Abstract
Most classification efforts for primary subtypes of lung adenocarcinoma (LUAD) have not yet been integrated into clinical practice. This study explores the feasibility of combining deep learning and pathomics to identify tumor invasiveness in LUAD patients, highlighting its potential clinical value in assisting junior and intermediate pathologists. We retrospectively analyzed whole slide image (WSI) data from 289 patients with surgically resected ground-glass nodules (GGNs). First, three ResNet deep learning models were used to identify tumor regions. Second, features from the best-performing model were extracted to build pathomics using machine learning classifiers. Third, the accuracy of pathomics in predicting tumor invasiveness was compared with junior and intermediate pathologists’ diagnoses. Performance was evaluated using the area under receiver operator characteristic curve (AUC). On the test cohort, ResNet18 achieved the highest AUC (0.956) and sensitivity (0.832) in identifying tumor areas, with an accuracy of 0.904; Random Forest provided high accuracy and AUC values of 0.814 and 0.807 in assessing tumor invasiveness. Pathology assistance improved diagnostic accuracy for junior and intermediate pathologists, with AUC values increasing from 0.547 to 0.759 and 0.656 to 0.769. This study suggests that deep learning and pathomics can enhance diagnostic accuracy, offering valuable support to pathologists.
Similar content being viewed by others
Introduction
Lung cancer is one of the cancers with the highest mortality rate in the world1. Adenocarcinoma is the most prevalent histopathological subtype of lung cancer, and accounts for approximately one-third to one-half of all lung cancer cases, which presents as ground glass nodules (GGNs) on computed tomography (CT) scans.2,3,4. According to the latest guideline of the 2021 World Health Organization (WHO), lung adenocarcinoma (LUAD) includes precursor glandular lesions (PGL), minimally invasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC). Within PGL, there are atypical adenomatous hyperplasia (AAH) and adenocarcinoma in situ (AIS). It is generally believed that the evolution of GGNs follows the sequence AAH \(\rightarrow\) AIS \(\rightarrow\) MIA \(\rightarrow\) IAC5. Previous studies reported that different subtypes of early-stage LUAD correspond to different responses to treatments and prognoses4,6. PGL and MIA are considered non-invasive and suitable for sublobar resection, with a good prognosis and a 5-year disease-free survival (DFS) rate of almost 100%. However, for IAC patients, lobectomy is more suitable but with a poor prognosis of only 40% to 85% 5-year DFS, and the risk of recurrence is higher as the degree of invasion increases4,6,7. Therefore, accurate assessment of the invasiveness of LUAD with GGNs is essential for clinical decision-making.
With the widespread application of CT in early screening of lung cancer, an increasing number of GGNs, especially sub-centimeter GGNs, are being detected and resected. Consequently, pathologists face multiple challenges in distinguishing AIS, MIA, and IAC, for example, determining the degree of invasion of MIA and IAC in the context of alveolar collapse, as well as distinguishing the adenoid structures formed in collapsed alveoli, requires pathologists to have a high level of expertise through long-term professional training8. Histopathological examination of tissue slides is considered to be the gold standard in the diagnosis of LUAD, and hematoxylin and eosin (H&E) staining technique can capture morphological features of tumor tissue via high-resolution images. However, the manual microscopic examination of H&E staining slides also relies on the expertise of pathologists, and is labor-intensive and prone to errors9. Therefore, it is urgent to develop a reliable, convenient, and fast auxiliary diagnostic tool to provide a reference for the invasiveness degree of LUAD, which is crucial to improving the diagnostic efficiency and accuracy of pathology practitioners, especially junior pathologists in the early stages of their careers.
