Abstract
To evaluate the value of increasing sequencing depths of non-invasive prenatal testing (NIPT) for fetal chromosomal aneuploidies based on the semiconductor sequencing platform. This study recruited a cohort of 59,800 singleton pregnancies from Guangdong Women and Children Hospital between January 2015 and December 2020, including 48,018 cases of NIPT and 11,782 cases of expanded NIPT. Cell-free DNA from plasma samples was sequenced at a sequencing depth of 0.15X for NIPT and 0.4X for expanded NIPT. Patients with positive NIPT results were offered fetal karyotyping or microarray analysis for confirmatory testing, and all pregnant women were followed up. A total of 892 cases were predicted as positive for chromosomal aneuploidies, including 682 cases of NIPT and 210 cases of expanded NIPT, with a positive rate of 1.42% and 1.78% (p = 0.0037), respectively. The positive predictive values (PPV) for trisomy 21/18/13 and other autosomal aneuploidies detected by NIPT were 84.80%, 69.23%, 25.00%, and 4.55%, respectively. For expanded NIPT, the positive predictive rates were 86.96%, 80.00%, 35.00%, and 8.77%, respectively. Both NIPT and expanded NIPT have the same PPV for detecting sex chromosome aneuploidies (approximately 50%). Additionally, one false-negative case was identified during the follow-up period. This was a trisomy 18 confirmed by prenatal diagnosis due to multiple ultrasound abnormalities during pregnancy. The study shows that increasing sequencing depth can significantly improve the PPV performance of aneuploidies, with a statistically significant difference, particularly in trisomy 18. The results may provide valuable guidance for clinical doctors during consultations.
Similar content being viewed by others
Introduction
Since 1997, the discovery of cell-free fetal DNA (cffDNA) has opened up a new approach for non-invasive prenatal testing(NIPT)1. NIPT, which uses massively parallel sequencing (MPS), has gradually developed into a first-line screening method for fetal chromosomal aneuploidies over the past decade due to a very high detection rate not only for trisomy 21, 18 and 132,3, but also an excellent predictive value for sex chromosomes and chromosome copy number variations (CNVs)4,5,6,7. In China, NIPT is recommended for screening trisomy 21 (T21), trisomy 18 (T18) and trisomy13 (T13) for patients with intermediate risk of serological screening results or serological screening in cases with a single marker abnormality of AFP, β-HCG or uE3 in the first or second trimester8, and with ultrasound soft marker abnormalities9. Some advanced-aged pregnant women voluntarily choose NIPT, especially after the implementation of the two-child policy in China because the number of advanced age pregnant women has gradually increased. Many of them refuse to undergo prenatal diagnosis, with the increased knowledge about NIPT and educational is popularisation more pregnant women actively choose NIPT10.
NIPT encompasses several technical platforms as it is derived from different high-throughput sequencing principles11,12,13. In recent years, expanded NIPT has gradually developed into a research focus for clinical applications. Expanded NIPT is a technique that expands the detection range of NIPT from trisomy 13,18 and 21 to all autosomal aneuploidies, sex chromosome aneuploidies, and sub-chromosomal microdeletions/microduplications by increasing sequencing depth14. More studies have demonstrated that expanded NIPT is feasible for the detection of CNVs15,16. However, few studies have compared the chromosomal aneuploidy detection efficiency using the same platform with different sequencing depths. The aim of the present study was to retrospectively compare the chromosomal aneuploidy detection efficiency of NIPT using the semiconductor sequencing platform (SSP) with a 0.15X depthand a data volume of 3 million reads for NIPT, and q 0.4X depthand a data volume of 8 million reads for expanded NIPT.
Methods
Participant recruitment
This study was approved by the Ethics Review Committee of Guangdong Women and Children Hospital (Guangzhou, China; No. 20251007). All clinical procedures and methods, including counseling, testing, were performed in strict accordance with the ‘Technical specifications for prenatal screening and diagnosis of fetal cell-free DNA in maternal peripheral blood’ in China9. Pregnant women who were willing to undergo NIPT were informed about the purpose, significance, accuracy, risks and limitations by the attending physicians and signed written informed consent.
