Abstract
The Middle Route of the South to North Water Diversion Project (MRP) and its water source, the Danjiangkou Reservoir (DJK), play a pivotal role in mitigating the chronic water scarcity challenges faced by northern China. Eukaryotic plankton are widespread in aquatic ecosystems, which are crucial for the water quality stability of DJK and MRP, yet comparative studies on their contemporaneous dynamics and assembly processes are scarce. In this study, amplicon sequencing was used to investigate the eukaryotic plankton communities. The results revealed that the similarity in community composition of DJK is significantly higher than that of MRP, exhibiting distance-decay patterns. Environmental heterogeneity exhibits significant differences between DJK and MRP, and it significantly influences community composition and alpha diversity. Additionally, the assembly processes of eukaryotic plankton in both DJK and MRP are predominantly influenced by stochastic processes. However, in comparison to DJK, deterministic processes have a more pronounced impact on MRP, accounting for 39.29% and 1.82%, respectively. The variations in total nitrogen (TN), chlorophy IIa (Chl.a), and conductivity (Spc) have led to a transition in the assembly of eukaryotic phytoplankton communities in MRP from a stochastic process to a deterministic process. This study extends insights into the dynamics and assembly processes of eukaryotic plankton communities in the large, engineered drinking water diversion project and its water source, which is also useful for the management and regulation of the DJK and MRP.
Similar content being viewed by others
Introduction
Microorganisms represent an essential component of ecosystems, exerting significant influence over material cycling processes and energy fluxes1. These organisms are crucial determinants of water quality and safety, particularly in potable water sources. Microbial communities possess the capacity to influence water quality, which may result in the generation of unpleasant odors and potential public health risks2. Research has shown that microorganisms are important producers of odor compounds and other metabolites, closely related to water quality3. Microorganisms present in potable water contribute significantly to the self-purification processes, which, in turn, have a profound impact on water quality and safety4. Furthermore, the succession of microbial communities could mirror alterations in water quality parameters, with their distribution intricately associated with the dynamic nature of local water quality attributes5. Microorganisms serve a crucial function as key indicators in assessing the condition of aquatic environments6. Accordingly, monitoring the dynamic fluctuations of microorganisms in drinking water is crucial for the early detection of microbial hazards and the effective management of drinking water safety.
Eukaryotic Plankton are passively floating organisms, broadly dispersed across oceans, lakes, rivers, and diverse aquatic ecosystems due to the influence of water currents. Eukaryotic plankton, which includes not only primary producers like algae but also consumers (e.g., protozoa) and decomposers (e.g., fungi), tend to be larger and more complex in structure compared to prokaryotic plankton. They often have more specific environmental requirements, such as particular temperature, nutrient, and light conditions, due to their higher metabolic demands and more intricate cellular structures7,8. Investigating eukaryotic plankton’s diversity and community structure offers an indirect assessment of the ecological environment water quality8. Presently, extensive researches have centered on the community composition, spatial distribution, and functional roles of phytoplankton, archaea, plants, bacteria, and animal taxa, as well as their associations with water quality9,10,11,12. Comprehending the dynamic shifts and fundamental assembly mechanisms of eukaryotic communities poses a significant challenge13, particularly given the complexity of these communities within extensive engineered water diversion canals, which exhibit distinct characteristics compared to natural ecosystems. Moreover, contrasting the variations in community dynamics and assembly mechanisms between diversion channels and source water is crucial for regulating and predicting drinking water quality. Consequently, it is imperative to investigate eukaryotic plankton within water diversion channels and source waters.