The emergence of digital pathology has made it possible to use artificial intelligence to perform quantitative analysis of H&E staining-based whole slide image (WSI), and has brought about a series of computer-aided diagnosis methods for pathological images10. For example, pathomics utilizes WSIs to quantitatively extract histological and cytological features, providing the high-throughput characterization of the tumor microenvironment11, e.g., pathomic features were used to reveal immune and molecular evolution from lung preneoplasia to invasive adenocarcinoma12. Unlike pathomics, which mainly includes lesion segmentation, feature extraction and screening, and machine learning classification, deep learning uses specific neural networks to build end-to-end classification models10,13. Previous studies have also demonstrated that deep learning-based histopathological analysis of LUAD has important potential for tumor segmentation, classification, and outcome prediction10,14,15. However, most classification efforts have focused on the main subtypes of LUAD, which have not yet been applied to clinical work, especially those related to the diagnosis of micropapillary subtypes and their prognosis. Moreover, there may be a risk of misleading pathologists if applied in clinical work, such as identifying invasive or non-invasive GGNs, especially for those with relatively limited experience.
Inspired by the success of artificial intelligence in helping the diagnosis of pulmonary nodules in CT scans16,17, we propose a simple and clinically practical method that combines deep learning and pathomics to detect the invasiveness of LUAD. This method can help pathologists identify abnormal tumor regions and preliminarily classify the invasiveness of GGNs from pathology sections, thereby reducing their workload. Unlike previous studies, the objective of this study was to help improve the diagnostic capabilities of junior and intermediate pathologists (those who diagnose fewer than 50 LUAD surgical resection specimens per year).
Materials and methods
Patients
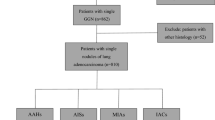
LUAD patients diagnosed with GGNs, from January 2019 to May 2022 at the Affilliated Zhongshan Hospital of Dalian University and from January 2019 to December 2019 at the Affilliated Xinhua Hospital of Dalian University, were enrolled retrospectively and consecutively. The inclusion criteria were: (i) patients aged over 18 years old; (ii) patients who had not been received radiotherapy, chemotherapy, or other treatments before surgery; (iii) CT images and pathological images are of the same GGN; (iv) patients with surgically resected and pathologically confirmed as LUAD. The exclusion criteria were: (i) solid nodules on CT; (ii) incomplete clinical or pathological data; (iii) combined with primary cancer in other locations. Finally, a total of 289 LUAD patients were included in the study cohort. All patients were randomly assigned to training and test cohorts in a ratio of 8:2. Each cohort was further categorized into invasive (ICA) and non-invasive (AAH, AIS, and MIA) groups. The patient enrollment flow chart of this study is shown in Fig. 1.
This study was carried out in accordance with the principles stated in the Declaration of Helsinki and approved by the Ethics Committee of Affiliated Zhongshan Hospital of Dalian University (No. 2021029, Dalian, China). Due to the retrospective nature of the study, the Ethics Committee waived the need of obtaining informed consent.
WSI collection
The postoperative pathological tissues were first immersed in 10% formalin. After embedding in paraffin, they were sectioned to 5\(\mu\)m-thick pathological slides, and then stained with hematoxylin and eosin (H&E) for pathological evaluation. The diagnosis was conducted by two pathologists with more than 10 years of expertise. In case of dispute, the decision was made by the higher-level chief physician. The histopathology slides were digitized using the fully automatic digital pathology whole-slide scanner (Easyscan model, Motic Medical Diagnostic Systems Co., Ltd., Xiamen, China) at 20\(\times\) magnification, with predefined pixel resolution ( 0.5\(\mu\)m/pixel) and an effective tissue area of 15 mm \(\times\) 15 mm. For each patient, two pathologists with more than ten years of clinical experience selected one specific WSI of the largest and most typical tumor tissue under a microscope. Finally, a total of 289 WSIs were collected in this study.
WSI processing
WSI annotation
All WSIs were manually delineated as tumor or non-tumor areas by two pathologists (with 10 and 7 years of diagnostic experience) using QuPath-0.3.2 software18. In case of disagreements during cross-examination, the final decision was made by the higher-level chief pathologist. During the annotation process, areas with folds, bleeding, and blurriness were excluded. Finally, the tumor areas were annotated and labelled as 0 (non-invasive: AAH, AIS, and MIA) or 1 (invasive: IAC) according to the pathological results, respectively. An example of WSI data annotation is illustrated in Fig. 2.