The retrospective study enrolled 59,800 consecutive high-risk singleton pregnancy women in Guangdong Woman and Children Hospital from January 2015 to December 2020, including 48,018 in NIPT group and 11,782 in expanded NIPT group. According to the Chinese fetal cell-free DNA testing guideline9, the pregnancy characteristics of these women were assigned to five group: (i) advanced maternal age (≥ 35 years, AMA); (ii) ultrasound soft marker abnormalities (including a thickened nuchal fold, rhizomelic limb shortening, mild fetal pyelectasis, echogenic bowel, echogenic intracardiac focus and choroid plexus cyst; (iii)serological screening for high or intermediate risks; (iv) serological screening for the single marker abnormality (AFP, β-HCG and uE3); and (v) Other types: history of pregnancies with fetuses having chromosomal aneuploidy abnormalities. Pregnant women within the scope of the indication chose NIPT or expanded NIPT according to their wishes.
Sample preparation and sequencing
For each patient, aperipheral blood sample (5 ml) was withdrawn from the cubital vein using EDTA-K2 tubes, and centrifuged at 4 ℃ and 1,600 × g for 10 min within 6 h first of all and then centrifuged at 4℃ and 16,000 × g fo 10 min to isolate plasma using an Eppendorf 5810R and 5424 centrifuge (Eppendorf) according to the JingXin Fetal Chromosome Aneuploidy (T21, T18, T13) Testing Kits (NMPA registration permit No. 0153400300). Samples were stored at -20℃as soon as possible until genomic DNA extraction. Whole-genome sequencing was performed using semiconductor sequencing technique on the Bioelectronseq 4000 sequencing platform (NMPA registration permit No. 20153400309). Following the DNA library construction, 9 ~ 23 libraries were pooled and then sequenced within ~ 200 bp reads as described previously17. Combined GC-correction and Z-score testing methods were used to identify fetal chromosome aneuploidy of trisomy 21, 18, and 13 as described previously18. Meanwhile, fetal and maternal chromosome aneuploidies were classified using our modified Stouffer’s Z score method as described previously18.
Prenatal diagnosis and pregnancy follow-up
Pregnant women at high risk for NIPT received genetic counselling and were fully informed about prenatal diagnosis based on fetal cells by amniotic fluid or cord blood puncture for fetal chromosomal analysis. Chromosomal detection techniques included karyotyping (G-banding resolution was 400 bands) and chromosomal microarray analysis (CMA) (CytoScanTM 750 K, available from Affymetrix, USA). To obtain information on neonatal outcomes and neonatal growth, all participants were followed up by telephone interviews.
Statistics
Excel and R language were used for data statistical analysis. Positive predictive values (PPVs) of fetal chromosomal aneuploidies detected by NIPT were calculated based on prenatal diagnosis results. Chi-square and Fisher’s exact probability tests were used to compare the PPVs of fetal chromosomal aneuploidy PPVs for NIPT between different groups. Results with p-values of less than 0.05 were considered statistically significant.
Results
Population profiles
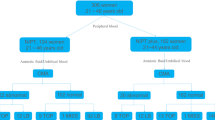
From January 2015 to December 2020, 59,800 cases were successfully recruited for NIPT detection in the Prenatal Diagnosis Center of Guangdong Women and Children Hospital.Among them, 48,018 patients who chose 3 million reads sequencing depth detection served as NIPT, and 11,782 patients who chose 8 million reads sequencing depth detection served as expanded NIPT. In NIPT group, the mean age of the pregnant women was 30.2 ± 12.5 years, and most of them (76.95%) were found to have abnormal serological screening. In expanded NIPT group, the mean age of the pregnant women was 30.1 ± 10.8 years, and most of them (60.64%) were found to have abnormal serological screening. Of these 59,800 pregnant women, 7609 were AMA women (age ≥ 35 years). In addition, there were 4020 in-vitro fertilization (IVF) pregnancies.Basic demographic and clinical characteristics are shown in Table 1, and the study flow is shown in Fig. 1.
NIPT results
A total of 892 positive and 58,908 negative cases of fetal chromosomal aneuploidy were detected in the cohort, including 682 positive cases in the NIPT group with a positive rate of 1.42% and 210 positive cases in the expanded NIPT group with a positive rate of 1.78%(1.42% vs. 1.78%, p = 0.0037). Of the positive pregnancies, 804 (90.13%) were followed up with amniocentesis and prenatal diagnosis, and the remaining 88 positive pregnancies (9.87%) declined prenatal diagnosis after genetic counselling. There were 270 cases of trisomy 21 (T21), 74 cases of trisomy 18 (T18) and 67 cases of trisomy 13 (T13). There were also 254 cases of rare autosomal aneuploidies and 227 cases of sex chromosome aneuploidies. In addition, pregnancy-related information was lost in 18 cases due to refusal of follow-up.