In recent years, community assembly mechanisms have risen to prominence as a central theme in ecological research. Vellend (2010) proposed a conceptual synthesis, categorizing the processes of community formation into four distinct mechanisms: selection, dispersal, ecological drift, and diversification14. Moreover, Stegen (2013) constructed an analytical framework aimed at quantifying the relative contributions of dispersal (encompassing dispersal limitation and homogenizing dispersal), selection (including heterogeneous and homogenizing selection), and ecological drift15. Employing this mechanistic predictive framework, the respective contributions of these processes have been elucidated in both lentic water bodies16,17 and marine ecosystems18, utilizing a phylogenetic null model that facilitates the quantification of various ecological forces shaping microbial community assembly15. Moreover, clarifying the mechanisms of biotic community formation and maintenance is a fundamental element in understanding spatial distribution patterns, with distance-decay relationships of microbial communities frequently reported in freshwater lakes, rivers18,19,20, oceans21,22, and intertidal zones23. While spatiotemporal distribution characteristics and assembly processes are critical for understanding eukaryotic plankton communities, information remains limited regarding the spatiotemporal dynamics and assembly processes of eukaryotic plankton in both water diversion channels and water sources.
The Danjiangkou Reservoir (DJK) is located on the Han River (the longest tributary of the Yangtze River in China), and it is the place where the Dan River flows into. Water in the DJK is mainly from the Han River and Dan River, and the total area is approximately 1023 km2, with a water storage capacity of 29 billion m3.24 Moreover, the DJK is the water source for the middle route of the South-to-North Water Diversion Project (MRP), which is the largest water transfer project in the world, spanning different watershed and different climatic zones, which has solved the water shortage problem in the northern four provinces and cities, namely Henan, Hebei, Beijing, and Tianjin25. Eukaryotic plankton, a pivotal determinant in sustaining water quality stability within the DJK and MRP, remains insufficiently explored. Additionally, comprehensive comparative studies on the assembly processes of eukaryotic plankton communities between water diversion channels and their source waters have yet to be thoroughly investigated. The MRP consists of reinforced concrete, facilitating a robust hydrodynamic exchange and comprising multiple hydraulic structures and engineered disturbances (e.g., inverted siphons, aqueducts, and tunnels) along its main channel26, thereby endowing its water characteristics with marked distinctions from those of natural water bodies27. Following the application of MRP, the rapid proliferation and biomass accumulation of Cladophora at specific times suggested that eukaryotic phytoplankton exhibit swift responses to particular habitat conditions, this phenomenon represents a potential ecological issue that may compromise water quality stability. Consequently, examining the community dynamics of eukaryotic plankton is critical to elucidating community variability, nutrient cycling processes, and the prevailing water quality conditions. In addition to those, it is critical to explore the dynamics of eukaryotic plankton communities and their relationship to water environment, both in terms of ecological significance and as an important issue for the management and regulation of the DJK and MRP.
To be specific, our research is committed to elucidating the following issues: (1) What are the spatial patterns of eukaryotic plankton community in the DJK and MRP? (2) What are the factors that influence the distribution of eukaryotic plankton communities? (3) What is the relative importance of stochastic and deterministic assembly processes of eukaryotic plankton community? Which environmental factors mediate community assembly processes? Our results might add to the understanding of the dynamics of eukaryotic plankton communities and their assembly processes between water diversion channels and their source waters.
Materials and methods
Sample collection and environmental factors analysis
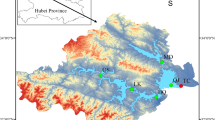
Surface water samples were collected in July 2022. Details of the sampling sites are shown in Fig. 1 and Table.S1. At each site, environmental factors, including water temperature (WT), pH, dissolved oxygen (DO), conductivity (Spc), salinity (Sal), and oxidation-reduction potential (ORP), were measured using a YSI Professional Plus meter (YSI, USA). Current water velocity (Vel) was recorded using an FL-KIT current meter. And mixed water samples of 2 L were collected at 0.5 m and 1 m depth below the water surface, which were divided into two parts for eukaryotic plankton community and chemical analyses, respectively. Eukaryotic plankton samples (1 L water samples) were filtered through 0.22 μm pore-size polycarbonate membrane filter (Whatman) to collect the eukaryotes and then frozen at -80 ℃ until DNA extraction. For measurements of chlorophy IIa (Chl.a) contens, 500 ml water samples were filtered using GF/C filters, and Chl a was then extracted with 90% acetone over 24 h and measured spectrophotometrically. 500 ml water samples were used for measuring total nitrogen (TN), ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3−-N), total phosphorus (TP), phosphate phosphorus (PO43−-P) and chemical oxygen demand (CODMn) according to the Monitoring and analysis methods of water and wastewater28.
