Abstract
Glaucoma poses a growing health challenge projected to escalate in the coming decades. However, current automated diagnostic approaches on Glaucoma diagnosis solely rely on black-box deep learning models, lacking explainability and trustworthiness. To address the issue, this study uses optical coherence tomography (OCT) images to develop an explainable artificial intelligence (XAI) tool for diagnosing and staging glaucoma, with a focus on its clinical applicability. A total of 334 normal and 268 glaucomatous eyes (86 early, 72 moderate, 110 advanced) were included, signal processing theory was employed, and model interpretability was rigorously evaluated. Leveraging SHapley Additive exPlanations (SHAP)-based global feature ranking and partial dependency analysis (PDA) estimated decision boundary cut-offs on machine learning (ML) models, a novel algorithm was developed to implement an XAI tool. Using the selected features, ML models produce an AUC of 0.96 (95% CI: 0.95–0.98), 0.98 (95% CI: 0.96–1.00) and 1.00 (95% CI: 1.00–1.00) respectively on differentiating early, moderate and advanced glaucoma patients. Overall, machine outperformed clinicians in the early stage and overall glaucoma diagnosis with 10.4 –11.2% higher accuracy. The developed user-friendly XAI software tool shows potential as a valuable tool for eye care practitioners, offering transparent and interpretable insights to improve decision-making.
Similar content being viewed by others
Introduction
Glaucoma, also known as glaucomatous optic neuropathy, encompasses a series of optic nerve diseases causing retinal ganglion cell impairment and death. In developed nations, it ranks as the second most common cause of irreversible blindness1. Early detection and treatment of glaucoma are crucial as the damage it causes is permanent and irreversible. A meta-analysis study suggests that the number of individuals diagnosed with glaucoma is projected to increase significantly over the next two decades, almost doubling from 76 million in 2020 to an estimated 112 million in 20402.
Currently, the diagnosis and management of glaucoma requires a comprehensive examination including tonometry3, visual field (VF) testing4,5 and fundoscopy6, within a routine ophthalmic assessment that aims to rule out other potential causes of optic nerve disease. Furthermore, ocular imaging, such as optical coherence tomography (OCT), has become a clinical standard for glaucoma assessment7. Although many of these tests can be competently conducted by trained technicians, the interpretation of the results requires clinical expertise for personalised care8 which therefore places a strain on the limited resources of the healthcare system.
The increase in the number of patients puts further pressure on healthcare systems and makes it difficult to diagnose and treat glaucoma in a timely manner using manual methods9,10. Reus et al.11 reported clinician variability in glaucoma diagnosis in 11 different counties (Austria, Belgium, Finland, France, Germany, Greece, Hungary, Italy, the Netherlands, Spain and the United Kingdom) based on the assessment of stereoscopic optic disc images, and found a significant difference between clinicians’ performance across countries (p < 0.0005). Based on the overall performance of clinicians on 243 total observations, the clinicians’ mean sensitivity was 74.7% (range 43.8–100%), specificity was 87.4% (25–100%) and accuracy was 80.5% (61.4–94.3%). This indicates that the human error rate can vary significantly between clinicians and result in misdiagnosis.
Due to the known variability across clinicians and in light of the above findings, there is emerging evidence that computer-aided methods may assist in improving the objectivity, efficiency and accuracy of glaucoma diagnosis, in a more accessible and less expensive manner.12 A recent review of artificial intelligence (AI)-based studies on glaucoma diagnosis revealed that most of them use colour fundus images from a public database and deep learning (DL) methods.12 While DL applied to fundus images can provide the user with an end-to-end diagnostic framework, such black-box13 approaches entail huge computational costs and hardware resources14, which may raise economic and environmental concerns15. Also, those approaches require huge datasets for training purposes, which has led some studies to include self-reported glaucoma patients16 and glaucoma suspects17,18 in the glaucoma group from public datasets, such as the UK biobank16. As such, those approaches have limited reliability, which hinders real-life clinical applicability. A traditional machine learning (ML) approach trained on a more reliable dataset of OCT images and producing more explainable predictions may better serve clinicians.
The existing handful of OCT-based studies using ML leave much room for improvement. First, most methods have focused on spatial domain features (extracted directly from images) and particular types of analysis, such as retinal nerve fibre layer (RNFL) thickness analysis19,20, but missed the frequency-domain patterns that can be extracted from the images and other changes in the retinal ganglion cell (GC) and macular (MC) area. Second, most studies did not perform any feature selection while diagnosing glaucoma21,22,23,24,25,26, leading to a redundant number of features supplied to the ML models21,23. Third, the focus of most studies on ML for diagnosing glaucoma has been on classifying clearly glaucomatous and normal eyes. However, it is important to identify the different severity levels of glaucoma, especially in the early stages and to track disease progression27. Fourth, past studies did not provide possible explanations or interpretations but used black-box AI models focusing only on performance metrics. Lastly, past studies were typically based on experimental Python or MATLAB code, and there is a scarcity of studies developing and releasing user-friendly AI apps for real-time diagnosis of glaucoma12.
In this study, we leverage spatial domain features, such as the RNFL temporal-superior-nasal-inferior-temporal (TSNIT) pattern, to extract frequency domain information28,29,30, aggregate GC inner plexiform layer (IPL) and MC thickness features, apply feature selection techniques, and assess the performance of ML models in diagnosing glaucoma stages for early, moderate and advanced glaucoma. Furthermore, we perform a sub-analysis to classify advanced glaucoma based on mean deviation (MD) and central field defects. The main contribution of this study is the use of explainable AI (XAI) techniques to uncover the black-box ML model to improve understanding, trustworthiness and reliability. To the best of our knowledge, this is the first study to include spatial domain RNFL, GC-IPL and MC thickness and frequency domain TSNIT features within an XAI framework, specifically SHapley Additive exPlanations (SHAP) analysis and partial dependency analysis (PDA), for OCT-based diagnosis of glaucoma. Leveraging the proposed XAI techniques, we also present a handy software tool to assist clinicians in glaucoma diagnosis.
Materials and methods
Data collection and categorisation
Ethics statement
This was a cross-sectional study using prospectively acquired data from the files of patients seen by the Centre for Eye Health (CFEH), University of New South Wales (UNSW), Sydney, Australia. Ethics approval was provided by the Human Research Ethics Committee of UNSW (HC210563). The study adhered to the tenets of the Declaration of Helsinki. All subjects provided their written informed consent for use of their de-identified clinical data for research purposes.
Data acquisition
The study collected data from patients seen between 2015 and 2021 at CFEH. The clinic is a referral-only service for patients with visual pathway diseases, including glaucoma. The glaucoma examination protocols of CFEH have been described previously31,32,33,34. In summary, the glaucoma examination protocols used at the clinic include a comprehensive assessment of the patient’s history, visual acuities, anterior segment examination, applanation tonometry, pachymetry, gonioscopy, dilated stereoscopic examination of the optic nerve head and the macula, standard automated perimetry, colour fundus photography of the optic disc and posterior pole and OCT imaging. The study excluded patients classified as "glaucoma suspect," and those with non-glaucomatous optic atrophy. As per the clinical protocols of CFEH34, the diagnoses were made by examining clinicians (experienced optometrists and ophthalmologists) and if required, reviewed remotely by senior clinicians, with further examination by a third expert for inclusion in the study. While phenotyping, any images with artefacts were flagged by the clinicians.
Data categorisation
We categorised the ocular diagnoses of eyes from glaucoma patients within the present cohort into three categories, based on the mean deviation of Visual Fields (VF). More specifically, we used the criteria as per Mills et al.35 to determine glaucoma severity levels (early glaucoma as MD up to -6 dB, moderate as -12 dB < MD ≤ -6 dB, and advanced as MD ≤ -12 dB) and our categories were healthy (n = 334 eyes); early glaucoma (n = 86 eyes); moderate glaucoma (n = 72 eyes); advanced glaucoma based on MD (n = 37 eyes); and advanced glaucoma based on central field defect (CFD)36 (n = 73 eyes). The advanced stage of glaucoma was defined on MD. The presence of a CFD that did not meet the MD criterion was not deemed as advanced. Overall, the group of consisted of 334 normal eyes and 268 glaucoma eyes. Normal individuals were matched to the ages of the glaucoma patients (Table 1). The reference labels determined by these criteria were also used for training the ML models (described in Table 1).
Feature extraction
Extraction of spatial domain features
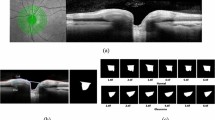
The spatial domain features, e.g., RNFL, GC-IPL and macular thickness, provide information about the distribution, thickness and variations of the specific retinal layers. Features from these scanning protocols are known to be useful for distinguishing between healthy, glaucoma suspect and glaucomatous eyes to various degrees and with levels of inter-parameter correlation37. These features refer to the characteristics and patterns observed within the two-dimensional space of the OCT image itself. Using the CIRRUS HD-OCT software (ZEISS), all the spatial domain features were extracted as numerical data in this study (Fig. 1(a)). The following analyses were performed with the CIRRUS software: optic nerve head (ONH) and RNFL oculus uterque (both eyes) (OU) Analysis, Ganglion Cell OU Analysis, and Macular Thickness OU Analysis (Supplementary Fig. 1; Supplementary Table 1). The software was used to extract the 256-point TSNIT patterns, plot them in the spatial domain and compute statistical features such as mean, median, standard deviation, skewness and kurtosis of the whole TSNIT (Supplementary Fig. 2), and each of the temporal, superior, nasal and inferior quadrants. Additionally, some information theory-based features were also derived from the TSNIT data, i.e., Shannon entropy, Fisher information and signal-to-noise ratio. To observe the effect of these features on visual fields, the structural–functional relationship was plotted (Supplementary Fig. 3).