Comparison of PPVs between NIPT and expanded NIPT for aneuploidies
A total of 892 cases at high risk of aneuploidy were detected by NIPT and expanded NIPT, and the overall positive rate was 1.49%. The positive predictive values (PPVs) for NIPT for detecting trisomy 21/18/13 were 84.80%, 69.23%, and 25.00%, respectively. In contrast, the PPVs for expanded NIPT for these conditions were 86.96%, 80.00%, and 35.00%, respectively.Expanded NIPT had increased the PPVs for detecting trisomy 21/18/13, especially for trisomy 18 (69.23% vs. 80.00%, p < 0.001) (Table 2; Fig. 1). The PPVs of NIPT and expanded NIPT for detecting other autosomal aneuploidies were 4.55% and 8.77%(p = 0.24) respectively, showing no significant difference. The PPVs of NIPT for sex chromosome aneuploidy were the same as expanded NIPT (50.00%).
Comparison of PPVs for aneuploidies between NIPT and expanded NIPT according to different pregnancy characteristics
At the same time, we analyzed and compared the PPVs of NIPT and expanded NIPT in different pregnancy characteristics groups. There was a significant difference in PPVs of ultrasound soft marker abnormalities between NIPT and expanded NIPT groups(p = 0.01), but there was no difference in the PPVs for any other group (Table 3).
Comparison of detection efficiency for rare trisomy
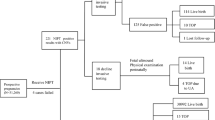
A total of 254 cases of rare trisomy aneuploidies were detected by NIPT and expanded NIPT (Fig. 2),and the positive rate was 0.51% for NIPT and 0.57% for expanded NIPT. The PPVs were similar between NIPT and expanded NIPT (4.55% vs. 8.77%,p = 0.24). Of the 254 positive cases, 12 true positives cases were confirmed by invasive prenatal diagnosis, including 2 cases of partial segment loss of heterozygosity, 9 cases of low-proportion mosaic trisomy, and 1 case of trisomy 20 (Table 4).
Follow-up low-risk pregnancies and pregnancies who declined prenatal diagnosis
According to the ‘Technical Specifications of Prenatal Screening and Diagnosis of Fetal Free DNA in Peripheral Blood of Pregnant Women’ promulgated by the China National Health and Family Planning Commission in 20169, all pregnant women were followed up until 3 months after newborn delivery, except one false-negative case of trisomy 1819, and 63 patients who refused prenatal diagnosis were also followed up. The rest of the pregnant women had given birth successfully, and no visible abnormality was found in the newborn screening.
Discussion
Previous research has mainly focused on assessing the performance of expanded non-invasive prenatal testing (NIPT) for the detection of copy number variations (CNVs). Our study is the first to evaluate the value of increasing sequencing depth for the detection of chromosomal aneuploidies using a semiconductor sequencing platform. We found no significant difference in detection performance between the two methods for aneuploidies overall. However, expanded NIPT showed a slightly higher total positive rate(1.78% vs. 1.42%;p = 0.0037). Despite these advantages, expanded NIPT did not significantly improve the detection of rare autosomal aneuploidies, highlighting a notable limitation of NIPT that needs to be acknowledged.
We used PPVs to evaluate the detection performance in this study. The PPVs of NIPT and expanded NIPT for all chromosome aneuploidies were 50.16% and 47.28%, respectively (p = 0.02,p < < 0.05). This result showed an improvement in the detection efficiency of expanded NIPT compared to NIPT, which benefited from increasing sequencing depth. The PPVs of NIPT for detecting T21/T18/T13 was 84.80%, 69.23%, 25.00% respectively. The results were basically consistent with Liu’s findings20. The PPVs of expanded NIPT for detecting T21/T18/T13 was 86.96%, 80.00%, and 35.00% respectively.The improvement of detection efficiency for T18 was especially marked(p < 0.001).The PPV for other chromosome aneuploidieswas 4.55% for NIPT and 8.77% for expanded NIPT. The difference was not statistically significant15. The PPV for detecting sex chromosome aneuploidy was 50% for NIPT, which was similar with that for expanded NIPT. This is an unexpected discovery. Similarly, Zheng’s paper also showed these results21. Sex chromosomes became the target disease. The awareness of preventing missed diagnoses directly affected the subjective judgement of the result reviewers.