Distribution of sampling sites in the Danjiangkou Reservoir and the channel of the middle route of South-to-North Water Diversion Project. (The base map was downloaded from the Standard Map Service System of the Ministry of Natural Resources of China (http://bzdt.ch.mnr.gov.cn/) and edited using Adobe Photoshop 2018 (https://www.adobe.com/products/photoshop.html)).
DNA extraction, PCR amplification, Illumina NovaSeq sequencing and data processing
Total genomic DNA was extracted from water samples using the TGuide S96 Magnetic Soil /Stool DNA Kit (Tiangen Biotech (Beijing) Co., Ltd.) according to manufacturer’s instructions. The quality and quantity of the extracted DNA were examined using electrophoresis on a 1.8% agarose gel, and DNA concentration and purity were determined with NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific, Wilmington, USA). DNA extracts were used as PCR templates, and barcoded primers targeting the V4 region of the 18S rRNA gene (TAReuk454FWD1: 5’-CCAGCASCYGCGGTAATTCC-3’ and TAReukREV3: 5’-ACTTTCGTTCTTGATYRA-3’) were used to construct sequencing libraries for each sample tested29,30. These primers have the Illumina sequencing primer sequences attached at their 5′ends. In the first amplification step, PCRs were carried out in a final volume of 12.5 µL, containing 1.25 µL of template DNA, 0.5 µM of the primers, 3.13 µL of Supreme NZYTaq 2x Green Master Mix (NZYTech, Portugal) and ultrapure water up to 12.5 µL31. The reaction mixture was incubated as follows: an initial denaturation step at 95℃ for 5 min, followed by 35 cycles of 95℃ for 30 s, 48℃ for 45 s, 72℃ for 45 s and a final extension step at 72℃ for 7 min. The oligonucleotide indices that are required for multiplexing different libraries in the same sequencing pool were attached in a second amplification step with identical conditions but only for 5 cycles and at 60℃ annealing temperature31. A negative control that contained no DNA (BPCR) was included in every PCR round to check for contamination during library preparation11. The amplified products were purified with Omega DNA purification kit (Omega Inc., Norcross, GA, USA) and quantified using Qsep-400(BiOptic, Inc., New Taipei City, Taiwan, ROC).The amplicon library was paired-end sequenced (2 × 250) on an Illumina novaseq 6000 (Beijing Biomarker Technologies Co., Ltd., Beijing, China).
The raw 18 S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.23.1 and merged by FLASH version 1.2.1132,33,34. Operational taxonomic units (OTUs) with a 97% similarity cutoff were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed35. The taxonomy of each representative sequence was analyzed by RDP Classifier version 2.2 against the 18 S rRNA database (Silva v138) using a confidence threshold of 0.736,37. Based on bioinformatics, Good’s coverage values of all samples were above 99.9%, confirming that the libraries could represent most of the OTUs in the samples.
Statistical analysis
The alpha diversity indices (including Shannon Wiener Diversity Index, Simpson’s Diversity Index, Chao1 Index and ACE Index) was calculated using QIIME version 1.9.139. One-way analysis of variance (ANOVA) was used to test the significance of the differences among the DJK and MRP based on the environmental factors and alpha diversity indices. The Fisher’s Least Significant Difference (LSD) test was applied to test for the difference in the environmental data and the homogeneity of variances (Levene’s test). Otherwise, the significance was tested using Tamhane’s in Post-hoc multiple comparisons. One-way ANOVA was completed by IBM SPSS Statistics for Windows (version 17, Chicago, IL, USA).