WSI preprocessing and augmentation
All WSIs were preprocessed as follows: (i) the WSIs were partitioned into non-overlapping patches of 512\(\times\)512 pixels to reduce the computer memory requirements; (ii) the patches were processed by thresholding method, followed by removing white regions and eliminating patches with bright pixel ratio exceeding 0.8 or without pathological tissues; (iii) color normalization was performed using Vahadane method to address issues of color variation, inconsistency, and deviation among pathological images19. After the preprocessing, 2,584,435 WSI patches were obtained. Then we adopted image augmentation to increase the diversity of training samples including random translation, random rotation, vertical flipping, and \(90^\circ\) rotation (an example of a patch with its augmented tiles can be found in Fig. S1 in the supplementary file), and image augmentation technique was selected randomly for each epoch, thereby addressing the issue of limited data volume and enhancing generalization ability of the model.
Model specification
Deep learning model
This study used three deep learning models, Resnet18, Resnet50, and Resnet101, to construct a model of tumor region recognition20,21. Only the last fully connected layer in the network used Softmax as the activation function, while the remaining layers used ReLU. The learning rate employed cosine decay learning rate algorithm, and other parameter settings were as follows: batch_size = 128, epoch = 32, initial learning_rate = 0.01, optimizer = Adam. Finally, the optimal model among the three was selected to extract deep learning features.
Feature selection
After feeding all patch-level WSIs into the deep learning model, labels and probabilities for each patch can be obtained. In this study, two multiple instance learning methods were employed to aggregate patch-level features to the WSI-level for image classification: patch likelihood histogram (PLH) and bag of words (BoW)22. PLH and BoW pipelines utilized prediction and probability histograms and term frequency-inverse document frequency features to characterize WSI features, respectively. Then, we used Spearman correlation to remove the features with high repeatability (one was retained if the correlation coefficient between the two pairs was larger than 0.9). Finally, for all features, the least absolute shrinkage and selection operator (LASSO) regression model was used for dimensionality reduction, and the optimal regulation weight (\(\lambda\)) was determined using a 10-fold cross-validation method on the training cohort.
Machine learning-based pathomics model
The features after dimensionality reduction were used to build pathomics models based on machine learning models, including random forest (RF), extremely randomized trees (ExtraTrees), eXtreme gradient boosting (XGBoost), and light gradient boosting machine(LightGBM). The classifier parameters were set as described in Table S1 in the supplementary file. By comparing the predictive performance of different models, the best machine learning model for identifying tumor invasiveness was selected as an assisted tool for pathologists.
Assisting model
The predictive results of pathomics for tumor invasiveness were compared with 10 pathologists (who lacked extensive experience in pathological diagnosis) before and after assistance, and the washout period was set to one month. Among them, 5 pathologists were junior physicians with 1-3 years of experience, while the other 5 pathologists were mid-level physicians with 6-9 years of experience. Inexperience was defined as pathologists reviewing less than 50 LUAD surgical specimens per year.
Performance metrics
Receiver operator characteristic (ROC) curves, the area under ROC curve (AUC), accuracy (ACC), sensitivity (SE), specificity (SP), positive predictive values (PPV), and negative predictive values (NPV) are used to evaluate the predictive performance of the model in the training and testing cohorts. Decision curve analysis (DCA) was employed to assess the clinical utility. A flowchart of pathological feature extraction and model establishment is shown as Fig. 3.