We also analysed the differences in PPV between groups with different pregnancy characteristics and between NIPT and expanded NIPT. The difference in ultrasound soft marker abnormalities of PPVs was significant different between NIPT and expanded NIPT (P = 0.01). The reason may be that more pregnant women with ultrasound soft markers chose to undergo extended NIPT testing (23.12% vs. 9.65%, P = 0), The detection rate of chromosomal abnormalities in fetuses with ultrasound soft markers is relatively higher compared to other groups in the study, and this conclusion has been reported in other literatures22,23. Whether it is in China or other countries and regions (“Screening for Fetal Chromosomal Abnormalities: ACOG Practice Bulletin, Number 226,” 2020)24, the applicable population of NIPT has been clearly defined. However, the PPV was slightly different in different clinical characteristics groups. This study provided aviable idea to improve the detection efficiencyof NIPT for clinicians. In addition, the differences in characteristics between the two groups were mainly due to the large differences in sample size between the two groups.
During recent years, other rare autosomal aneuploidies (RAAs) were often reported in clinical practice as the unexpected findings of NIPT. The PPVs for other chromosome aneuploidies were at depressed 8.77% and similar with those reported in Chen’s paper15. But compared with Liang’s paper4, it was slightly lower. One possible reason is the lack of data for the NIPT’s analysis model which caused by the pretty low incidence of RAAs, and the frequent early spontaneous abortions of the RAAs25. Another important reasonis that these aneuploidies had high rates of confined placental mosaicism (CPM)26,27. Ascell-free fetal DNA was mainly derived from apoptotic placental trophoblasts, the result of NIPT actually shows the characteristic of the placental trophoblasts. That makes the Confined placental mosaicism led to the false positive results of NIPT28,29. Among all the rare autosomal aneuploidies, Trisomy 7 was the most common, but PPV was still low like other rare autosomal aneuploidies. Simultaneously, the prognosis of perinatal infants was very good30. It is worth noting that the 2 true positive cases were confirmed to be loss of heterozygosity. It was suspected to be a uniparental diploid of chromosome 16 (mixed type) and a uniparental diploid of chromosome 14 respectively.Intrauterine growth retardation occurred in two fetuses, and intrauterine death occurred in the case of uniparental diploid of chromosome 16, and the woman with uniparental diploid of chromosome 14 had a normal delivery and live birth.
Follow-up played a crucial role in Non-Invasive Prenatal Testing (NIPT). With the exception of those who declined follow-up, all other patients were monitored for pregnancy outcomes. In accordance with the guidelines set forth by the National Health Commission of the People’s Republic of China, follow-up commenced at the 12th week post-delivery. The scope of follow-up encompassed any complications experienced during pregnancy, the outcomes of the pregnancies, and the health status of the newborns. It is worth noting that a limitation of this study was the absence of post-delivery verification for false-positive cases concerning the placenta. Another limitation of this study is that the detection performance of NIPT for RAA is affected by aneuploidy types and the potential for mosaicism, and further analysis with large samples is still needed. In addition, There are some tendencies among pregnant women who choose either NIPT or expanded NIPT. We compared the PPVs for for whole chromosome aneuploidies of NIPT and expanded NIPT according to different pregnancy characteristics. We found that pregnancies with ultrasound soft-marker abnormalities had higher PPVs with both NIPT and expanded NIPT than the other 4 groups of women, and the difference was statistically significant between the NIPT and expanded NIPT groups ac cording to pregnancy characteristics.Since pregnant women chose NIPT or expanded NIPT nonrandomly, some deviach was a limitation of the present study. Therefore, a large-scale NIPT study of dif ferent pregnancy characteristics is a direction of our fur ther research.
Conclusion
The results showed that increasing sequencing depth can significantly improve the PPV performance of aneuploidies, with a statistically significant difference, particularly in Trisomy 18. The results may provide some guidance for clinical doctors when consulting.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- NIPT:
-
Noninvasive prenatal testing
- PPV:
-
Positive predictive values
- RAAs:
-
Rare autosomal aneuploidy
- CNV:
-
Copy number variations
- cffDNA:
-
Cell free fetal DNA
- MPS:
-
Massively parallel sequencing
- CMA:
-
Chromosome microarray analysis
- NGS:
-
Next generation sequencing
- SSP:
-
Semiconductor sequencing platform
- AFP, β-HCG and uE3:
-
Alpha fetal protein, β human chorionic gonadotropin, and unconjugated estriol
- AMA:
-
Advanced maternal age
References
Lo, Y. et al. Presence of fetal DNA in maternal plasma and serum. Lancet 350(9076), 485–487. https://doi.org/10.1016/s0140-6736(97)02174-0 (1997).