Beta diversity of eukaryotic plankton was assessed using the Bray-Curtis similarity coefficient and Nonmetric Multidimensional Scaling (NMDS)39. The ANOSIM statistic (R) was used to compare mean ranks between groups, with values ranging from − 1 to + 1, where 0 indicates random grouping40. Statistical significance (α = 0.05) was assessed with 999 permutations. Eukaryotic plankton data were clustered using Ward’s Minimum Variance Method based on Euclidean distance41. Linear discriminant analysis (LDA) effect size (LefSe) was applied to identify significant differences among abundant modules, with a threshold of 4.0 on the logarithmic LDA score for discriminative functional biomarkers. LefSe analysis was performed using the Galaxy framework 2.042.
All environmental factors, except pH, were standardized to improve normality and homoscedasticity (Xst =(X-‾X)/SD)43. Principal component analysis (PCA) was conducted based on the methods outlined by Jolliffe and Cadima44. Preliminary detrended correspondence analysis (DCA) of the eukaryotic data indicated that the longest gradient was less than 3. Therefore, redundancy analysis (RDA) was employed to explore the relationship between community structures and environmental factors. Variance inflation factors were kept below 10 to minimize multicollinearity among environmental factors. Before the RDA analysis, the ‘ordiR2step’ function in the R ‘vegan’ package was used for forward selection to filter out significantly affecting variables (P < 0.05)45. Influential factors were selected using 999 Monte Carlo permutation tests at the P < 0.05 significance level. In addition, relative importance of environmental factors was assessed using variation partitioning analysis (VPA)46. The Mantel test was also used to further investigate correlations between environmental factors and Bray-Curtis similarity of communities47. Bray-Curtis similarity between samples and geographical distances were calculated using the R packages ‘vegan’ and ‘geosphere’48. Euclidean distances of environmental factors were also analyzed to detect whether they correlated with geographical distance. Regressions were generated with functions available in the ‘ggplot2’ package49.
The Stegen null model was applied to quantify the relative importance of stochastic and deterministic processes50. Phylogenetic and taxonomic diversity variations were assessed using β-diversity metrics based on null models (βNTI and RCbray). Specifically, |βNTI| < 2 indicates the dominance of stochastic processes, while the relative contributions of variable and homogeneous selection were determined by the percentage of pairwise comparisons with βNTI > 2 and < -2, respectively51. The Bray-Curtis-based Raup Crick index (RCbray) was used to investigate pairwise comparisons not attributed to deterministic processes. By integrating |RCbray| values, we identified underlying community assembly processes such as homogenizing dispersal (|βNTI| < 2 and RCbray < -0.95), dispersal limitation (|βNTI| < 2 and RCbray > + 0.95), and undominated processes (e.g., weak selection, weak dispersal, diversification, and drift), where |βNTI| < 2 and |RCbray| < 0.9551. The null community for all samples was randomized 999 times to derive average null expectations.
Clustering, NMDS, PCA, DCA, RDA, Mantel test, and stochastic and deterministic processes were analyzed using the ‘vegan’, ‘stats’, ‘ggcor’, and ‘picante’ packages of R (version 4.0.3, R Foundation for Statistical Computing, Vienna, Austria).
Results
Community composition and diversity
A total of 2395 eukaryotic OTUs were identified from 1,493,024 valid sequences. The rarefaction curves for the OTUs detected and post-filtering sequencing data in this study are displayed in Supplementary Table. S2 and Fig. S1. Cluster analysis divided the 19 sampling sites into four groups (Fig. 2a). Overall, the clustering identified two large groups–the DJK (D1-D11) and the MRP (Z12-Z19). The former was further divided into two subgroups (Hanku: D1 to D5, Danku: D6 to D11), the latter was also further divided into two subgroups (Henan: Z12 to Z17, Hebei: Z18 and Z19) (Fig. 2a). The two-dimensional NMDS analysis showed a clear difference in species composition between the DJK and the MRP. Results with a stress of 0.07 are shown in Fig. 2b. ANOSIM analysis recorded a significance level of 0.001 of all the compared groups (Fig. S2), indicating significant differences between the eukaryotic plankton communities of the two groups.