Results
Patient characteristics
This study included 289 patients with LUAD, ranging in age from 26 to 80 years (57.73±10.87). Among them, 196 cases (67.82%) were in the invasive group and 93 cases (32.18%) were in the non-invasive group, including 52 cases of MIA, 32 cases of AIS, and 9 cases of AAH. The training cohort consisted of 230 patients with the mean age of 57.59±10.87, 23.48% males and 76.52% females. 59 cases in the testing cohorts had the mean age of 58.25±10.97 years, with 37.29% males and 62.71% females. Statistical analysis showed the significant difference in age, volume, and maximum diameter between the invasive and non-invasive groups (\(p < 0.001\)), whereas no significant differences were observed between the two groups in terms of CT value, gender, and GGN location (\(p > 0.05\)). Table 1 summarizes the demographic information and sematic CT features of the patients. Out of a total of 2,584,435 WSI patches, 2,135,006 WSI patches were used for the training cohort, 449,429 for the test cohort.
Model evaluation of deep learning
As shown in Table 2, ResNet50 achieved the highest AUC value of 0.973 on the training cohort, while ResNet18 reached the highest AUC value of 0.956 on the testing cohort. Higher AUC values indicate good predictive performance of the model in distinguishing between tumor and non-tumor regions. Regarding ACC, ResNet101 performed well in both the training and test cohorts, achieving 0.924 and 0.904, respectively. Notably, ResNet18 also reached an ACC of 0.904 on the test cohort. In terms of SE, ResNet50 performed the best in the training cohort with a value of 0.836, while in the test cohort, ResNet18 had the highest SE of 0.832. Higher SE values suggest that the model can more accurately identify tumor regions (including invasive regions). Therefore, we selected the ResNet18 model as the best model for its excellent ACC and AUC values, as well as relatively high SE and SP. Figure 4 shows the comparison of ROC curves of three deep learning models for recognizing tumor regions.
Feature selection
After training the patch-level model, we obtained predictions and corresponding probabilities for each patch. Using two pipelines, PLH and BoW, the results at the patch-level were aggregated into WSI-level features. The PLH method yielded a total of 101 probability features and 2 prediction result features, while the BoW similarly produced 101 probability features and 2 prediction result features. Therefore, 206 WSI-level features were then subjected to dimensionality reduction. First, 70 of 206 features were retained via screening the features with high repeatability using Spearman correlation. Second, applying the least absolute shrinkage and selection operator (LASSO) regression model, 7 features with non-zero coefficient (comprising 6 probability features and 1 prediction result feature) were retained for model creation, and the parameter (\(\lambda\)=0.0222) in the LASSO model was selected via 10-fold cross validation based on Mean Squared Error (MSE) value, as shown in Fig. 5.
Moreover, Fig. 6 shows the distribution of values for the retained 7 features across different groups. There are significant differences in the values among the groups (p<0.01), which proves that the features obtained through multiple instance learning have excellent discriminative ability for identifying the invasiveness of LUAD.
Model evaluation of pathomics
Table 3 and Fig. 7 give the predictive results of machine learning-based pathomics model for the invasiveness of LUAD. The RF model showed higher ACC and AUC on both cohorts. Specifically, RF achieved an ACC of 0.817 and an AUC of 0.897 on the training cohort, with a high SE of 0.795, SP of 0.865, PPV of 0.925 and NPV of 0.667. On the testing cohort, RF achieved an ACC of 0.814 and an AUC of 0.807, showing a slight decrease in performance compared to the training cohort. Figure 8 illustrates that, within most threshold probability ranges, the RF model yields high clinical net benefit on both the training and testing cohorts, as shown by the distribution of sample prediction and DCA curves.
Performance comparison of pathomics in assisting junior and intermediate pathologists
Table 4 and Fig. 9 show the performance of the pathomics model, pathologists, and pathology-assisted pathologists in discriminating the invasiveness of LUAD. In the test cohort, the ACC value of RF-based pathomics reached 81.4%, which was significantly higher than the independent review levels of pathologists (junior pathologists at an average of 56.6%, intermediate pathologists at 69.5%). After a one-month washout period, with the assistance of pathomics, the average ACCs of two-level pathologists improved to 79.7% and 80.7%, with increases of 0.231 and 0.112. Additionally, the average AUCs increased by 0.212 and 0.113 for the junior and intermediate pathologists, respectively. In the training cohort, ACCs of RF-based pathomics, junior and intermediate pathologists reached 81.7%, 55.0%, and 71.6%, respectively, whereas with pathomics assisted, ACCs for junior and intermediate pathologists reached 77.3% and 79.9%, respectively. Meanwhile, AUCs for junior and intermediate pathologists increased to 77.2% and 76.0%, respectively.