Samura, O. Update on noninvasive prenatal testing: A review based on current worldwide research. J. Obstet. Gynaecol. Res. 46(8), 1246–1254. https://doi.org/10.1111/jog.14268 (2020).
Van der Meij, K. R. M. et al. TRIDENT-2: National implementation of genome-wide non-invasive prenatal testing as a first-tier screening test in the Netherlands. Am. J. Hum. Genet. 105(6), 1091–1101. https://doi.org/10.1016/j.ajhg.2019.10.005 (2019).
Liang, D. et al. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet. Med. 21(9), 1998–2006. https://doi.org/10.1038/s41436-019-0467-4 (2019).
Li, W., Zhang, Y., Gao, J. & Hu, J. The predictive value of noninvasive prenatal screening for copy number variations: A cohort study and a systematic meta-analysis. Expert Rev. Mol. Diagn. 23(8), 713–722. https://doi.org/10.1080/14737159.2023.2233415 (2023).
Zou, Y. et al. Performance of expanded non-invasive prenatal testing for fetal aneuploidies and copy number variations: A prospective study from a single center in Jiangxi province, China. Front. Genet. 13(13), 1073851. https://doi.org/10.3389/fgene.2022.1073851 (2023).
Yang, J. et al. Noninvasive prenatal detection of fetal sex chromosome abnormalities using the semiconductor sequencing platform (SSP) in Southern China. J Assist. Reprod. Genet. 38(3), 727–734. https://doi.org/10.1007/s10815-020-02056-2 (2021).
McCullough, R. M. et al. Non-invasive prenatal chromosomal aneuploidy testing–clinical experience: 100,000 clinical samples. PLoS ONE 9(10), e109173. https://doi.org/10.1371/journal.pone.0109173 (2014).
Health, N. & Office, F. P. C. Technical specifications for prenatal screening and diagnosis of fetal cell-free DNA in maternal peripheral blood. Bull. Natl. Health Fam. Plan. Commiss. People’s Republic of China 10, 52–68 (2016).
Zhu, W., Ling, X., Shang, W. & Huang, J. The knowledge, attitude, practices, and satisfaction of non-invasive prenatal testing among chinese pregnant women under different payment schemes: A comparative study. Int. J. Environ. Res. Public Health 17(19), 66. https://doi.org/10.3390/ijerph17197187 (2020).
Benn, P. & Cuckle, H. Overview of noninvasive prenatal testing (NIPT) for the detection of fetal chromosome abnormalities; differences in laboratory methods and scope of testing. Clin. Obstet. Gynecol. 66(3), 536–556. https://doi.org/10.1097/GRF.0000000000000803 (2023).
Shi, P. et al. The potential of expanded noninvasive prenatal screening for detection of microdeletion and microduplication syndromes. PrenatDiagn https://doi.org/10.1002/pd.6002 (2021).
Xie, M. et al. Noninvasive prenatal testing of rare autosomal aneuploidies by semiconductor sequencing. DNA Cell Biol. 37(3), 174–181. https://doi.org/10.1089/dna.2017.4075 (2018).
Li, C. et al. Clinical potential of expanded noninvasive prenatal testing for detection of aneuploidies and microdeletion/microduplication syndromes. Mol. Diagn. Ther. 27(6), 769–779. https://doi.org/10.1007/s40291-023-00674-x (2023).
Chen, Y. et al. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 42,910 single pregnancies with different clinical features. Hum. Genomics 13(1), 60. https://doi.org/10.1186/s40246-019-0250-2 (2019).
Srinivasan, A., Bianchi, D. W., Huang, H., Sehnert, A. J. & Rava, R. P. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am. J. Hum. Genet. 92(2), 167–176. https://doi.org/10.1016/j.ajhg.2012.12.006 (2013).
Yin, A. et al. Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA. Proc. Natl. Acad. Sci. USA 112(47), 14670–14675. https://doi.org/10.1073/pnas.1518151112 (2015).
Liao, C. et al. Noninvasive prenatal diagnosis of common aneuploidies by semiconductor sequencing. Proc. Natl. Acad. Sci. USA 111(20), 7415–7420. https://doi.org/10.1073/pnas.1321997111 (2014).
