillance in cattle, as only 36% of breakdowns yielded sequenceable isolates across the study area. Previous evidence has demonstrated the contemporaneous circulation of different strains within badgers and cattle, showing the value in surveying both species to entirely evaluate the disease landscape25.
The apparent prevalence of NTM in this study was 7.0%, far greater than a previously reported value of 0.9% (424/45,705) from archived UK cull data (recorded as M. avium and other mycobacteria), but similar to that reported in Spain from a small opportunistic study61. Unlike M. bovis, NTMs were distributed relatively evenly between the sexes, likely related to the environmental nature of these bacteria contributing to even exposure risks. Also, in contrast to M. bovis, the likelihood of detectable NTM infection did not increase with age, though the small number of juveniles in this study likely limited our ability to detect any such effects.
Estimates of prevalence from studies such as ours should be treated with caution as biases can arise as a result of the relatively small sample sizes, the unknown absolute population size, local clustering of infection, potential behavioural correlates of susceptibility to becoming a RTA, and the limitations of the diagnostic methods employed. However, the sampling approach and culture-based methodology in the present study are broadly consistent with those shared by several other badger RTA surveys, allowing for some level of comparison whilst recognising the known limitations23,33,34,35. Furthermore, the collection of found dead badgers provides a geographically broad estimate of prevalence, which would otherwise be unachievable, and the collection of epidemiologically valuable WGS data to assist in disease tracking. There were underlying concerns that the apparent prevalence may be susceptible to collector bias, but when considered alongside other factors such as the county level prevalence, it was thought unlikely that the submissions were biased – with conservation/vaccination-affiliated collectors submitting a greater percentage of positive carcasses than farming affiliated collectors.
To conclude, the present study has provided the first estimates of M. bovis prevalence in badger populations in the southern EA. In addition to identifying county-level heterogeneity, the use of WGS has added significant value to the surveillance data available for cattle by uncovering additional clades in badgers that were not detected in local herds and has confirmed infection in wildlife where it was previously unstudied. This study has provided further evidence for the link between infection in badgers and cattle at the edge of the area of TB endemism in England. Further investigation of WGS data from this area may shed light on the relative importance of cattle or badgers in driving herd breakdowns and whether (as seems likely) cattle movements may have seeded infection in both local cattle and badger populations. This may, in some locations, facilitate the circulation of infection in the local badger population and risk spill-back to cattle. Understanding the proximal drivers of infection in cattle across the EA may inform management options which could include enhanced cattle measures, badger vaccination and targeted badger culling. In some parts of the EA it may not be too late to prevent infection spilling over into this wildlife host before it becomes endemic in both populations.
Materials and methods
Carcass collection and storage
A network of collectors was established across the five counties of the southern Edge Area with the aim of collecting 100 carcasses from each county. A dedicated phoneline and e-mail address were used for communications, and the study was promoted at events, through veterinarians, and on the TB Hub website. Interested parties were provided with instructions and kits for the safe collection of found-dead badgers as well as a small financial reward. Collectors placed carcasses in three double thickness PVC bags and sealed them with pre-labelled tags. Useful metadata was recorded on the submission form by the collector, including the date and time, tag identification code, location, type of submission (e.g., road traffic accident) and any overnight storage conditions if the courier was unable to collect the same day. Carcasses were not frozen, but when storage was required, collectors were asked to place in a chiller if available, or else in a cool and dry area. If the courier was unable to transport the carcass to the PME lab (University of Nottingham, Sutton Bonington Campus) the same day, then it would be stored in a chiller overnight or over the weekend until it could be safely received. The median time between carcass collection and PME was 3 days (range: 0–12). Carcass collection took place from April 2021 to April 2023 inclusive, with collections ceasing at the county level once 100 carcasses had been sampled.
Post-mortem examination
Carcasses were deemed suitable for PME if they had not suffered significant damage, were not autolysed, and arrived with the form linking them to a location. The sex, weight, length, age, (adult or juvenile), estimated from dentition, size, and pelage, condition as inferred from weight and body fat, and probable cause of death were recorded. In some cases, these data are not recorded due to the condition of the carcass, but sampling was possible, thus test data exists that cannot be related to certain population demographics. The tissue sampling protocol for isolation of mycobacteria followed that of the Northern Edge Study35, based on the protocols used by 33 and 36, modified by34. Carcasses were examined externally and internally for TB-like lesions, and if present these were harvested for tissue processing and bacterial culture. Selected lymph nodes and lung tissue were harvested and pooled (see Table 3). Harvested tissues were stored at 4 °C for up to 48 h before processing for bacterial culture.