Discussion
GGNs on CT images usually represent a series of pathological progression, including AAH, AIS, MIA, and IAC. Considering their biological behaviour, AAH, AIS, and MIA are classified as non-invasive with a favorable prognosis compared to IAC.23. Accurate postoperative pathological diagnosis of the invasiveness of GGNs is crucial for subsequent treatment and follow-up. This study focused on postoperative patients with LUAD manifesting as GGNs on CT images. We retrospectively compared the pathological features of patients with invasive and non-invasive LUAD and established a deep learning model based on ResNet18 for predicting tumor regions. We found that the ResNet18 model had high predictive efficiency in identifying tumor regions of LUAD. For pathologists with less experience in hospitals with limited surgical cases, the incorporation of pathomics-assisted diagnosis has the potential to enhance diagnostic efficiency. We initially opted not to conduct a detailed subtype analysis to ensure clarity for pathologists with varying levels of experience. We aim to contribute to both work efficiency and diagnostic assistance, addressing the challenges faced by pathologists with high workload and potential misdiagnoses. We aspire to provide pathologists with positive prompts during the diagnostic process. Our contributions in this study are as follows: (1) Three end-to-end ResNet deep learning models in this study are applied and to accurately identify tumor regions; (2) Using the features from deep learning, we built four kinds of pathomics based on machine learning models to assess the invasiveness of lung adenocarcinoma; (3) We built a simple and clinically practical tool using deep learning and pathomics for tumor invasiveness, and explore its convenience in future clinical applications and its potential value in improving the diagnostic capabilities of junior and intermediate pathologists.
Significance of ResNet model in identifying tumor regions
In this study, convolutional neural network (CNN) based ResNet models were employed to classify WSIs into tumor and non-tumor regions, aiming to enhance the efficiency of pathologists in identifying tumor regions of LUAD and reducing diagnostic omissions. The ResNet model was initially proposed in 2015 by He et al., introducing residual learning units to solve the issues of vanishing gradients when increasing the network depth20. Although there are deeper ResNet variants such as ResNet50, ResNet101, and ResNet152, ResNet18 had satisfactory performance in the current task. In this study, we found the performance of the ResNet model does not change significantly with the increase in the number of layers in identifying the tumor regions. The AUC values on the training and testing cohorts were 0.957 and 0.956, demonstrating superior generalization performance of the ResNet18 model.
Furthermore, the ResNet18 model achieved an ACC of 90.4% on the test cohort, showing excellent SE, SP, PPV, and NPV values in identifying invasive tumor regions. These results indicate that despite the limited amount of tumor tissues, especially when GGNS are smaller than 1cm on CT images, we can still accurately localize tumor regions. Therefore, utilizing the ResNet18 model in identifying tumor regions of LUAD in WSIs is an effective and feasible strategy. It not only enhances the efficiency of pathologists in recognizing and evaluating tumor region, but also helps reduce diagnostic omissions and provides a valuable auxiliary tool for future clinical pathological diagnoses. While it would also be valuable to compare different CNN structures, such as VGG, Inception, or DenseNet, exploring a single family of architectures in depth can provide detailed insights into their specific advantages and limitations in the given context, thereby offering a robust foundation for future comparisons with other CNN structures.