(a) Experimental procedure of this study. The key steps include raw data phenotyping, feature extraction in the spatial and frequency domain, feature selection, classification and model interpretation using XAI. (b) Flow-chart of the implementation of the explainable diagnostic tool based on SHAP and PDA (Excel tool). Blue shaded boxes represent explainable analysis. Note: CFEH: Centre for Eye Health, GC: Ganglion Cell, MC: Macular, RNFL: Retinal Nerve Fibre Layer, TSNIT: Temporal-Superior-Nasal-Inferior-Temporal, MD: Mean Deviation.
Derivation of frequency domain features
The 256-RNFL values extracted from the RNFL thickness analysis form the TSNIT pattern, which is a typical 'double hump pattern’30, representing the superior and inferior in the first and second hump respectively. In line with Essok et al.30, we applied the fast Fourier transform (FFT) to the OCT TSNIT pattern to transform the spatial domain into the frequency domain and extracted several frequency-domain features, including the power spectral density (PSD) of the TSNIT pattern (overall and RNFL quadrants) using the ‘Welch’ and ‘FFT’ method, the PSD of the TSNIT pattern after discrete wavelet transformation (DWT) (levels 1 to 4), the slopes of the harmonics of the PSD, and the spectral entropy (Supplementary Table 1).
Feature imputation
For some patients, the data contained artefacts such as image truncation, inaccurate delineation of the cup and/or disc margin, media opacities and segmentation error, affecting either RNFL, GC-IPL or MC measurements. We included any patients having at least one artefact-free parameter (e.g., RNFL). The parameters containing artefacts were considered as missing data, amounting to 3.65% of the total samples (e.g., GC-IPL), and multiple imputation using chained equations (MICE)38 imputation was performed to impute the missing data points based on other existing artefact-free data (e.g., RNFL & MC). MICE is a statistically robust imputation technique that preserves the inherent relationships between features for a specific patient39,40. Unlike single imputation methods, MICE generates multiple plausible datasets while accounting for the uncertainty associated with missing features (e.g., RNFL) by leveraging information from other available features40 (e.g., GC-IPL and MC). Patients with at least one artefact-free parameter were included, and artefact-affected parameters were treated as missing data. MICE imputation used the existing artefact-free data to reconstruct these values. However, when all the RNFL, GC-IPL or Macular thickness data were affected by artefacts, the sample was removed from the dataset.
Feature selection
Eliminating irrelevant features is critical as they can hinder the accuracy of classification models41. To address this problem in our study, a feature selection process was carried out on the spatial-frequency domain features derived from the OCT-based analysis. Based on the literature, filter methods in feature selection are independent of any learning algorithms and are effective in various real-world datasets42,43,44. Given the computational complexity of feature selection and previous studies on the subject42,43, this study concurrently employed two univariate filter-based feature selection techniques: analysis of variance (ANOVA) F Test and Pearson’s correlation-based feature selection. The ANOVA F-test was chosen because it captures inter-class variability by identifying features with high variation between group means relative to class labels45,46. As a univariate feature selection method, it evaluates each feature independently, making it an effective base for ranking features with higher discriminative power. Pearson’s correlation-based method operates on the hypothesis that a feature is valuable if it demonstrates relatively low correlation with other features46,47. In this process, highly correlated features (r > 0.98) were identified using Pearson’s correlation, retaining only the top-ranking feature with the highest F-score from each group of correlated features. The remaining features were ranked by their F-scores, and the ‘SelectKBest’ scikit-learn module48 was applied, selecting the K highest scored features with the wrapper approach49. In this approach, a tuned classification model with five-fold cross-validation is used to determine the optimal cut-off by observing validation accuracy with an increasing number of top-ranking features from the spatial-frequency domains.
Classification models
To date, there is no standardised or generalised model for the classification of glaucoma stages. Due to the distinct computational cost and complexity50,51 and based on current literature, three supervised learning models were employed for classification: K-Nearest Neighbour (KNN), Support Vector Machines (SVM), and Random Forests (RF) classifier. The hyperparameters of each classifier were adjusted using an iterative process to obtain the best accuracy after cross-validation (Supplementary Table 2). For the KNN classifier, different values of 'k' (range: \(1\le k \le 40\)) were used to tune the model, and the 'k' value that corresponded to the best training accuracy was selected (optimal value of k = 7 for overall glaucoma vs normal). For SVM, the Gaussian radial basis function (RBF) kernel was utilised, and the 'C' (optimal value: 10 for overall glaucoma diagnosis) and 'γ' (optimal value: 0.01 for overall glaucoma diagnosis) parameters were tuned using the grid-search algorithm to achieve optimal parameter values for best accuracy after cross-validation. For the RF classifier, the number of trees in the forest (‘n_estimate’) (optimal value: 100 for overall glaucoma) and the maximum depth of the tree ('max depth’) (optimal value: 3 for overall glaucoma) were tuned through a grid-search algorithm. Feature scaling was performed before supplying the features to the KNN and SVM models (min–max scaling for KNN and standardisation for SVM52); however, we did not perform any feature scaling for RF as it does not require scaling given the nature of the algorithm52, which further helped us explain the features using XAI.
In our dataset, disease-positive cases (i.e., the number of glaucomatous eyes) are usually less frequent than normal or healthy cases. In medical diagnosis, the cost of misclassifying a disease-positive case is typically much higher than misclassifying a healthy case. To address this, the synthetic minority over-sampling technique (SMOTE)53 was used in our study to overcome data imbalance (imbalance ratio for glaucoma : normal = 0.802 : 1). SMOTE was selected because it generates synthetic samples by interpolating between minority class samples rather than simply duplicating existing data, which helps to reduce the risk of overfitting54. This approach effectively increases the representation of the minority class (glaucoma in our case) while preserving variability within the dataset53,54. It has been widely used in retinal disease diagnostic studies, including retinal image diagnosis, like glaucoma55,56 and age-related macular degeneration57 and preeclampsia57.
We note that SMOTE and MICE-based augmented data were not used in the feature selection and explainable analysis. Additionally, we ran a sub-analysis with all the classification models using artefact-free data only (without MICE-based imputation and SMOTE-based oversampling) to maintain clinical transparency and reliability of the results. All models were implemented, trained and tested using Python version 3.7 in the Google Colab58 platform. Performance evaluation, patient-level splitting and cross-validation have been described in supplementary methods.
Explainable machine learning
Shapley additive analysis
SHapley Additive exPlanations (SHAP) is a method for explaining the output of ML models by attributing the prediction to the features that contributed to it59. It does this by using the concept of Shapley values from game theory, for fairly distributing the “credit” for a prediction among the contributing features. Shapley values are useful to calculate the importance of each feature in a model’s prediction and have been used to interpret ML models in medical60 and other domains61,62. Generally, the Shapley value of a feature i, denoted \({\varphi }_{i}\left(val\right)\), indicates the contribution of a feature value (\(val\)) in a payout. It sums over all possible feature combinations (\(S\)) and measures the difference in feature values (\(val\left(S\cup [i]\right)-val(S)\)), adjusting for the size of the combinations63 (Eq. 1).
Here, i is the feature of interest, m represents the total number of features excluding the feature i, while \(S\) is the subset of the features excluding feature i, \(val\left(S\cup [i]\right)\) represents the total value generated by the coalition that includes feature i, and \(val(S)\) represents the value of the coalition S, which is the total value generated by the coalition that does not include feature i.
While Shapley values are a concept from cooperative game theory, SHAP is a unified framework that generalises Shapley values and typically refers to the aggregated ‘Shapley values’ as ‘SHAP values’63. This study used SHAP-based feature importance, SHAP dependence plot and SHAP interaction plot as a part of the SHAP analysis, both in global and local scopes63.
Partial dependency analysis
Partial dependency analysis (PDA) is another technique used for interpreting and explaining the output of ML models64. PDA helps to understand the relationship between a single feature in relation to the model’s output, holding all other features constant. The resulting plot from PDA representing the relationship between feature values and the model’s output is called a partial dependency plot (PDP). PDPs help understand the marginal effect of the features on the predicted probability of the ML models and have been widely used in interpreting real-world problems61,62, specifically for identifying the decision boundary65,66,67 in ML-based classification problems. The partial dependence function63 (Eq. 2) is defined as:
Here, \(i\) is the feature of interest, for which we want to know the effect on the prediction, \({\widehat{f}}_{i}\left(i\right)\) represents the predicted outcome or response variable for the specific feature i, based on a ML model, S is the set of the other features. \({E}_{S}\) stands for expected or average value of the function \({\widehat{f}}_{i}\left(i, S\right)\) over all possible values of the set of features \(S\). \({\widehat{f}}_{i}\left(i, S\right)\) is the predicted outcome or response variable for i, based on a ML model, which explicitly indicates the dependence of the feature of interest i on the set of other features S. \(d{\mathbb{P}}(S)\) represents the probability density function (PDF) / probability distribution over the set of features \(S\).