Tissue processing
Tissue processing and microbiological examination was performed in the Containment Level 3 (CL3) facility at the University of Nottingham, according to the methods used in previous studies34,35. Briefly, lesioned tissue or pools of target tissues were gently ground with sterile sand in phosphate buffered saline (PBS). Samples were mixed 1:1 with 5% oxalic acid and incubated at room temperature for 10 min, before 200 µL of each pool was inoculated onto Stonebrink Selective agar + PACT (BD Diagnostics) and onto Middlebrook 7H11 slopes supplemented by PANTA (BD Diagnostics). Media were incubated at 37 °C for a minimum of 12 weeks with checking at regular intervals for putative mycobacterial colonies.
Characterisation of mycobacteria
After a minimum of 12 weeks incubation, putative mycobacterial colonies were heat killed at 80 °C for 30 min. DNA was extracted by crude lysis; heat killed colonies were frozen and subsequently heated to 95 °C for 5 min in sterile distilled water (SDW), centrifuged at 13,000 x g for 3 min to remove cellular debris and the resultant supernatant aliquoted for molecular analyses.
Isolates were screened by PCR for IS6110, an insertion sequence unique to the MTC, and hsp65 sequencing which also identified non-tuberculous mycobacteria (NTM). Confirmed members of the MTC were subjected to whole genome sequencing (WGS; performed at APHA laboratories, Weybridge). Sequences were compared to M. bovis reference sequence AF2122 for final confirmation and assigned to one of 30 lineages known to be in circulation in Great Britain (pipeline: https://github.com/APHA-CSU/btb-seq)62. Major groups are denoted by the initial ‘B’ number denoting ‘bovis’ with a range of 1–6. The second number is hierarchical and demonstrates relatedness within the major group, i.e. B6-61 is more closely related to B6-62 than it is to B6-41, but all of these are more closely related than they are to another major group (e.g. B3-11)63.
Cattle data
APHA conducts routine surveillance and testing of cattle herds for bTB. In the HRA and EA cattle are tested annually or six-monthly, with increased frequency of testing following detection of infection or after high-risk cattle movements. Testing is routinely conducted in the UK using the single intradermal comparative cervical tuberculin test (SICCT) with subsequent removal of reactors (animals exhibiting a positive result). Herds that are on schedule with their testing routine and have no reactors are classed as ‘Officially bTB Free’ (OTF). If at least one reactor (a bovine with one positive or two inconclusive results) was detected, then a TB incident (herd breakdown) is declared and the OTF status is suspended (OTF-S). At PME, if lesions that are typical of TB are identified in a reactor, or a positive bacterial culture or (as of March 2022) positive PCR test is obtained from a TB test reactor, or a non-reactor animal presenting with suspected tuberculous lesions at routine slaughter, then the OTF status of the affected herd is withdrawn (OTF-W). The APHA held dataset includes the breakdown identifier, herd identifier, case reference, herd location, breakdown status, the test result and WGS clade if determined. For the present study, the data for each herd was combined across the study period and any herd that had been OTF-S or OTF-W was classed as positive.
Statistical analyses
Analyses were conducted in R version 4.2.2, with graphs generated using the ‘ggplot2’ and ‘patchwork’ packages. The ‘sf’ package was used to analyse spatial data and QGIS 3.8.1 to visualise it. Statistical comparisons of count, percentage and distance data were performed within this study; data were tested for normality using the Shapiro-Wilk test. Significance testing of count and distance data was performed using the following: students T-test for normal and non-percentage data otherwise a Chi-squared when sufficient data were available, else the Fisher’s Exact Test. Paired data was tested using a T-test for normal data or Wilcoxon Signed Rank Test when the data were not normal. Multiple comparisons were controlled using the Benjamini-Hochberg method to reduce the false discovery rate.