Significance of machine learning-based pathomics in predicting the invasiveness
The morphological evaluation criteria for tissue specimens in lung cancer diagnosis are often considerably subjective, Thunnissen et al. pointed out that alveolar collapse and extensive epithelial proliferation were the main causes of interobserver variability24. The combination of digital pathology and artificial intelligence is expected to provide objective support for decision-making13. This study employed machine learning-based pathomics approaches to predict the invasiveness of GGN-type LUAD. First, the LASSO model was applied to reduce 206 WSI-level features to 7 significant features. Second, machine learning models including RF, ExtraTrees, XGBoost, and LightGBM were utilized to construct the predictive models. Among them, RF and ExtraTrees both achieved ACC value of 0.814 on the test cohort, while RF had a higher AUC value of 0.807. To the best of our knowledge, there is limited research specifically addressing deep learning-based pathomics for lung adenocarcinoma invasiveness classification, and this gap shows the novelty and significance of our study. While direct comparisons are rare, there are several studies in related areas that provide valuable context. Yu et al. combined traditional thresholding and image processing techniques with machine learning methods, achieving an AUC of 0.85 for distinguishing normal and tumor tissues using WSI data, and an AUC of 0.75 for differentiating LUAD from squamous cell carcinoma25. Wang et al. subdivided WSI slides of LUAD into 300\(\times\)300-pixel patches and employed a CNN model to classify malignant or non-malignant, with an overall classification accuracy of 89.8%26. In another study, the authors generated a deep learning pathomics (DLP) with WSI features for the automated segmentation of tissue, and achieved an AUC value of 0.802 to preoperatively distinguish between luminal and non-luminal breast cancers at early stages21.
Studies have shown that distinguishing MIA from invasive adenocarcinoma is often challenging when the maximum diameter of tumor invasiveness on frozen sections approaches 5mm27. This difficulty also occurs in postoperative paraffin sections, compounded with the tendency of pathologists to overestimate the diseases, leading to inevitable human errors28. Therefore, it is significantly important to accurately assess the extent of invasiveness. Furthermore, there is a lack of consistency in the diagnosis of LUAD histological subtypes, especially in the diagnosis of invasive and non-invasive cancer subtypes, evaluated by different pathologists or even the same pathologist at different times29,30,31. These practical clinical problems provide potential opportunities for artificial intelligence to make a significant contribution to diagnostic standardization and homogenization process in the future. Although the model demonstrated high accuracy in classifying non-invasive and invasive LUAD, we acknowledge its limitation in distinguishing the invasive component of MIA, i.e., the model failed to classify the invasive component of MIA as invasive. This may be attributed to the fact that although pathomic divides digitized pathological slides into multiple patches, the model is trained to differentiate between invasive and non-invasive lesions based on feature patterns derived from the entire WSIs. While MIA may contain small invasive components, our model primarily focuses on larger regions of invasion that exceed this threshold, as these have a more significant impact on prognosis. Therefore, the model does not classify the minor invasive component of MIA as invasive. To address this limitation, the refinement of the training process and the incorporation of multi-scale analysis or auxiliary modalities may be required to better differentiate between small-scale invasiveness in cases like MIA.
Significance of pathomics in assisting junior and intermediate pathologists
Traditional pathological diagnosis suffers from high subjectivity, low efficiency, and prolonged time consumption, and relies heavily on the practical experience and diagnostic proficiency of pathologists. These problems are particularly pronounced in hospitals that perform fewer surgeries and lack subspecialty pathologists. Therefore, this study combined deep learning and pathomics as an assistant tool to enhance the efficiency and accuracy of pathological diagnosis. The results demonstrated that pathologists with assistance can effectively adjust their diagnostic levels, thereby improving diagnostic accuracy. In addition, the deep learning model can help pathologists rapidly interpret tumor regions. Even though the accuracy of deep learning models may not be entirely ideal, pathologists can make effective adjustments to tumor regions by evaluating the model results.
This study focuses on how pathomics can assist pathologists in interpreting the invasiveness of LUAD, thereby improving work efficiency and reducing misdiagnosis and missed diagnosis. Fortunately, with the help of pathomics, both pathologists obtained improvements in ACC, AUC, and other metrics. Wei et al. built a deep learning model to distinguish subtypes of LUAD, and the model’s performance was comparable to pathologists, suggesting that it can be used clinically to help pathologists classify LUAD14. Pan et al. utilized artificial intelligence-based grading to predict disease-free survival and consistently improved prognosis in stage I tumors to further assist pathologists8. Unlike these studies, we aim to help improve the diagnostic performance of junior and intermediate pathologists (those who diagnose fewer than 50 surgical resection specimens of LUAD per year). The pathomics model performed better than the pathologists, whose ACC and AUC values both improved with pathomics’ assistance.