The partial function is approximated using the Monte Carlo method63, wherein averages are computed based on the training data (Eq. 3):
Here, \({\widehat{f}}_{i}\left(i\right)\) is the average marginal effect on the prediction / partial dependence function for feature of interest (i), \({S}^{(j)}\) is the set of all possible combinations of the other features in the ML model, in which we are not interested, where j ranges from 1 to n (the number of samples in the dataset).
Diagnostic tool
Based on the SHAP-based global feature importance and PDA-based predicted probabilities for each feature, we developed two software tools to assist clinicians to calculate the glaucoma likelihood of patients based on basic CIRRUS OCT data. The first one is an Excel (Microsoft 365 version-2019) based tool, where the clinicians can input relevant features, categorised into a major and a minor group. At the backend, the PDA-based predicted probabilities and estimated feature cut-offs at the decision boundary were used to make a decision based on the outcome defined with major and minor criteria (Fig. 1(b)). If the maximum of the major criteria is fulfilled, then the glaucoma likelihood score is based on the major criteria. Otherwise, the algorithm checks for the maximum minor criteria to be satisfied to produce a likelihood based on the minor criteria. If the second condition is not fulfilled, the likelihood score is calculated as the mean of major and minor criteria altogether.
The second tool is a web-based app, where a RF model has been deployed. This tool uses the most important features based on SHAP analysis; however, the key difference from the Excel app is that here we let the model decide on using the features based on the RF model rather than defining major and minor criteria and weighted likelihood scores. To validate the likelihood score produced by the apps, the glaucoma likelihood scores (Excel app) and predicted probabilities (Web app) were plotted corresponding to the MD values of each sample (excluding the advanced glaucoma cases based upon central VF loss). The web app was developed using the Flask framework and a Python web framework, and incorporates HTML templates for the user interface. It follows a client–server architecture, where the client interacts with the web application through a web browser, and the server handles the requests and provides responses.
We developed two distinct sets of machine learning models: non-explainable models (leveraging both spatial and frequency domain features) and explainable models (utilising a small set of explainable spatial domain features). The initial non-explainable models were built using features derived from RNFL, GC-IPL, and MC thickness in the spatial domain, as well as the frequency domain features. These models (the white-shaded boxes in Fig. 1a) are best suited for integration with OCT machine software (e.g., CIRRUS HD-OCT). Here, the availability of 256 TSNIT data points enables the derivation of frequency domain components, which can be concatenated with spatial domain features to enhance glaucoma diagnosis.
In contrast, the explainable models are specifically designed for deployment on edge devices (e.g., mobile phones, tablets, or web apps) that are not directly connected to the OCT machine or its software. In this scenario, the 256 TSNIT data points required to compute frequency domain features are unavailable. Instead, the spatial domain features represent global measures derived from the TSNIT data points (e.g., RNFL superior, nasal, inferior, and temporal thicknesses are calculated as the mean of the first 64, second 64, third 64, and fourth 64 data points, respectively). Additionally, the clinical interpretability of frequency domain features is limited due to the lack of studies exploring their significance. To ensure practical utility and alignment with the needs of clinicians, the explainable models were re-trained exclusively with spatial domain features only.
Results
TSNIT plots and statistical analysis
A TSNIT plot was constructed showing the spatial domain TSNIT pattern for early, moderate and advanced glaucoma for both right (Supplementary Fig. 2 (a)) and left (Supplementary Fig. 2 (b)) eyes separately. The plots indicate that for both left and right eyes, the RNFL thickness value in the inferior and superior quadrant is significantly reduced (p < 0.05) when a patient progresses from early to moderate and finally advanced glaucoma. Additionally, scatter plots of the RNFL/GC-IPL thickness versus MD of visual fields to visualise the structural–functional relationship (Supplementary Fig. 2) were similar to previous published works68.
Selected features
Feature selection was performed on the 67 spatial and 64 frequency domain features (total features: 131) extracted from the OCT-based analysis (Supplementary Table 1). Feature selection was performed concurrently using ANOVA F Test and Pearson’s correlation method (see Feature Selection in Materials and Methods). Highly correlated features were excluded (r > 0.98) retaining the best one having highest F-score as some of the features within the spatial domain, frequency domain, or cross-domain had higher correlation between themselves. Including these highly correlated features would not add information, while increasing redundancy of the feature set, adding complexity to the model’s prediction and reducing interpretability. For example, within the spatial domain features, ILM-RPE thickness central subfield thickness has a higher correlation with ILM-RPE centre-foveal thickness (r = 0.986). Additionally, ILM-RPE thickness-volumetric cube and ILM-RPE thickness-average cube are highly correlated (R = 0.998). As all those four features are correlated, only ILM-RPE thickness-average cube was retained among the four, as it has the highest F-score among the correlated features.
Once the highly correlated features were excluded (keeping the topmost with highest F-score), the K-highest ranked features were selected to observe the validation accuracy wrapping with an RF classifier (see Feature Selection in Materials and Methods). Adding features with F-scores below 30 did not contribute to improved validation accuracy; consequently, a cut-off F-score of 30 was established, corresponding to approximately 6% of the highest-ranked feature’s F-score (502). This resulted in the selection of 66 features (45 spatial domain and 21 frequency domain) out of the initial 131 features (Supplementary Table 3), representing nearly 50% of the total features. The feature ranking shows that the topmost features are obtained from the RNFL analysis, with RNFL symmetry and RNFL inferior having the highest ANOVA F score (Fig. 2 (a). Among the frequency domain features, PSD values at the inferior quadrant appeared to be the highest-ranked feature by the feature selection methods.
(a) Illustration of top 30 features selected by feature selection techniques. (b) Human versus Machine (RF) performance comparison for Glaucoma stage diagnosis. (c) Overall glaucoma diagnosis. Two optometrists and one glaucoma specialist were provided with RNFL and GC-IPL thickness data with masking (only keeping the numerical thickness values that were fed to the ML models). All the normal and glaucoma patients were randomly assigned, so the clinicians could not predict the order in which they appeared. Once they completed the grading, sensitivity and specificity were calculated (from TP, FP, TN, FN) and plotted in the ROC plot generated for the RF model (using the same data with one-vs one approach) for comparison. Reported AUC = mean ± standard error, standard error = standard deviation/ √n , n = 5 (for five-fold cross validation). Note: RNFL: Retinal nerve fibre layer, GC-IPL: ganglion cell–inner plexiform layer, ILM-RPE: Inner limiting membrane-retinal pigment epithelium, TSNIT: temporal-superior-nasal-inferior-temporal, std: standard deviation.
Differential diagnosis
Glaucoma staging
The separation of the different stages of glaucoma was performed based on two approaches: one versus rest (OvR) and one versus one (OvO) model training using three different classifiers, namely KNN, SVM and RF using the RNFL, GC-IPL and MC thickness features (see Table 2). In the OvR approach, all stages of glaucoma and normal were used for training, considering four possible two-class classification problems: early versus the rest (moderate, advanced, normal), moderate versus the rest (early, advanced, normal), advanced versus the rest (early, moderate, normal), and normal versus the rest (early, moderate, advanced). The performance of the diagnostic model was evaluated using multiple metrics, i.e., sensitivity, specificity, accuracy, AUC and F1-score. Higher sensitivity with a lower specificity may cause higher false positives and thus result in overdiagnosis; however, AUC best summarises diagnostic performance in terms of both sensitivity and specificity, so we have focused on that for comparative analysis. From the comparative performance of the three classification models (Table 2), it is evident that the RF classifier outperformed the others in terms of AUC, with an AUC of 0.84 (95% CI: 0.78–0.89), 0.88 (95% CI: 0.85–0.91), 0.96 (95% CI: 0.96–0.99) for early, moderate and advanced glaucoma patients (Supplementary Fig. 4(a), Table 2).
On the other hand, in the OvO approach, three separate binary class problems were considered: early versus normal, moderate versus normal, and advanced versus normal. From the comparative performance of the three classification models (Table 2, and Supplementary Fig. 4(b)), it is evident that SVM and RF outperformed the KNN classifier in terms of AUC. For early glaucoma, we achieved the best AUC of 0.96 (95% CI: 0.95–0.98) using SVM; for moderate glaucoma, an AUC of 0.98 using both RF (95% CI: 0.96–1.00) and SVM (95% CI: 0.96–1.00); and for advanced glaucoma, an AUC of 1.00 (95% CI: 1.00–1.00) using both RF and SVM (Supplementary Fig. 4(b), Table 2). A sub-analysis was performed with advanced glaucoma identified based on CFD and MD of visual fields (Supplementary Fig. 5).