A total of 185 orders of eukaryotic plankton were detected in the 19 samples. The taxa richness of the four subgroups (Danku, Hanku, Henan, Hebei) of eukaryotic planktons was 91, 90, 144, and 74, respectively. Thirty-four common species occurred in all four subgroups. Particular species (occurring only in Henan) had the highest (n = 45) taxa richness, although Henan shared 67, 69, and 69 species with Hebei, Danku, and Hanku, constituting 36.22%, 37.29%, and 37.29% of their total order number, respectively (Fig. 3a).
(a) Venn diagram of the taxa richness of the eukaryotic plankton orders; (b-c) Composition of the major eukaryotic plankton phyla and order in the DJK and the MRP (Relative abundance of the top 15); Number of OTUs (d), Chao 1 index (e), Simpson index (f) and Shannon index (g) in all samples. Significant differences (***P < 0.001, **P < 0.01, *P < 0.05) between categories are indicated by asterisk, statistical analysis is ANOVA with Tamhane’s with post hoc multiple comparisons.
The structures and relative abundances of the identified eukaryotic plankton at the phylum and order levels are shown in Fig. 3b and c, respectively. The most abundant phyla across all samples were Cryptophyta, Chlorophyta, Mollusca, Intramacronucleata, and Arthropoda, accounting for 28.97%, 11.06%, 10.37%, 10.16%, and 8.84% of all sequences, respectively (Fig. 3b). Cryptophyta was more abundant in the DJK (D1 to D11) than in the MRP. The relative abundance of Mollusca was higher in the Henan section compared to other sections, while Chytridiomycota was more abundant in the Hebei section (Fig. 3b). At the order level, Cryptomonadales, Sphaeropleales, and Calanoida were dominant in the DJK. Mytiloida was more abundant in the Henan section than in other sections (Fig. 3c).
The number of OTUs showed more significant differences in both the DJK and the MRP (P < 0.001) (Fig. 3d). For diversity, significant differences in the Chao1 index were found in both the DJK and the MRP (Fig. 3e, P < 0.001). In addition, the Simpson index and Shannon index of the MRP were significantly higher than those in the DJK (P < 0.05) (Fig. 3f-g).
LefSe analysis was performed to identify significant differences in the abundance of eukaryotic plankton taxa. The cladogram revealed distinct differences between the DJK and the MRP (Fig. 4a). The plot from LefSe analysis displays LDA scores of eukaryotic plankton taxa with significant differences between the DJK and the MRP (Fig. 4b). At the order level, the biomarkers showing significant differences between the DJK and the MRP included Sphaeropleales, Chlorellales, Cryptomonadales, Ochromonadales, Pyrocystales, Chytridiales, and Mytiloida. The relative abundance of Chlorellales, Ochromonadales, Pyrocystales, Chytridiales, and Mytiloida was significantly higher in the MRP compared to the DJK.
(a) The cladogram diagram shows the eukaryotic plankton order displaying significant differences in the two groups. Red and green indicate different groups, species classification being shown at the level of phylum, class, order from the inside to the outside. (b) Species exhibiting significant difference with an LDA score greater than the estimated value; the default score is 4.0. The length of the histogram represents the LDA score; i.e., the degree of influence of species with significant differences between groups.