While achieving some anticipated results in this study, there are still some limitations and future directions. First, the sample sizes of both the training and testing cohorts were relatively small, potentially impacting the generalizability and robustness of the learning model to a certain extent, and next we plan to expand the cohort size and incorporate external datasets or to implement nested cross-validation for more comprehensive validation. Second, the WSIs were manually annotated by pathologists, in future work, we will explore weakly supervised or unsupervised methods for more objective classification. Third, this study only utilized the ResNet model, next we will try to conduct comparative studies with other models, such as AlexNet, VGGNet, InceptionNet, EfficientNet, ViT, HIPT, and GigaPath etc., to further improve and validate the diagnostic efficiency. Furthermore, we acknowledge that a more detailed subtype analysis could provide valuable insights, and this will be one of our future work.
Conclusion
This study retrospectively included 289 patients of lung adenocarcinoma (LUAD) with GGNs on CT and established a predictive model for tumor invasiveness based on deep learning and pathomics. The ResNet18 network can accurately locate tumor regions, while pathomics can effectively distinguish the invasiveness of LUAD using WSIs. We also confirmed the efficacy of the model in assisting inexperienced pathologists in identifying tumor regions and determining tumor invasiveness. Our future work will focus on enhancing the model’s accuracy and generalizability by incorporating a larger, multi-center dataset and exploring more state-of-the-art deep learning architectures. We plan to integrate multi-modal imaging data and clinical information to further improve the predictive capabilities of our model, and we hope to advance the capabilities of AI-assisted diagnosis on LUAD.
Data availability
The data generated during the current study are not publicly available due to patient privacy but are available from the corresponding author on reasonable request.
References
Tran, K. B. et al. The global burden of cancer attributable to risk factors, 2010–19: a systematic analysis for the global burden of disease study 2019. Lancet 400, 563–591 (2022).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. Ca Cancer J Clin 73, 17–48 (2023).
Wang, C. et al. Deep learning for predicting subtype classification and survival of lung adenocarcinoma on computed tomography. Transl Oncol 14, 101141 (2021).
Ding, H. et al. Ct-based deep learning model for invasiveness classification and micropapillary pattern prediction within lung adenocarcinoma. Front Oncol 10, 1186 (2020).
Weichert, W. & Warth, A. Early lung cancer with lepidic pattern: adenocarcinoma: in situ: minimally invasive adenocarcinoma, and lepidic predominant adenocarcinoma. Curr Opinion Pulmonary Med 20, 309–316 (2014).
Hattori, A. et al. Distinct clinicopathologic characteristics and prognosis based on the presence of ground glass opacity component in clinical stage ia lung adenocarcinoma. J Thoracic Oncol 14, 265–275 (2019).
Travis, W. D. et al. The iaslc lung cancer staging project: proposals for coding t categories for subsolid nodules and assessment of tumor size in part-solid tumors in the forthcoming eighth edition of the tnm classification of lung cancer. J Thoracic Oncol 11, 1204–1223 (2016).
Pan, X. et al. The artificial intelligence-based model anorak improves histopathological grading of lung adenocarcinoma. Nat Cancer 1–17 (2024).
Zhang, Y. et al. Histopathology images-based deep learning prediction of prognosis and therapeutic response in small cell lung cancer. NPJ Digital Med 7, 15 (2024).
Wang, S. et al. Artificial intelligence in lung cancer pathology image analysis. Cancers 11, 1673 (2019).
Gupta, R., Kurc, T., Sharma, A., Almeida, J. S. & Saltz, J. The emergence of pathomics. Curr Pathobiol Rep 7, 73–84 (2019).