Overall glaucoma
Overall, binary classification was performed between whole glaucoma (early, moderate, and advanced) and normal using five-fold cross-validation (Supplementary Table 4, Supplementary Fig. 6). The outcomes indicate that SVM outperformed the other ML models and produced a mean sensitivity of 0.91 (95% CI: 0.87–0.94), specificity of 0.97 (95% CI: 0.95–0.99), accuracy of 94.0% (95% CI: 0.91–0.97) and AUC of 0.97 (95% CI: 0.95–0.99). On five-fold cross-validation, performance was robust across all the folds (Supplementary Table 4, Supplementary Fig. 6). For further verification, the data was shuffled five times (five-times five-fold cross-validation) and the classifier still produced an AUC of 0.97 which confirms the robustness of the model in glaucoma diagnosis across all the folds (Supplementary Fig. 6). The RF model performed close to the SVM with an AUC of 0.97 (95% CI: 0.95–0.99), while the KNN model provided the least performance for the diagnosis of overall glaucoma with an AUC 0.92 (95% CI: 0.90–0.95). A sub-analysis using the original artefact-free data without MICE-based imputation and SMOTE-based oversampling showed no significant difference in performance (see Supplementary Results and corresponding Supplementary Tables 5 and 6).
Sub-analysis using RNFL, GC-IPL and MC thickness feature sets
A sub-analysis was performed to compare the subset of selected features obtained from RNFL, GC-IPL and MC thickness analysis (Supplementary Fig. 11). The subset of features consisted of the RNFL feature set (49 features), GC-IPL feature set (8 features) and MC thickness feature set (9 features). Dual and triple combinations of the feature sets were also compared, i.e. RNFL + GC-IPL (57 features), RNFL + MC thickness features (58 features), and all features RNFL + GC-IPL + MC (66 features). Using a RF classifier, the sub-analysis comparing the singular feature sets showed that RNFL performs better (AUC: 0.92, 95% CI: 0.88–0.96) than GC-IPL (AUC: 0.89, 95% CI: 0.88–0.90) and MC thickness (AUC: 0.80, 95% CI: 0.78–0.83) feature sets, with MC thickness being the worst performer (AUC: 0.80). Comparing the dual and triple combination of the feature sets, we observed that the RNFL + GC-IPL combination of feature sets produced similar performance (AUC: 0.97, 95% CI: 0.95–0.99) as the triple combination using MC (RNFL + GC-IPL + MC) (AUC: 0.97, 95% CI: 0.95–0.99), with GC-IPL + MC being the worst performer (AUC: 0.86, 95% CI: 0.84–0.87). Considering this, RNFL + GC-IPL feature sets were used to train the other models and compare the machine performance with the human expert.
Comparing human vs machine performance
A sub-analysis was performed to compare human performance with ML (Table 3 and Fig. 2 (b–c)). In this case, only RNFL and GC-IPL data was used, as the inclusion of MC thickness data does not change the machine performance significantly (Supplementary Fig. 11). The performance of the clinicians was evaluated on the analysed image dataset using CIRUS HD OCT (RNFL and GC-IPL analysed data containing both numbers and images) in three ways by three clinicians based at CFEH: first, the deviation maps and colour OCT maps were masked except for the thickness values (numbers) used by the machine (masked, Supplementary Fig. 7(a)). Second, all the relevant information including colour maps and deviation maps were revealed to grade on the images (unmasked-sequential, Supplementary Fig. 7(b)). Finally, one of the clinicians was provided with unmasked data with random distribution again (unmasked-random, Supplementary Fig. 7(c)). The performance metrics were calculated for the clinicians and compared with the machine performance (Table 3 and Supplementary Table 7). It is important to note that CFEH is a referral-only, optometry-ophthalmology service, providing assessment of patients who are suspected of having eye disease of the visual pathways, or those who have a disease including glaucoma. The clinicians are highly trained in ocular assessment and interpreting imaging. The three clinicians who graded the images had 12, 11 and 6 years of experience at CFEH.
The sensitivity and specificity obtained from the clinicians (Table 3) are plotted in the ROC curve to compare with machine performance (Fig. 2). It shows that with the masked data, the clinicians had a sensitivity ranging from 44.4–57.1% and specificity ranging from 86.4%–89.3% for early glaucoma, sensitivity of 71.4%–80.0% and specificity of 93.6–96.6% for moderate glaucoma, and sensitivity of 100.0% and specificity of 98.3–100.0% for advanced glaucoma. While diagnosing overall glaucoma by the clinicians, they have a sensitivity ranging from 79.7–86.3% and specificity ranging from 83.8–84.2% using the masked data.
Overall, ML outperforms mean human performance while diagnosing early and moderate glaucoma by 12.3% (SVM)–12.6% (RF) and 3.1% (RF)–3.5% (SVM) in terms of accuracy. However, the performance for advanced glaucoma is somewhat similar for both human and machine. For overall glaucoma diagnosis, ML accuracy was 10.4% (RF)–11.2% (SVM) and 9.2% (RF)–10.0% (SVM) higher than the human expert using the masked and sequential unmasked data respectively. For both glaucoma staging and overall glaucoma diagnosis, the clinician’s performance for sequentially unmasked and random unmasked data was somewhat close; however, it was substantially lower than the machine’s performance, especially while diagnosing early glaucoma (Supplementary Fig. 8 & Supplementary Fig. 9).
Explainable ML results
SHAP analysis
SHAP analysis was performed on the dataset containing the included spatial domain features used to train the RF model. The goal was to generate a global feature rank based on the mean of absolute SHAP values per feature, corresponding to all samples from each class and sorted in descending order (Fig. 3). The SHAP-based global feature ranking (Fig. 3 (a)) and summary plot (Fig. 3 (b)) show that RNFL symmetry, RNFL thickness at the inferior quadrant, RNFL clock-11 (part of RNFL superior), and RNFL superior thickness have a higher impact on the classification. The SHAP summary plot (Fig. 3 (b)) further indicates that the lower the RNFL symmetry, the higher the chances of being classified as glaucoma. Similarly, the lower the RNFL inferior/ RNFL clock-11 /superior thickness, the higher the chances of being classified as glaucoma.
SHAP analysis results. (a) SHAP feature ranking and (b) SHAP summary plot. The top-ranked features are RNFL symmetry, RNFL inferior, RNFL clock-11 (part of the superior quadrant) and RNFL-superior thickness. The blue colour represents lower magnitudes of feature values while the red colour represents higher values. positive SHAP values on the right side push the model to predict a win (in this case the probability of being classified as glaucoma), while negative SHAP values on the left push the model to predict a loss (probability of being classified as normal). (c) SHAP dependence plot (RNFL symmetry). Positive SHAP values on the vertical axis mean that the corresponding feature values have a positive impact on the prediction, while negative SHAP values mean that the feature values have a negative impact. Points close to the zero line can be useful for identifying instances where a feature did not contribute to the model’s prediction. (d) SHAP dependence plot with interaction visualisation (interaction between RNFL inferior and RNFL symmetry). For (c), the data points in red above the horizontal dashed line represent the samples classified as glaucoma, while those in blue below the line represents the samples classified as normal. In (d), the red points corresponding to higher RNFL inferior thickness indicate the presence of higher RNFL symmetry, which generates negative SHAP values and shows a lower chance of being classified as glaucoma. RNFL clock hours-2,6,7,11 represents RNFL thickness measured in clock-hour sectors. Each sector is measured in degrees, usually in increments of 30 degrees, creating a 360-degree circle around the optic nerve head.
To understand the impact of each feature in the model’s prediction for every data sample, a SHAP dependence plot was generated, illustrating the relationship between the feature’s magnitude and its contribution to the model prediction. The SHAP dependence plot was created for the two top-ranked OCT features, i.e., RNFL symmetry and RNFL inferior thickness (Fig. 3(c, d)). The SHAP dependence plot (Fig. 3 (c)) confirms that lower RNFL symmetry increases the chance of a sample being classified as glaucoma (points corresponding to positive SHAP values on the vertical axis), whereas higher RNFL symmetry increases the chance of being classified as normal (points corresponding to the negative SHAP values), with a crossover point of around 71. While RNFL Symmetry is the highest-impact feature, the SHAP interaction visualisation plot between RNFL inferior thickness and RNFL symmetry further clarifies the interpretation (Fig. 3(d)). In the cases where the RNFL inferior thickness is lower, a lower RNFL symmetry (i.e., higher asymmetry) increases the chances of the patient being diagnosed with glaucoma. Conversely, higher symmetry reduces the chances of the instances being classified as glaucoma.
Partial dependency analysis
To understand the marginal effect of the feature values on the predicted output of the classification model, both individual component expectation (ICE) and partial dependence plot (PDP) plots were created (Fig. 4 (a–f)). The decision boundaries for early, moderate and advanced glaucoma patients are illustrated in the combined PDPs for early, moderate and advanced glaucoma patients (Fig. 4 (a-b)) for the top features ranked by SHAP analysis: RNFL symmetry and RNFL inferior thickness. The relative comparison shows that the cut-off for the decision exhibits a trend from right to left, i.e., the lower the symmetry and the lower the inferior quadrant thickness, the higher the chances of disease severity.