Influencing factors of community variations
PCA demonstrated that samples showed a significant environmental heterogeneity between the DJK and the MRP (P < 0.001, Fig. S3). Fourteen measured environmental factors were included in the statistical analyses (Fig. 5). The nutrient-related measures of the MRP such as TN, NO3−-N, NH4+-N and CODMn were significantly higher than the DJK (P < 0.05). Also, WT and pH differed, being higher in the DJK, respectively, than in the MRP (P < 0.05). The values of Spc, Sal, ORP, Chl.a, and Phosphorus (including TP and PO43−-P) showed no differences between the two groups (P > 0.05). The F and P values determined by one-way ANOVA are shown in Table. S3. Vel of the MRP are shown in Table. S4. There was fluctuating variation in the spatial variation of the environmental factors, the Euclidean distances calculated for studied environmental factors showed significant positive correlation with geographical distances (Fig. S4).
Environmental factors in the Danjiangkou Reservoir and the channel of the middle route of South-to-North Water Diversion Project. Significant differences (***P < 0.001, **P < 0.01, *P < 0.05) between categories are indicated by asterisk, statistical analysis is ANOVA with LSD with post hoc multiple comparisons.
The negative correlations between community similarity and both geographic distance and environmental distance were significant in eukaryotic plankton communities (Fig. 6a-b). The eigenvalues of the first two RDA axes (γ1 = 0.1632, γ2 = 0.0589) explained 29.44% and 10.63% of the variation in the species-environment relationships, respectively (Fig. 6c). The RDA scores showed strong relationships between the environmental factors (i.e., TN, TP, WT, ORP, Spc, CODMn, Chl.a and Vel) and community variation. The DJK was strongly correlated with ORP, Spc and WT, while the MRP had strong correlation with TN, TP, Vel, CODMn and Chl.a. The Henan section had a high content of TN and TP, while the content of CODMn and Chl.a in Hebei section were higher than other sections. The relationship between dominant biological communities of the eukaryotic plankton communities and environment factors were analyzed by the Mantel test (Fig. S5). The results showed that WT, CODMn, Chl.a and ORP were the main environmental factors affecting the eukaryotic plankton community.
Bray-Curtis similarity of the eukaryotic plankton community against geographical distance (a) and environmental distance (b), RDA ordinations of sampling sites based on environmental factors (c). Danku, Hanku, Henan and Hebei are indicated by different symbols as mentioned in Fig. 2(a).
Community assembly processes
The range of variation in βNTI of the two groups (The DJK and The MRP) was mainly concentrated in |βNTI| <2 and βNTI < 2, respectively (Fig. 7a). In the DJK and the MRP, the stochastic processes explained a higher proportion of community variation than deterministic processes (60.72–98.19% vs. 1.82–39.29%). As shown in Fig. 7b, the communities at the DJK were mainly controlled by undominated processes and homogenizing dispersal. By contrast, the communities at the MRP were mainly driven by homogeneous selection, undominated processes, homogenizing dispersal and dispersal limitation. Homogeneous selection played a more important role in the assembly of the MRP (39.29%) than that in the DJK (1.82%).
The relationships between βNTI and the variation of environmental factors were also examined (Fig. S6 and Fig. 8). The βNTI values in the communities at the DJK were not significantly and positively related to environmental factors, but the βNTI values were still in the region of the stochastic process (-2 < βNTI < 2) (Fig. S6f-i). Pairwise comparisons of βNTI for the communities at the MRP were significantly and negatively correlated with Chl.a, Spc and TN (Fig. 8), indicating increasing variations in Chl.a, Spc and TN led to a shift from primarily stochastic community assembly (-2 < βNTI < 2) to homogeneous selection (βNTI < -2).