Chen, P. et al. Pathomic features reveal immune and molecular evolution from lung preneoplasia to invasive adenocarcinoma. Modern Pathol 36, 100326 (2023).
Bishnoi, V. & Goel, N. A color-based deep-learning approach for tissue slide lung cancer classification. Biomed Signal Process Control 86, 105151 (2023).
Wei, J. W. et al. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep 9, 3358 (2019).
Kim, P.-J. et al. A new model using deep learning to predict recurrence after surgical resection of lung adenocarcinoma. Sci Rep 14, 6366 (2024).
Hendrix, W. et al. Deep learning for the detection of benign and malignant pulmonary nodules in non-screening chest ct scans. Commun Med 3, 156 (2023).
Gao, J. et al. Artificial-intelligence-based computed tomography histogram analysis predicting tumor invasiveness of lung adenocarcinomas manifesting as radiological part-solid nodules. Front Oncol 13, 1096453 (2023).
Bankhead, P. et al. Qupath: Open source software for digital pathology image analysis. Sci Rep 7, 1–7 (2017).
Vahadane, A. et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging 35, 1962–1971 (2016).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778 (2016).
Huang, Y. et al. Deep learning radiopathomics based on preoperative us images and biopsy whole slide images can distinguish between luminal and non-luminal tumors in early-stage breast cancers. EBioMedicine 94 (2023).
Cao, R. et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in colorectal cancer. Theranostics 10, 11080 (2020).
Travis, W. D. et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thoracic Oncol 6, 244–285 (2011).
Thunnissen, E. et al. Defining morphologic features of invasion in pulmonary nonmucinous adenocarcinoma with lepidic growth: A proposal by the international association for the study of lung cancer pathology committee. J Thoracic Oncol 18, 447–462 (2023).
Yu, K.-H. et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 7, 12474 (2016).
Wang, S. et al. Comprehensive analysis of lung cancer pathology images to discover tumor shape and boundary features that predict survival outcome. Sci Rep 8, 10393 (2018).
He, P., Yao, G., Guan, Y., Lin, Y. & He, J. Diagnosis of lung adenocarcinoma in situ and minimally invasive adenocarcinoma from intraoperative frozen sections: an analysis of 136 cases. J Clin Pathol (2016).
Sakamoto, T. et al. A narrative review of digital pathology and artificial intelligence: focusing on lung cancer. Transl Lung Cancer Res 9, 2255 (2020).
Thunnissen, E. et al. Reproducibility of histopathological subtypes and invasion in pulmonary adenocarcinoma. an international interobserver study. Modern Pathol 25, 1574–1583 (2012).
Warth, A. et al. Interobserver variability in the application of the novel iaslc/ats/ers classification for pulmonary adenocarcinomas. Eur Resp J 40, 1221–1227 (2012).
Shih, A. R. et al. Problems in the reproducibility of classification of small lung adenocarcinoma: an international interobserver study. Histopathology 75, 649–659 (2019).
Acknowledgements
This work was supported by National Natural Science Foundation of China (No.82071911), Natural Science Foundation of Inner Mongolia (No.2023MS08031), Dalian Outstanding Young Technology Talents Program (No.2023RY034). The authors thank all the staffs from the Departments of Pathology, Affiliated Zhongshan Hospital of Dalian University and Affiliated Xinhua Hospital of Dalian University, for their help in collecting the data.
Author information
Authors and Affiliations
Contributions
HD: Conceptualization, Study design, Formal analysis, Investigation, Writing-Original draft. XW: Study design, Formal analysis, Methodology, Visualization, Writing-Review & Editing. KW, QA, JS, and RZ: Data acquisition, Data analysis and interpretation, Writing-Review & Editing. JW: Conceptualization, Supervision, Project administration, Funding acquisition, Writing-Review & Editing
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Du, H., Wang, X., Wang, K. et al. Identifying invasiveness to aid lung adenocarcinoma diagnosis using deep learning and pathomics. Sci Rep 15, 4913 (2025). https://doi.org/10.1038/s41598-025-87094-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87094-5