(a) Partial dependency plot (PDP) comparison of early, moderate, and advanced glaucoma for RNFL Symmetry (b) PDP for RNFL Inferior (c) PDP and individual conditional expectation (ICE) Plots for RNFL symmetry (estimated cut-off = 71%), (d) PDP and ICE plots for RNFL inferior (estimated cut-off = 88.13 μm), (e) two-way numerical PDP for RNFL symmetry and RNFL inferior (f) 3D feature interaction plot for RNFL symmetry and RNFL inferior. In all cases (a–f), the y-axis represents the predicted probability of a machine learning model, and the x-axis presents the magnitude of feature values. For Individual Conditional Expectation (ICE) plots (b,d), the thin separate curves show the dependency of the prediction on the feature (individual dependence from each sample) and the thick curves represent the average effect (mean partial dependence).
To better understand the overall dependency, plots were drawn for overall glaucoma diagnosis, considering mean prediction (PDP) and prediction for every sample (ICE). From the ICE and PDP plots (Fig. 4 (c–d)), it can be observed that there is a clear descending slope between 60–80% and 85–90 μm respectively for RNFL symmetry and RNFL inferior thickness feature values, where the higher probability (left) indicates higher chances of being classified as glaucoma and lower probability (right) represents chances of being classified as normal. The comparison of the magnitude of the slope indicates that RNFL inferior thickness (strong slope, Fig. 4d) has a higher contribution towards classification than RNFL superior (moderate slope, Supplementary Fig. 10e). Similarly, GC-IPL thickness at the inferotemporal area has a higher contribution than the supertemporal area (Supplementary Fig. 10g,i). Based on the predicted probability of each feature by PDPs, a horizontal line was drawn at the decision boundary and thus cut-off values were estimated at the intersection point of the decision boundary and the PDPs (Supplementary Fig. 10).
A two-way interaction plot was created to understand the dependency of the predicted outcome on multiple top-ranked features and interaction between themselves, e.g., RNFL symmetry and RNFL inferior (Fig. 4 (e)). From the two-way interaction plot (Fig. 4e), we observe that if the RNFL symmetry is below about 71%, this feature is dependent on RNFL inferior thickness, and their combined contribution creates a higher probability of patients being classified as glaucoma (predicted probability > 0.58). Above RNFL symmetry 71% (vertical contour), the RNFL inferior thickness is independent of RNFL symmetry and there is a higher possibility of patients being classified as normal (predicted probability < 0.51). The 3D feature interaction plots (Fig. 4 (f)) show the combined effect of each of the two features on the prediction outcome, i.e., the dependency of RNFL symmetry on RNFL inferior thickness.
App development
Using the explainable results, two tools were developed to assist clinicians in glaucoma diagnosis. The first one is an Excel-based tool, utilising SHAP feature importance and PDA-based cut-off and estimated probabilities. The second one is an RF model deployed as a web app, which is trained with SHAP-based most important features and allows the user to input those feature values.
Glaucoma likelihood calculator as an excel app
To develop the Excel app, the feature ranking provided by the SHAP analysis was categorised into major and minor criteria, based on previous literature and the clinicians’ experience. Major features included those suggested by the SHAP analysis: RNFL symmetry, inferior RNFL thickness and superior RNFL thickness. Minor criteria included the inferotemporal and supertemporal GC-IPL thickness, inferior and superior GC-IPL thickness, and average RNFL and GC-IPL thickness. RNFL clock hours (e.g., RNFL clk-11, RNFL clk-6 & 7) were excluded as they are already part of superior and inferior RNFL thickness. The decision boundary cut-off for the given features were estimated using partial dependency plots (Supplementary Fig. 10). The numerical cut-off and estimated probabilities by PDA corresponding to each feature were provided in the back end, and the Excel program operates based on weighted outcomes on major and minor criteria (Fig. 1 (b)). At the front end, the clinicians can see a live scale (e.g., very low, low, intermediate, high, very high) with the average likelihood of glaucoma in percentage (%) (Fig. 5 (a)). The plot shows that using the likelihood probability score cut-off 0.25 (considering probability > 25% as glaucoma and probability < 25% as normal), the tool generates a sensitivity of 89.3%, specificity of 87.9% and accuracy of 88.5%. This implies that above the 0.25 probability cut-off, the app identifies around 90% of the glaucoma patients as glaucoma and 88% normal patients as normal. Moving the threshold cutoff higher (upwards) causes lower sensitivity and higher specificity; on the other hand, moving the threshold downwards produces a higher sensitivity and lower specificity.
(a) Snapshot of the Glaucoma likelihood OCT calculator (GLOC) based on XAI (partial dependency analysis-based predicted probabilities and SHAP-based feature ranking using the RF classifier). (b) Plot of glaucoma likelihood scores versus mean deviation of visual fields. The % values on the right indicate the % of glaucoma cases above that threshold, and the % of normal cases below that threshold. (c) Snapshot of the Glaucoma likelihood OCT web app based on a RF model deployed on the web. (d) Plot of predicted probabilities for glaucoma versus mean deviation of visual fields. The % values on the right indicate the % of glaucoma cases above that threshold and the % of normal cases below that threshold. (e) Correlation between glaucoma likelihood score by the XAI-based Excel app and predicted probability by the RF deployed web app. Demographics: 195 glaucoma eyes (early:86, moderate:72, advanced based on MD:37), 261 normal eyes. The other normal subjects’ MD values were unavailable.
Ml-Model deployment as a web app
To validate our proposed XAI-based Excel app, a second, web-based application was developed, where an RF model was deployed using the SHAP-ranked most important features (all major and minor features described above). The key difference of this web-based app from the Excel-based app is the method of prediction of glaucoma likelihood. While the Excel-based app produces a likelihood score based on a weighted outcome of major and minor criteria based on PDA, in the web app we allow the machine learning model to predict the probability based on the same inputs (Fig. 5(d)). The plot shows a better probability distribution classifying ‘glaucoma’ and ‘normals’ at a probability cut-off 0.5 (sensitivity = 89.3%, specificity = 97.4%, accuracy = 90.1%); however, for most of the early and moderate glaucoma samples, the predicted probabilities range from 60–100%.
It is worth mentioning that the web app not only generates predicted probabilities with classification, it produces local explainable results based on SHAP-analysis (for the test sample), which shows the top three features and their percentage of contributions towards the classification. This functionality enables clinicians to revisit and scrutinise specific features (e.g., RNFL Symmetry, RNFL Inferior and GC-IPL Inferotemporal thickness in the example of Fig. 5c, with predicted probabilities in Fig. 5 (d)), which significantly affect the classification or diagnosis. Unlike the SHAP-based global feature importance, this local feature importance may vary for different test patients.
A correlation analysis was performed on the glaucoma likelihood using our proposed Excel-based tool and predicted probabilities produced by the web-based ML-app (Fig. 5e). The correlation analysis produces a correlation coefficient of r = 0.92, which implies a strong positive correlation between our proposed Excel-app and ML-deployed explainable web app and validates its performance with the given data (Fig. 5 (e)). Additionally, a correlation analysis was conducted between the predicted probability scores of the non-explainable (trained with 66 spatial-frequency domain features) and SHAP-based explainable machine learning RF model deployed on the web (trained with the limited 9 spatial domain features). The analysis revealed a correlation coefficient of r = 0.98, indicating a strong correlation between the explainable and non-explainable models.
Discussion
The present study shows a comparative analysis of glaucoma staging and overall glaucoma diagnosis of human and machines. The results obtained from the study can be discussed from three angles: performance of differential diagnosis, implications of the research findings, and the study limitations and possible future research directions. First, using the same dataset, the analysis shows that machine performance surpasses human performance by 10.4 (RF)–11.2 (SVM)%, 12.3 (SVM)–12.6% (RF) and 3.1 (RF)–3.5 (SVM)% in terms of accuracy when diagnosing overall, early and moderate glaucoma respectively; however, the performance of advanced glaucoma diagnosis shows identical performance for both human and machines. The clinicians faced challenges in distinguishing early glaucoma from normal cases, where subtle differences in retinal features are not visually discernible. This makes detecting early glaucoma particularly critical, yet subjective and prone to variability among clinicians. In contrast, the machine learning model excelled in these cases, demonstrating its potential to assist clinicians by providing consistent and reliable support in decision-making.