Discussion
Eukaryotic plankton community dynamics
Our analyses indicated a significant environmental heterogeneity between the DJK and the MRP, the spatial heterogeneity among various sampling sites within the MRP was higher than that in the DJK (Fig. S3). The MRP passes through subtropical monsoon climate zones and warm temperate monsoon climate zones. The long-distance water conveyance project has obvious differences in water temperature, climate, and environment, resulting in significant regional differences in water quality along the entire route52. In this study, we found that the concentrations of CODMn and Chl.a in the water of the MRP in Hebei section are higher than those in Henan section, while the concentrations of NH3-N, NO3−-N, and TN are lower than those in Henan section (Fig. 5). Previous research has shown that the density of algae cells in the MRP shows an increasing trend from south to north, and is significantly higher after the Yellow River crossing project than before53. CODMn represents the concentration of organic and inorganic substances in water that can be oxidized by permanganate54. The release of proteins and microbial residues into the water during autogenous processes such as algae and microbial activities in the water body increases the CODMn concentration of the water body55. Some studies have shown that nutrients are necessary for the growth and reproduction of plankton, and the reason for the massive proliferation of algae in early spring is the rapid absorption and utilization of nitrogen56. In our study, the lower concentrations of dissolved nitrogen (NO2−-N, NH3-N, NO3−-N) in the water of the MRP in Hebei section may be caused by the massive reproduction of plankton. The complex environmental and hydrological hydrodynamic conditions along the MRP indirectly affect the migration and transformation laws of substances, leading to changes in the growth rate and distribution pattern of plankton, and ultimately affecting the changes in the physicochemical characteristics of the water body, resulting in higher spatial heterogeneity of the water environment in the MRP.
In this study, we found that Chlorophyta and Cryptophyta play an important role in the DJK, and Cryptophyta, Mollusca, Diatomea and Arthropoda were the dominant taxa in the Henan section, while the Hebei section were dominated by Intramacronucleata, Chlorophyta, Chytridiomycota and Cercozoa, and the relative abundance of Cryptophyta decreased over long distances of water diversion in the MRP(Figs. 3 and 4). These results were highly in accordance with the previous analysis of planktonic eukaryotic in the DJK8,57 and the MRP53,58. Certain Cryptomycota species are recognized as primary parasites of Chlorophyta59, utilizing phagocytosis to absorb nutrients, thereby directly inhibiting phytoplankton growth60. Previous research has indicated that when the relative abundance of Cryptomycota was highest, the abundance of phytoplankton, particularly Chlorophyta, decreased significantly, which was similar to that of our study (Fig. 3b)61. Furthermore, it has been reported that Cryptophyta often grow in low flow velocity, eutrophic environments62 and that reducing nutrients and increasing flow velocity could control the proliferation of Cryptophyta63. Consequently, the observed decline in Cryptophyta could be attributed to adverse environmental shifts, including diminished nutrient availability and elevated flow velocities in the channel relative to the source water reservoir, conditions that may ultimately undermine their competitive dominance64,65. Generally, Chlorophyta and Cryptophyta are better adapted for growth in quiescent or low-disturbance aquatic environments, while Diatomea have a competitive advantage in terms of proliferation in flowing or turbulent aquatic environments66. An unusually high species richness of Cryptomycota and Chlorophyta was noted in this study. Chlorophyta exhibited dominance across diverse habitats, likely attributable to their autotrophic mode of nutrition, capacity for mixed acid fermentation67, and resilience in the face of environmental stressors68. Cryptomycota also demonstrated considerable genetic diversity and exhibited efficient mechanisms for transferring carbon and energy through trophic levels69. Hence, the still or low-disturbance water bodies in the DJK allowed Cryptomonas and Chlorophyta to be the important dominant species. Additionally, the higher CODMn concentration in the Hebei section of the MRP indicates that this water body has a large amount of organic matter, which can provide sufficient food sources for Chytridiomycota and Arthropoda. In addition, the appearance of Mollusca also indicates that the MRP has some attributes of the natural river channel.