Data availability
Data from cattle and badgers will be available on figshare (10.6084/m9.figshare.27226140). Due to the sensitivity of the badger data, these locations are only available to within 1 km. For any queries regarding the dataset or its availability please contact the corresponding author.
References
Brosch, R. et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U S A. 99, 3684–3689. https://doi.org/10.1073/pnas.052548299 (2002).
Bespiatykh, D., Bespyatykh, J., Mokrousov, I. & Shitikov, E. A comprehensive map of Mycobacterium tuberculosis complex regions of difference. mSphere 6, e0053521. https://doi.org/10.1128/mSphere.00535-21 (2021).
O’Reilly, L. M. & Daborn, C. J. The epidemiology of Mycobacterium bovis infections in animals and man: A review. Tuber. Lung Dis. 76 (Suppl 1), 1–46. https://doi.org/10.1016/0962-8479(95)90591-x (1995).
Murhead, R. H. & Burns, K. J. Tuberculosis in wild badgers in gloucestershire: Epidemiology. Vet. Rec. 95, 552–555. https://doi.org/10.1136/vr.95.24.552 (1974).
Delahay, R. J. et al. Bovine tuberculosis infection in wild mammals in the South-West region of England: A survey of prevalence and a semi-quantitative assessment of the relative risks to cattle. Vet. J. 173, 287–301. https://doi.org/10.1016/j.tvjl.2005.11.011 (2007).
Delahay, R. J., De Leeuw, A. N. S., Barlow, A. M., Clifton-Hadley, R. S. & Cheeseman, C. L. The status of Mycobacterium bovis infection in UK wild mammals: A review. Vet. J. 164, 90–105. https://doi.org/10.1053/tvjl.2001.0667 (2002).
Foyle, K. L., Delahay, R. J. & Massei, G. Isolation of Mycobacterium bovis from a feral wild boar (Sus scrofa) in the UK. Vet. Rec. 166, 663–664. https://doi.org/10.1136/vr.c2681 (2010).
Lee, J., Hanna, R., Hill, R., McCormick, C. M. & Skuce, R. A. Bovine tuberculosis in an eurasian otter. Vet. Rec. 164, 727–728. https://doi.org/10.1136/vr.164.23.727-a (2009).
Broughan, J. M. et al. Mycobacterium bovis infections in domesticated non-bovine mammalian species. Part 1: Review of epidemiology and laboratory submissions in Great Britain 2004–2010. Vet. J. 198, 339–345. https://doi.org/10.1016/j.tvjl.2013.09.006 (2013).
Defra Next steps for the strategy for achieving bovine tuberculosis free status for England. The government’s response to the strategy review, 2018. https://assets.publishing.service.gov.uk/media/5e60ad0de90e077e3d2678d2/bovine-tb-strategy-review-government-response.pdf (2020).
Defra Tuberculosis (TB) in cattle in Great Britain. Department for Environment, Food and Rural Affairs. https://www.gov.uk/government/statistical-data-sets/tuberculosis-tb-in-cattle-in-great-britain (2024).
APHA. Bovine TB testing intervals. https://www.gov.uk/guidance/bovine-tb-testing-intervals (2020).
Gilbert, M. et al. Cattle movements and bovine tuberculosis in Great Britain. Nature 435, 491–496. https://doi.org/10.1038/nature03548 (2005).
Brooks-Pollock, E., Roberts, G. O. & Keeling, M. J. A dynamic model of bovine tuberculosis spread and control in Great Britain. Nature 511, 228–231. https://doi.org/10.1038/nature13529 (2014).
ISG. (ed. Food and Rural Affairs Department for Environment) (Defra, 2007).
Phillips, C. J., Foster, C. R., Morris, P. A. & Teverson, R. The transmission of Mycobacterium bovis infection to cattle. Res. Vet. Sci. 74, 1–15. https://doi.org/10.1016/s0034-5288(02)00145-5 (2003).
Garnett, B. T., Delahay, R. J. & Roper, T. J. Use of cattle farm resources by badgers (Meles meles) and risk of bovine tuberculosis (Mycobacterium bovis) transmission to cattle. Proc. Biol. Sci. 269, 1487–1491. https://doi.org/10.1098/rspb.2002.2072 (2002).