Second, the present study outperforms recent ML and DL-based studies using a limited number of features (Table 4). Among the ML-based studies, Wu et al.21 used an SVM classifier for glaucoma staging using 114 OCT features (Spectralis OCT) and achieved an AUC of 0.78, 0.89 and 0.93 respectively for early, moderate and advanced glaucoma. Kooner et al.69 used 64 OCTA and other clinical features using an XGBoost classifier and achieved an overall accuracy of 71.3% in differentiating early, mild, moderate and advanced glaucoma eyes, and 83.9% accuracy in differentiation in glaucoma versus normal. Similarly, Xu et al.70 used 3D circumpapillary RNFL (cpRNFL) thickness (4 quadrants) measurements plus 68 super-pixel-based features and achieved AUCs of 0.71–0.86 differentiating normal subjects from glaucoma subjects and 0.87–0.90 differentiating glaucoma versus normal. They reported no significant difference in differentiating glaucoma versus normal subjects and glaucoma suspects versus normal subjects. Among the DL-based studies, George et al.71 used 3D-OCT features extracted using convolutional neural networks (CNNs) and obtained an AUC of 0.87 using a baseline CNN and 0.94 using their proposed 3D CNN using their attention-guided component. Juneja et al.72 used 513 features, including Statistical (18), Grey Level Co-occurrence Matrix (GLCM) (23) and Run Length Matrix (GLRM) (16) and wavelet filter features (456)) from 3D OCT and obtained a precision ranging from 0.91–0.95 and sensitivity ranging from 0.91–0.97 differentiating glaucoma versus normal cases. Panda et al.73 used raw Spectralis OCT volume scans applied to an autoencoder-based deep model with principal component analysis and obtained a sensitivity of 90% and an accuracy of 92% with a five-fold cross-validation. Conversely, we have used only 66 OCT spatial-frequency domain features and achieved an AUC of 0.97 (95% CI: 0.95–0.99), 0.96 (95% CI: 0.95–0.98) (SVM), 0.98 (95% CI: 0.96–1.00) (both SVM and RF), and 1.00 (95% CI: 1.00–1.00) (both RF and SVM) for differentiating overall, early, moderate and advanced glaucoma from normal subjects, which is higher compared to the aforementioned studies. However, we acknowledge that direct comparisons with existing machine and deep learning-based studies are limited by differences in datasets, input types, and experimental settings. A benchmarking study is using the same dataset and evaluation protocol, would be required for a more definitive assessment of relative performance.
Third, our study also outperformed clinicians’ performance in glaucoma staging and overall glaucoma diagnosis across multiple trials of clinicians (see clinician’s variability, Supplementary Table 8). For example, in terms of glaucoma staging, Reus et al.11 reported that a group of ophthalmologists achieved a sensitivity of 61.9%, 66.2% and 96.8% and a specificity of 95.1% in discriminating early, moderate and severe glaucoma patients respectively. Conversely, we achieved a sensitivity of 90.9%, 93.6% and 100% and a specificity of 92.0%, 97.0% and 99.8% using OCT data, and discriminated early, moderate and severe glaucoma patients using an RF classifier.
The present study varies from other ML-based studies on glaucoma diagnosis in terms of both feature engineering and glaucoma likelihood prediction. First, this study utilised frequency-domain features, transferring the spatial domain TSNIT pattern (local measures) using a fast Fourier transform. The comparison of trained models using RNFL, GC-IPL and MC thickness features sets shows that RNFL outperforms the other, which contains the spatial as well as TSNIT-derived frequency domain features. As such, the frequency domain features such as power spectral density and slope of the harmonics of power spectral density show promise in the diagnosis, which represents the utility of TSNIT data. Second, for glaucoma likelihood prediction, we leveraged the SHAP feature ranking (global explanation) to identify major and minor criteria and predicted probability scores by PDA to decide on glaucoma likelihood and developed an Excel-based app. Last, the local feature importance (local explanations) at the inference phase by SHAP analysis enables clinicians to revisit and scrutinise specific features, which significantly affect the classification or diagnosis. It instils confidence and trust in clinicians regarding the AI-generated results and reduces the time and effort required to review the whole OCT data, which contains hundreds of feature values. We found it was useful for them to look at the specific area of the TSNIT pattern (e.g., RNFL inferior thickness) to quickly identify the changes due to glaucoma. The clinicians gave more attention to the RNFL symmetry being the most crucial feature as flagged by the machine. In the past, clinicians used to give less priority to this information.
However, several limitations should be considered while interpreting the results. First, the model is limited to the CIRRUS and not generally applicable to all OCT machines. Second, the glaucoma cohort in this study was small (imbalance ratio: 0.802). However, SMOTE-based oversampling53 was performed to overcome this issue. Additionally, some of the patients had artefacts in CIRRUS RNFL analysis, while GC-IPL and macular thickness data were available and vice-versa. Instead of removing a patient’s whole data when one parameter is artefact-free (e.g., RNFL) and another parameter has an artefact (e.g., GC-IPL), we utilised the MICE38 algorithm to impute missing values (3.65% of total samples). The performance metrics with and without the augmented data showed a non-significant difference, which suggests that in our case the original data was already diverse enough and use of imputation and oversampling was not necessary to improve the model’s performance. As imputation and oversampling add computational complexity to the ML pipeline, it may be omitted for a reasonably well-balanced dataset. Also, although imputation and oversampling have been used widely in medical machine learning, a recent study has raised concerns about the clinical validity of using SMOTE in medical data, highlighting the need for further research into advanced modelling techniques74. Using a large dataset and exploring alternative imputation techniques such as KNN imputation75 and oversampling techniques such as Adaptive Synthetic Sampling (ADASYN)76 could be considered as possible future work.
Second, regarding the interpretation techniques used, both SHAP and PDA are computationally demanding. Also, feature independence62 is a limitation of PDA with a single feature as observation. However, SHAP addresses this limitation by using a game theoretical approach and considering the feature interaction of multiple high-ranked features. Therefore, being a mixed approach and taking advantage of the decision on multiple major and minor criteria, our proposed multi-feature diagnostic tool overcomes the limitations. Minor variations in the top-ranked features (e.g., RNFL symmetry) are unlikely to significantly affect the model’s classification outcome for several reasons. First, in our Excel-based explainable tool (Fig. 1b and Fig. 5a), decisions are based on a weighted scoring system that combines major and minor criteria. This approach inherently reduces the sensitivity to small variations in any single feature, including RNFL symmetry, by aggregating the contributions of multiple features. Second, the web-based app employs an RF classifier using SHAP-ranked features. RF models, by design, aggregate predictions from multiple decision trees, each of which evaluates features hierarchically based on metrics like the Gini index77. This structure ensures that the model’s decisions are robust to minor fluctuations in individual features, such as RNFL symmetry. Last, the web app provides local explanations for individual predictions using SHAP, highlighting the contribution of each feature during the inference/test phase. This transparency allows clinicians to examine specific features, such as RNFL symmetry, and identify any potential biases.
There is an inherent trade-off between explainability and model performance, and achieving a balance is crucial78. Using 66 spatial-domain features with an RF-based classifier, we achieved an accuracy of 93.2%. However, when limited to 9 explainable features, the accuracy was 88.2% with the Excel tool and 90.1% with the web app—a difference of 3.1–5%. Despite this slight reduction in performance, our explainable tools successfully balance accuracy with interpretability. By providing both global and local explanations, they enhance trust, reliability, and user-friendliness, making them highly practical for real-world glaucoma diagnosis. However, our focus on ML explainability was limited to using model-agnostic explainable methods (i.e., SHAP59 and PDA64). It is important to acknowledge other explainable approaches such as Local Interpretable Model-Agnostic Explanations (LIME)79. Future research could focus on developing model-specific explainable methods for more robust explanations80.
Third, we acknowledge that current versions of the explainable applications are limited to glaucoma diagnosis only (i.e., glaucoma vs normal subjects). The primary reason for excluding glaucoma staging from the app is that the SHAP-based diagnostic approach and PDA are inherently suited for binary classification tasks (e.g., distinguishing between glaucoma and non-glaucoma). Extending these methods to multiclass problems like glaucoma staging introduces significant challenges. SHAP requires separate value computations for each class independently, leading to higher computational complexity and reduced interpretability when combining insights across multiple classes63,81. The current implementation of our SHAP-based model focuses on binary classification, which allows a straightforward identification of features critical for diagnosing glaucoma versus non-glaucoma cases. Extending this approach to glaucoma staging would require developing and validating separate SHAP analyses for each stage, which was beyond the scope of this study. Conversely, PDA visualises the effect of each feature on the predicted probability, making it well-suited for binary outcomes where a clear decision boundary can be defined64,82. For multiclass staging, PDA would need to account for the interaction of features with multiple decision boundaries, which complicates the interpretation and reduces clinical usability. Future iterations of the app could explore integrating staging capabilities, potentially using advanced methods using hierarchical classification frameworks to design the explainable model for multiclass tasks.
There is scope to extend this work to make it even more useful for future clinical practice. First, the proposed tool simply uses CIRRUS OCT-based RNFL and GC-IPL thickness analysis, which can be used offline for possible interpretation of major and minor important features in glaucoma diagnosis. Future work may include integration of the web-based app with the CIRRUS software or using it as an external application to suggest a predicted probability based on XAI. Second, this study did not consider some important OCT based features, such as cup-to-disc ratio and rim thickness (because CIRRUS does not offer precise measurement); clinical features such as age, gender, medications and intraocular pressure (IOP); or visual field features such as PSD and MD as input features, though some recent studies83,84 have shown the utility of these features in diagnosis. Future work may involve fusing these clinical features as functional measures with the structural OCT features. Third, the machine learning models developed in this study do not account for glaucoma suspects and non-glaucomatous optic nerve disease. Future studies should be performed to differentiate glaucoma suspects from glaucoma and normal. For robust detection, multimodal fusion can be performed with colour fundus images and the existing OCT features, which may further improve diagnostic accuracy. In conclusion, the XAI-based developed software tools demonstrated in this study show promise in assisting clinicians and improving differential glaucoma diagnosis with extra reliability.