Yang, J. et al. A case of placental trisomy 18 mosaicism causing a false negative NIPT result. Mol. Cytogenet. 10, 40. https://doi.org/10.1186/s13039-017-0341-5 (2017).
Liu, Y. et al. Clinical performance of non-invasive prenatal served as a first-tier screening test for trisomy 21, 18, 13 and sex chromosome aneuploidy in a pilot city in China. Hum. Genomics 14(1), 21. https://doi.org/10.1186/s40246-020-00268-2 (2020).
Zheng, Y. et al. Clinical experience regarding the accuracy of NIPT in the detection of sex chromosome abnormality. J. Gene Med. 22(8), e3199. https://doi.org/10.1002/jgm.3199 (2020).
Hu, T. et al. Prenatal chromosomal microarray analysis in 2466 fetuses with ultrasonographic soft markers: a prospective cohort study. Am. J. ObstetGynecol. 224(5), 516.e511-516.e516. https://doi.org/10.1016/j.ajog.2020.10.039 (2021).
Yuan, X., Yong, W., Dai, L., Wang, W. & Wu, L. The role of non-invasive prenatal testing and ultrasound in prenatal screening of fetal chromosomal abnormalities in singleton: a retrospective study. Ann. Transl. Med. 11(2), 111. https://doi.org/10.21037/atm-22-6343 (2023).
Screening for Fetal Chromosomal Abnormalities: ACOG Practice Bulletin, Number 226. ObstetGynecol. 136(4), e48–e69. https://doi.org/10.1097/aog.0000000000004084 (2020).
Li, F. et al. Detection of chromosomal abnormalities in spontaneous miscarriage by low-coverage next-generation sequencing. Mol. Med. Rep. 22(2), 1269–1276. https://doi.org/10.3892/mmr.2020.11208 (2020).
Qi, Y. et al. The significance of trisomy 7 mosaicism in noninvasive prenatal screening. Hum. Genomics 13(1), 18. https://doi.org/10.1186/s40246-019-0201-y (2019).
Lund, I. C. B. et al. Use of cell-free non-invasive prenatal testing in pregnancies affected by placental mosaicism. Prenat. Diagn. https://doi.org/10.1002/pd.6558 (2024).
Raymond, Y. et al. Placental, maternal, fetal, and technical origins of false-positive cell-free DNA screening results. Am. J. Obstet. Gynecol. 230(4), 381–389. https://doi.org/10.1016/j.ajog.2023.11.1240 (2024).
Bonanni, G. et al. Case report: Challenges of non-invasive prenatal testing (NIPT): A case report of confined placental mosaicism and clinical considerations. Front. Genet. 12(13), 881284. https://doi.org/10.3389/fgene.2022.881284 (2022).
Gou, L. et al. Clinical management of pregnancies with positive screening results for rare autosomal aneuploidies at a single center. J. Int. Med. Res. 48(11), 300060520966877. https://doi.org/10.1177/0300060520966877 (2020).
Acknowledgements
We thank all the authors for their contributions and support. We are grateful to all the pregnant women in the study who provided blood samples. We would also like to thank the hospital staff who contributed to the data collection for this study.
Funding
This work was supported in part by The National Nature Science Foundation of China (grant no. 82171856) and Guangzhou Science and Technology Planning Project (grant no. 202103000047) and Guangdong Province Basic and Applied Basic Research Funding Project (grant no. 2020A1515110779) .
Author information
Authors and Affiliations
Contributions
All authors have materially participated in the study and manuscript preparation.D.-m. W. collected all clinical data and drafted the manuscript; F.-f. G. and Y.-p. H. carried out all the molecular genetic analyses; H.-s. P. and T.-t. H participated in the data analysis; J.-x. Y.designed the work and revised the manuscript. All authors have approved the final article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Guangdong Women and Children Hospital. This study was conducted in accordance with the Declaration of Helsinki, and written informed consent was obtained from all participants before their inclusion.All clinical procedures and methods, including counseling, testing, were performed in strict accordance with the ‘Technical specifications for prenatal screening and diagnosis of fetal cell-free DNA in maternal peripheral blood’ in China .
Consent for publication
The authors declare that they have no competing interests and the patients in this case report had provided their consent for publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, D., Guo, F., Hou, Y. et al. The value of increasing sequencing depth for noninvasive prenatal screening for whole chromosomal aneuploidy. Sci Rep 15, 2673 (2025). https://doi.org/10.1038/s41598-025-87218-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-87218-x