Benham, P. F. J. & Broom, D. M. Interactions between cattle and badgers at pasture with reference to bovine tuberculosis transmission. Br. Vet. J. 145, 226–241. https://doi.org/10.1016/0007-1935(89)90075-4 (1989).
Woodroffe, R. et al. Badgers prefer cattle pasture but avoid cattle: Implications for bovine tuberculosis control. Ecol. Lett. 19, 1201–1208. https://doi.org/10.1111/ele.12654 (2016).
Jenkins, H. E. et al. The prevalence, distribution and severity of detectable pathological lesions in badgers naturally infected with Mycobacterium bovis. Epidemiol. Infect. 136, 1350–1361. https://doi.org/10.1017/S0950268807009909 (2008).
Woodroffe, R. et al. Spatial association of Mycobacterium bovis infection in cattle and badgers Meles meles. J. Appl. Ecol. 42, 852–862. https://doi.org/10.1111/j.1365-2664.2005.01081.x (2005).
Krebs, J. et al. (eds) Fisheries and Food Ministry of Agriculture (MAFF, 1997). bovinetb.info/docs/krebs.pdf.
Schroeder, P. et al. Temporal and spatial Mycobacterium bovis prevalence patterns as evidenced in the all Wales Badgers Found Dead (AWBFD) survey of infection 2014–2016. Sci. Rep. 10, 15214. https://doi.org/10.1038/s41598-020-72297-9 (2020).
Milne, G. et al. Mycobacterium bovis population structure in cattle and local Badgers: Co-localisation and Variation by Farm Type. Pathogens 9. https://doi.org/10.3390/pathogens9070592 (2020).
Crispell, J. et al. Combining genomics and epidemiology to analyse bi-directional transmission of Mycobacterium bovis in a multi-host system. Elife 8. https://doi.org/10.7554/eLife.45833 (2019).
Rossi, G. et al. Identifying likely transmissions in Mycobacterium bovis infected populations of cattle and badgers using the Kolmogorov Forward equations. Sci. Rep. 10, 21980. https://doi.org/10.1038/s41598-020-78900-3 (2020).
Akhmetova, A. et al. Genomic epidemiology of Mycobacterium bovis infection in sympatric badger and cattle populations in Northern Ireland. Microb. Genom. 9. https://doi.org/10.1099/mgen.0.001023 (2023).
APHA. (ed. Food and Rural Affairs Department for Environment) (Defra, https://www.gov.uk/government/statistics/incidence-of-tuberculosis-tb-in-cattle-in-great-britain/quarterly-tb-in-cattle-in-great-britain-statistics-notice-december-2023, 2024).
N.E. (ed. Natural England). https://www.gov.uk/government/publications/bovine-tb-summary-of-badger-control-monitoring-during-2022/summary-of-2022-badger-control-operations#natural-england-chief-scientists-advice-on-the-outcome-of-badger-control-operations-2022 (2023).
N.E. (ed Natural England). https://www.gov.uk/government/publications/bovine-tb-summary-of-supplementary-badger-control-monitoring-during-2022/summary-of-2022-supplementary-badger-control-operations (2023).
N.E. (ed Natural England). https://www.gov.uk/government/publications/bovine-tb-summary-of-badger-control-monitoring-during-2023/summary-of-2023-badger-control-operations#:~:text=The%20results%20from%202023%20indicate,high%20standard%20of%20public%20safety (2024).
N.E. (ed Natural England). https://www.gov.uk/government/publications/bovine-tb-summary-of-supplementary-badger-control-monitoring-during-2023/summary-of-2023-supplementary-badger-control-operations (2024).
Goodchild, A. V., Watkins, G. H., Sayers, A. R., Jones, J. R. & Clifton-Hadley, R. S. Geographical association between the genotype of bovine tuberculosis in found dead badgers and in cattle herds. Vet. Rec. 170, 259. https://doi.org/10.1136/vr.100193 (2012).
Barron, S. et al. A study of tuberculosis in road traffic-killed badgers on the edge of the British bovine TB epidemic area. Sci. Rep. 8 (2018).