Data availability
The patients provided approval for data to be used as part of our study (UNSW human research ethics approval number: HC210563). Consequently, the raw data are not available for public access, but the statistically analysed OCT data in tabular format for a group of patient cohorts (normal, glaucoma/glaucoma stages) may be accessible upon reasonable request to the corresponding author.
References
Fingert, J. H. et al. No association between variations in the WDR36 gene and primary open-angle glaucoma. Arch. Ophthalmol. 125, 434–436 (2007).
Tham, Y.-C. et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090 (2014).
Stamper, R. L. A history of intraocular pressure and its measurement. Optometry Vis. Sci. 88, E16–E28 (2011).
Wu, Z. & Medeiros, F. A. Recent developments in visual field testing for glaucoma. Curr. Opinion Ophthalmol. 29, 141–146 (2018).
Phu, J. et al. The value of visual field testing in the era of advanced imaging: clinical and psychophysical perspectives. Clin. Exp. Optometry 100, 313–332 (2017).
Michelessi, M. et al. Optic nerve head and fibre layer imaging for diagnosing glaucoma. Cochrane Database Syst. Rev. 2020, 1–313 (2015).
Ly, A., Phu, J., Katalinic, P. & Kalloniatis, M. An evidence-based approach to the routine use of optical coherence tomography. Clin. Exp. Optometr. 102, 242–259 (2019).
Phu, J., Agar, A., Wang, H., Masselos, K. & Kalloniatis, M. Management of open-angle glaucoma by primary eye-care practitioners: toward a personalised medicine approach. Clin. Exp. Optometry 104, 367–384 (2021).
Burr, J. M. et al. The clinical effectiveness and cost-effectiveness of screening for open angle glaucoma: a systematic review and economic evaluation. Health Technol. Assess. 11, 3–190 (2007).
Nayak, B. K., Maskati, Q. B. & Parikh, R. The unique problem of glaucoma: under-diagnosis and over-treatment. Indian J. Ophthalmol. 59, S1-2 (2011).
Reus, N. J. et al. Clinical assessment of stereoscopic optic disc photographs for glaucoma: the European Optic Disc Assessment Trial. Ophthalmology 117, 717–723 (2010).
Hasan, M. M., Phu, J., Sowmya, A., Meijering, E. & Kalloniatis, M. Artificial intelligence in the diagnosis of glaucoma and neurodegenerative diseases. Clin. Exp. Optometry 130–146, 1–15 (2024).
Charng, J. et al. Deep learning: applications in retinal and optic nerve diseases. Clin. Exp. Optometry 106, 466–475 (2022).
Thompson, N. C., Greenewald, K., Lee, K. & Manso, G. F. The computational limits of deep learning. arXiv preprint arXiv:2007.05558, 1–33 (2020).
Strubell, E., Ganesh, A. & McCallum, A. Energy and policy considerations for modern deep learning research. Proc. AAAI Confer. Artif. Intell. 34, 13693–13696 (2020).
Mehta, P. et al. Automated detection of glaucoma with interpretable machine learning using clinical data and multimodal retinal images. Am. J. Ophthalmol. 231, 154–169 (2021).
Medeiros, F. A., Jammal, A. A. & Mariottoni, E. B. Detection of progressive glaucomatous optic nerve damage on fundus photographs with deep learning. Ophthalmology 128, 383–392 (2021).
Thakoor, K. A. et al. Strategies to improve convolutional neural network generalizability and reference standards for glaucoma detection from oct scans. Transl. Vis. Sci. Technol. 10, 16–16 (2021).
Maetschke, S. et al. A feature agnostic approach for glaucoma detection in OCT volumes. PloS One 14, e0219126 (2019).
Kim, S. J., Cho, K. J. & Oh, S. Development of machine learning models for diagnosis of glaucoma. PloS One 12, e0177726 (2017).
Wu, C. W., Chen, H. Y., Chen, J. Y. & Lee, C. H. Glaucoma detection using support vector machine method based on Spectralis OCT. Diagnostics 12, 391 (2022).
Sharma, P. et al. A lightweight deep learning model for automatic segmentation and analysis of ophthalmic images. Sci. Rep. 12, 1–18 (2022).
Wu, C. W., Shen, H. L., Lu, C. J., Chen, S. H. & Chen, H. Y. Comparison of different machine learning classifiers for glaucoma diagnosis based on spectralis OCT. Diagnostics 11, 1–14 (2021).
Zhou, W., Gao, Y., Ji, J. H., Li, S. C. & Yi, Y. G. Unsupervised anomaly detection for glaucoma diagnosis. Wireless Commun. Mobile Comput. 2021, 1–4 (2021).
Fernandez Escamez, C. S., Martin Giral, E., Perucho Martinez, S. & Toledano Fernandez, N. High interpretable machine learning classifier for early glaucoma diagnosis. International Journal of Ophthalmology 14, 393–398 (2021).
Mukherjee, R., Kundu, S., Dutta, K., Sen, A. & Majumdar, S. Predictive diagnosis of glaucoma based on analysis of focal notching along the neuro-retinal rim using machine learning. Pattern Recognition Image Anal. 29, 523–532 (2019).
Susanna, R., De Moraes, C. G., Cioffi, G. A. & Ritch, R. Why do people (still) go blind from glaucoma?. Transl. Vis. Sci. Technol. 4, 1–1 (2015).
Gunvant, P. et al. Analysis of Retinal Nerve Fiber Layer Data Obtained by Optical Coherence Tomograph Using Fourier Based Analysis. Investig. Ophthalmol. Visual Sci. 47, 3338–3338 (2006).
Hsieh, M.-H., Chang, Y.-F., Liu, C.J.-L. & Ko, Y.-C. Fourier analysis of circumpapillary retinal nerve fiber layer thickness in optical coherence tomography for differentiating myopia and glaucoma. Sci. Rep. 10, 1–9 (2020).
Essock, E. A., Sinai, M. J., Fechtner, R. D., Srinivasan, N. & Bryant, F. D. Fourier analysis of nerve fiber layer measurements from scanning laser polarimetry in glaucoma: emphasizing shape characteristics of the’double-hump’pattern. J. Glaucoma 9, 444–452 (2000).
Jamous, K. F. et al. Clinical model assisting with the collaborative care of glaucoma patients and suspects. Clin. Exp. Ophthalmol. 43, 308–319 (2015).
Huang, J., Hennessy, M. P., Kalloniatis, M. & Zangerl, B. Implementing collaborative care for glaucoma patients and suspects in Australia. Clin. Exp. Ophthalmol. 46, 826 (2018).
Phu, J., Hennessy, M. P., Spargo, M., Dance, S. & Kalloniatis, M. A collaborative care pathway for patients with suspected angle closure glaucoma spectrum disease. Clin. Exp. Optometry 103, 212–219 (2020).
Wang, H. & Kalloniatis, M. Clinical outcomes of the Centre for Eye Health: an intra-professional optometry-led collaborative eye care clinic in Australia. Clin. Exp. Optometry 104, 795–804 (2021).
Mills, R. P. et al. Categorizing the stage of glaucoma from pre-diagnosis to end-stage disease. Am. J. Ophthalmol. 141, 24–30 (2006).
White, A. et al. Guidelines for the collaborative care of glaucoma patients and suspects by ophthalmologists and optometrists in Australia. Clin. Exp. Ophthalmol. 42, 107–117 (2014).
Phu, J. et al. Visualizing the consistency of clinical characteristics that distinguish healthy persons, glaucoma suspect patients, and manifest glaucoma patients. Ophthalmol. Glaucoma 3, 274–287 (2020).
White, I. R., Royston, P. & Wood, A. M. Multiple imputation using chained equations: issues and guidance for practice. Stat. Med. 30, 377–399 (2011).
Austin, P. C., White, I. R., Lee, D. S. & van Buuren, S. Missing data in clinical research: a tutorial on multiple imputation. Can. J. Cardiol. 37, 1322–1331 (2021).
Azur, M. J., Stuart, E. A., Frangakis, C. & Leaf, P. J. Multiple imputation by chained equations: what is it and how does it work?. Int. J. Methods Psychiatr. Res. 20, 40–49 (2011).
Chandrashekar, G. & Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 40, 16–28 (2014).
Das, S. Filters, wrappers and a boosting-based hybrid for feature selection. 2001 International Conference on Machine Learning (ICML) 1, 74–81 (2001).
Suto, J., Oniga, S. & Sitar, P. P. Comparison of wrapper and filter feature selection algorithms on human activity recognition. 2016 6th International Conference on Computers Communications and Control (ICCCC), 124–129 (2016).
Hasan, M. M., Watling, C. N. & Larue, G. S. Physiological signal-based drowsiness detection using machine learning: Singular and hybrid signal approaches. J. Safety Res. 80, 215–225 (2021).