Swift, B. M. C. et al. Tuberculosis in badgers where the bovine tuberculosis epidemic is expanding in cattle in England. Sci. Rep. 11, 20995. https://doi.org/10.1038/s41598-021-00473-6 (2021).
Byrne, A. W. et al. Spatial and temporal analyses of metrics of tuberculosis infection in badgers (Meles meles) from the Republic of Ireland: Trends in apparent prevalence. Prev. Vet. Med. 122, 345–354. https://doi.org/10.1016/j.prevetmed.2015.10.013 (2015).
Jenkins, H. E. et al. Effects of culling on spatial associations of Mycobacterium bovis infections in badgers and cattle. J. Appl. Ecol. 44, 897–908. https://doi.org/10.1111/j.1365-2664.2007.01372.x (2007).
Davies, M., Roper, J., Sheperdson, D. J. & T. J. & Seasonal distribution of road kills in the European badger (Meles meles). J. Zool. 211, 525–529 (1987).
Anderson, R. M. & Trewhella, W. Population dynamics of the badger (Meles meles) and the epidemiology of bovine tuberculosis (Mycobacterium bovis). Philos. Trans. R. Soc. Lond. B Biol. Sci. 310, 327–381. https://doi.org/10.1098/rstb.1985.0123 (1985).
Macdonald, D. W. & Newman, C. Population dynamics of badgers (Meles meles) in Oxfordshire, U.K.: Numbers, density and cohort life histories, and a possible role of climate change in population growth. J. Zool. 256, 121–138 (2002). https://doi.org/10.1017/S0952836902000158
Rogers, L. M., Cheeseman, C. L., Mallinson, P. J. & Clifton-Hadley, R. The demography of a high-density badger (Meles meles) population in the west of England. J. Zool. 242, 705–728. https://doi.org/10.1111/j.1469-7998.1997.tb05821.x (1997).
Graham, J. et al. Multi-state modelling reveals sex-dependent transmission, progression and severity of tuberculosis in wild badgers. Epidemiol. Infect. 141, 1429–1436. https://doi.org/10.1017/S0950268812003019 (2013).
McDonald, J. L., Smith, G. C., McDonald, R. A., Delahay, R. J. & Hodgson, D. Mortality trajectory analysis reveals the drivers of sex-specific epidemiology in natural wildlife-disease interactions. Proc. Biol. Sci. 281 https://doi.org/10.1098/rspb.2014.0526 (2014).
Harris, S., Cresswell, W. J. & Cheeseman, C. L. Age determination of badgers (Meles meles) from tooth wear: The need for a pragmatic approach. J. Zool. 228, 679–684. https://doi.org/10.1111/j.1469-7998.1992.tb04467.x (1992).
Macdonald, D. W., Newman, C., Nouvellet, P. M. & Buesching, C. D. An analysis of eurasian badger (Meles meles) population dynamics: Implications for regulatory mechanisms. J. Mammal. 90, 1392–1403. https://doi.org/10.1644/08-mamm-a-356r1.1 (2009).
Delahay, R. J., Langton, S., Smith, G. C., Clifton-Hadley, R. S. & Cheeseman, C. L. The spatio-temporal distribution of Mycobacterium bovis (bovine tuberculosis) infection in a high-density badger population. J. Anim. Ecol. 69, 428–441. https://doi.org/10.1046/j.1365-2656.2000.00406.x (2000).
Gallagher, J. & Clifton-Hadley, R. S. Tuberculosis in badgers; A review of the disease and its significance for other animals. Res. Vet. Sci. 69, 203–217. https://doi.org/10.1053/rvsc.2000.0422 (2000).
Newman, C., Buesching, C. D. & Macdonald, D. W. in Biology and Conservation of Musteloids (eds. Macdonald, D. W., Newman, C. & Harrington, L. A.) (2017).
Kendon, M. et al. State of the UK climate 2021. Int. J. Climatol. 42, 1–80. https://doi.org/10.1002/joc.7787 (2022).
Kendon, M. et al. State of the UK climate 2022. Int. J. Climatol. 43, 1–83. https://doi.org/10.1002/joc.8167 (2023).
Kruuk, H. & Parish, T. Feeding specialization of the European Badger Meles meles in Scotland. J. Anim. Ecol. 50, 773–788. https://doi.org/10.2307/4136 (1981).
Kruuk, H. Foraging and spatial organisation of the European Badger. Meles meles Behav. Ecol. Sociobiol. 4, 75–89 (1978).
Nguyen, M. V. H. et al. Factors associated with sputum culture-negative vs culture-positive diagnosis of pulmonary tuberculosis. JAMA Netw. Open. 2, e187617–e187617. https://doi.org/10.1001/jamanetworkopen.2018.7617 (2019).
Achkar, J. M. & Jenny-Avital, E. R. Incipient and subclinical tuberculosis: Defining early disease states in the context of host immune response. J. Infect. Dis. 204 (Suppl 4), 1179–1186. https://doi.org/10.1093/infdis/jir451 (2011).
Salina, E. G. et al. Resuscitation of dormant non-culturable Mycobacterium tuberculosis is characterized by Immediate Transcriptional Burst. Front. Cell. Infect. Microbiol. 9, 272. https://doi.org/10.3389/fcimb.2019.00272 (2019).
Mukamolova, G. V., Turapov, O., Malkin, J., Woltmann, G. & Barer, M. R. Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum. Am. J. Respir. Crit. Care Med. 181, 174–180. https://doi.org/10.1164/rccm.200905-0661OC (2010).
Bachmann, N. L. et al. Key transitions in the evolution of Rapid and slow growing mycobacteria identified by comparative genomics. Front. Microbiol. 10 https://doi.org/10.3389/fmicb.2019.03019 (2020).
Mohammed Adam, M. A., Ebraheem, R. S. M. & Bedri, S. A. Statistical investigation of High Culture Contamination Rates in Mycobacteriology Laboratory. Front. Microbiol. 13, 789725. https://doi.org/10.3389/fmicb.2022.789725 (2022).
APHA. (ed Animal and Plant Health Agency). https://www.gov.uk/government/publications/bovine-tb-epidemiology-and-surveillance-in-great-britain-2022 (2023).
APHA. Year end descriptive epidemiology report. Bovine TB in the Edge Area of England 2023. County: East Sussex. https://assets.publishing.service.gov.uk/media/67167e36d100972c0f4c9b3b/EdgeTBReport_2023_EastSussex.pdf (2024).
Balseiro, A. et al. Infection of eurasian badgers (Meles meles) with Mycobacterium avium complex (MAC) bacteria. Vet. J. 188, 231–233. https://doi.org/10.1016/j.tvjl.2010.05.003 (2011).
McCormack, J. Horizon scanning: What next for bovine TB control in England? Ir. Veterinary J. 76 https://doi.org/10.1186/s13620-023-00242-z (2023).
APHA. Bovine tuberculosis in Great Britain in 2023. Explanatory Supplement to the annual TB surveillance reports. September 2024. https://assets.publishing.service.gov.uk/media/66eaba069975b7a980b304e1/ExplanatorySupplement_FINAL.pdf (2024).
Acknowledgements
This project was funded by DEFRA (project APHATBOR1093). We are extremely grateful to the collectors who both promoted the study and actively participated in badger carcass collection, without whom this project would not have been possible. We also thank the pathology and microbiology teams at the University of Nottingham, Sutton Bonington Campus, and the sequencing team at APHA Weybridge.
Author information
Authors and Affiliations
Contributions
S.M.P. was the project manager during the final year of the project, collated and analysed the data, produced the map and figures, and wrote the main manuscript. N.D. was the project manager prior to S.M.P. and collated data, conducted preliminary analyses, and reviewed the manuscript. M.B. conducted PMEs and reviewed the manuscript. B.W. conducted tissue processing, bacterial culture, and isolate screening. E.W. conducted clade assignment and reviewed the manuscript. A.R. assisted with the preliminary analyses and reviewed the manuscript. G.C.S. was involved in project conception and reviewed the manuscript. R.J.D. led on the project conception and contributed to preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Powell, S.M., Dessi, N., Bennett, M. et al. Tuberculosis in found dead badgers at the edge of the expanding bovine tuberculosis epidemic. Sci Rep 15, 10547 (2025). https://doi.org/10.1038/s41598-025-86930-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-86930-y