Abraham, A. et al. Machine learning for neuroimaging with scikit-learn. Front. Neuroinf. 8, 71792 (2014).
Hasan, M. M. Biomedical signal based drowsiness detection using machine learning: Singular and hybrid signal approaches, Queensland University of Technology, (2021).
Yu, L. & Liu, H. Feature selection for high-dimensional data: A fast correlation-based filter solution. In Proceedings of the 20th international conference on machine learning (ICML-03). 856–863.
Scikit-learn developers. sklearn.feature_selection.SelectKBest, <https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.SelectKBest.html> (2024).
Kohavi, R. & John, G. H. The wrapper approach. In Feature Extraction, Construction and Selection: A Data Mining Perspective 33–50 (Springer, 1998).
Mårtensson, H., Keelan, O. & Ahlström, C. Driver sleepiness classification based on physiological data and driving performance from real road driving. IEEE Trans. Intell. Trans. Syst. 20, 421–430 (2018).
Persson, A., Jonasson, H., Fredriksson, I., Wiklund, U. & Ahlström, C. Heart rate variability for classification of alert versus sleep deprived drivers in real road driving conditions. IEEE Trans. Intell. Trans. Syst. 22, 3316–3325 (2020).
Raschka, S. Python machine learning. (Packt Publishing Ltd, 2015).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. J. Art. Intell. Res. 16, 321–357 (2002).
Elreedy, D. & Atiya, A. F. A comprehensive analysis of synthetic minority oversampling technique (SMOTE) for handling class imbalance. Inform. Sci. 505, 32–64 (2019).
Tékouabou, S. C. K., Chabbar, I., Toulni, H., Cherif, W. & Silkan, H. Optimizing the early glaucoma detection from visual fields by combining preprocessing techniques and ensemble classifier with selection strategies. Expert Syst. Appl. 189, 115975 (2022).
Mamo, B. & Tsegaw, T. Application of Data Mining Techniques to Developing A Classification Model for Glaucoma Type Identification. Abyssinia J. Eng. Comput. 3, 27–33 (2023).
Martinez-Velasco, A., Martínez-Villaseñor, L. & Miralles-Pechuán, L. Addressing Class Imbalance in Healthcare Data: Machine Learning Solutions for Age-Related Macular Degeneration and Preeclampsia. IEEE Latin Am. Trans. 22, 806–820 (2024).
Bisong, E. & Bisong, E. Google colaboratory (Apress, 2019).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inform. Process. Syst. 30, 1–10 (2017).
Das, A. & Rad, P. Opportunities and challenges in explainable artificial intelligence (XAI): A survey. arXiv preprint arXiv:2006.11371, 1–24 (2020).
Yang, R. Who dies from COVID-19? Post-hoc explanations of mortality prediction models using coalitional game theory, surrogate trees, and partial dependence plots. MedRxiv, 1–19 (2020).
Linardatos, P., Papastefanopoulos, V. & Kotsiantis, S. Explainable AI: A review of machine learning interpretability methods. Entropy 23, 1–45 (2020).
Molnar, C. Interpretable machine learning: A guide for making black box models explainable. 2nd edn, (Leanpub, 2022).
Friedman, J. H. Greedy function approximation: a gradient boosting machine. Annals Stat. 29, 1189–1232 (2001).
Elhenawy, M., Larue, G. S., Masoud, M., Rakotonirainy, A. & Haworth, N. Using random forest to test if two-wheeler experience affects driver behaviour when interacting with two-wheelers. Trans. Res. 92, 301–316 (2023).
Komol, M. M. R. et al. Crash severity analysis of vulnerable road users using machine learning. PloS One 16, e0255828 (2021).
Sohns, J. T., Garth, C. & Leitte, H. Decision boundary visualization for counterfactual reasoning. Comput. Graphics Forum 42, 7–20 (2023).
Rao, H. L. et al. Structure-function relationship in glaucoma using spectral-domain optical coherence tomography. Arch. Ophthalmol. 129, 864–871 (2011).
Kooner, K. S. et al. Glaucoma diagnosis through the integration of optical coherence tomography/angiography and machine learning diagnostic models. Clin. Ophthalmol. 16, 2685–2697 (2022).
Xu, J. et al. Three-dimensional spectral-domain optical coherence tomography data analysis for glaucoma detection. PloS One 8, e55476 (2013).
George, Y. et al. Attention-guided 3D-CNN framework for glaucoma detection and structural-functional association using volumetric images. IEEE J. Biomed. Health Inform. 24, 3421–3430 (2020).
Juneja, M. et al. Fused framework for glaucoma diagnosis using Optical Coherence Tomography (OCT) images. Expert Systems with Applications 201 (2022).
Panda, S. K. et al. Describing the Structural Phenotype of the Glaucomatous Optic Nerve Head Using Artificial Intelligence. Am. J. Ophthalmol. 236, 172–182. https://doi.org/10.1016/j.ajo.2021.06.010 (2022).
Gholampour, S. Impact of Nature of Medical Data on Machine and Deep Learning for Imbalanced Datasets: Clinical Validity of SMOTE Is Questionable. Machine Learning Knowledge Extraction 6, 827–841 (2024).
Pujianto, U., Wibawa, A. P. & Akbar, M. I. K-nearest neighbor (k-NN) based missing data imputation. In 2019 5th International Conference on Science in Information Technology (ICSITech). 83–88 (IEEE).
He, H., Bai, Y., Garcia, E. A. & Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence). 1322–1328 (IEEE).
Ferdowsi, M., Hasan, M. M. & Habib, W. Responsible AI for cardiovascular disease detection: Towards a privacy-preserving and interpretable model. Comput. Methods Progr. Biomed. 254, 108289 (2024).
Herm, L.-V., Heinrich, K., Wanner, J. & Janiesch, C. Stop ordering machine learning algorithms by their explainability! A user-centered investigation of performance and explainability. Int. J. Infor. Manag. 69, 102538 (2023).
Biecek, P. & Burzykowski, T. Local interpretable model-agnostic explanations (LIME). Explanatory Model Analysis Explore Explain Examine Predictive Models 1, 107–124 (2021).
Belle, V. & Papantonis, I. Principles and practice of explainable machine learning. Frontiers Big Data 4, 688969 (2021).
Huang, X. & Marques-Silva, J. Updates on the Complexity of SHAP Scores. In Proceedings of the Thirty-Third International Joint Conference on Artificial Intelligence (pp. 403-412).
Hasan, M. M., Watling, C. N. & Larue, G. S. Validation and interpretation of a multimodal drowsiness detection system using explainable machine learning. Comput. Methods Progr. Biomed. 243, 107925 (2024).
Oh, S., Park, Y., Cho, K. J. & Kim, S. J. Explainable machine learning model for glaucoma diagnosis and its interpretation. Diagnostics 11, 510 (2021).
Akter, N. et al. Glaucoma diagnosis using multi-feature analysis and a deep learning technique. Sci. Rep. 12, 8064 (2022).
Acknowledgements
This work was supported by a UNSW Tuition Fee Scholarship (TFS), Australian Government Research Training Program (RTP) Fees Offset, plus a stipend for MMH from EM’s Academic Start-Up Fund for Biomedical Image Computing. This work was also supported in part by a National Health and Medical Research Council of Australia Ideas Grant to MK and JP (NHMRC 1186915), and a Beta Sigma Kappa Fellowship to JP. Clinical services at the Centre for Eye Health are primarily supported by Guide Dogs NSW/ACT. The funding bodies had no role in the conceptualisation or decision to disseminate the present work. Additionally, the authors would like to sincerely thank Janelle Tong for her image grading to evaluate human vs machine performance.
Author information
Authors and Affiliations
Contributions
M.M.H extracted the raw data and contributed to the conceptualisation, methodology and software, conducted the experiment(s) and formal analysis, visualisation and validation, and wrote the original draft. J.P, H.W. and M.K. contributed to data curation and cleaning and clinical validation of the experimental results. E.M., A.S. and M.K. supervised the project, contributed to the research design, methodology and conceptualisation, visualisation, validation and provided resources. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
Commercial Relationship disclosure: Md Mahmudul Hasan, Jack Phu, Henrietta Wang, Arcot Sowmya, Michael Kalloniatis, Erik Meijering have submitted a patent application (#2024900616) in the name of “NewSouth Innovations Pty Limited” and “Deakin University” which is currently pending. They declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hasan, M.M., Phu, J., Wang, H. et al. OCT-based diagnosis of glaucoma and glaucoma stages using explainable machine learning. Sci Rep 15, 3592 (2025). https://doi.org/10.1038/s41598-025-87219-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87219-w
Keywords
This article is cited by
-
Machine learning with multitype functional connectivity uncovers whole-brain network disruption in primary angle-closure glaucoma
Brain Informatics (2026)
-
Predicting HFA 30–2 Visual Fields with Deep Learning from Multimodal OCT–Fundus Feature Fusion and Structure–Function Discordance Analysis
Journal of Imaging Informatics in Medicine (2026)
-
Evaluating Machine Learning Classifiers for Automated Glaucoma Detection Using Fundus Images
Biomedical Materials & Devices (2025)