Our results indicated that there are significant differences in the composition of eukaryotic plankton communities between the DJK and the MRP. Moreover, the similarity of community compositions among sampling sites in the MRP was relatively low, while the similarity among sampling sites in the DJK was relatively high (Fig. 2). Notably, environmental heterogeneity was significantly different between the DJK and the MRP (P < 0.001, Fig. S3), and significantly influenced communities and alpha diversity (P < 0.05, Fig. 3d-g). It is clear that local diffusion will make the microbial community structure tend to be homogenized in a connected water habitat without a preferred water flow direction70. The previous study has demonstrated that disordered hydraulic forces can increase the similarity of phytoplankton communities between interconnected water bodies71, and indeed, similar results have also been found in planktonic bacterial communities72. It has been reported that the wind-induced turbulence in the DJK may play a role in enhancing water column mixing, thereby exerting a certain influence on the distribution and community structure of plankton57. Furthermore, the previous investigation into the spatial patterns of the bacterioplankton in lake Taihu revealed that the disordered hydraulic mixing effect can promote the spatial diffusion of planktonic bacteria, thereby weakening the spatial difference of the planktonic bacterial community structure73. Thereby, the high similarity of the eukaryotic plankton communities in the DJK may be due to the disordered wind and wave disturbance. In contrast, it has been reported that the community structure of prokaryotic plankton generally exhibited significant spatial differences in a large spatial span72,74 or between regions with geographical isolation75,76. The Yellow River Crossing Project of the MRP (dark conditions and high flow velocity) leads to significant differences in the community structure of prokaryotic microorganisms77 and the density of phytoplankton cells53 before and after the Yellow River Crossing Project of the main canal. In addition, it has been reported that the physical and chemical factors also have an important impact on the community structure and distribution of plankton78. In the present study, the large geographical span of the sampling sites and the higher spatial heterogeneity of the water environment in the MRP may have led to the low similarity of the community structure of eukaryotic plankton.
The RDA showed that WT, SPC, and ORP were the significant water environment factors affecting the community distribution of planktonic eukaryotes in the DJK, while the MRP was mainly significantly influenced by Vel, TN, TP, CODMn, and Chl.a (Fig. 6c). Water quality, water temperature, and hydrological factors affected the phytoplankton community structure79. Some studies have shown that temperature can directly affect the growth of phytoplankton by influencing its metabolic process80, and it will also affect the growth of zooplankton and fish as well as other physical and chemical conditions, resulting in changes in the density and community structure of phytoplankton81. The flow velocity of the MRP ranges from 0.1 to 2.1 m·s-1, which is relatively faster than that of other water bodies82. Previous research has indicated that the cell density of cyanobacteria and Chlorophyta in the MRP is significantly negatively correlated with the flow velocity, and the relative abundance of Diatomea is positively correlated with the flow velocity53. The high flow velocity of the MRP enhances the dominant position of Diatomea, which was similar to that of our study. Moreover, previous research on phytoplankton in the lower reaches of the Xijiang River showed that increased flow rate and flow velocity are beneficial to the increase of species richness83. Consistent with these previous results, we found that the number of OTUs and alpha diversity index of eukaryotic plankton in the MRP were higher than those in the DJK (Fig. 3). Additionally, studies have shown that the plankton community is affected by the water body type and its physical and chemical properties such as pH, temperature, electrical conductivity, and nitrogen, phosphorus and organic matter84,85,86.
Moreover, eukaryotic plankton communities demonstrated analogous spatial variability, with community similarities displaying a pronounced distance-decay pattern (Fig. 6a-b). Prior research has revealed that spatial variation within small eukaryotic communities is predominantly regulated by dispersal constraints and/or the breadth of their ecological niches87. On one hand, expanding geographical distance may amplify community heterogeneity by intensifying dispersal limitation effects88. Comparable findings have been reported in studies of loosely connected soil ecosystems as well as certain interconnected lake systems72,89. Conversely, greater geographical distances can also foster habitat variability, as indicated by the Euclidean distances between environmental factors and geographical separation (Fig. S4), ultimately restricting the likelihood of microbial dispersal across multiple sites90. Consequently, habitat heterogeneity arising from increased geographical distances, coupled with dispersal limitation, may collectively shape the spatial distribution patterns of eukaryotic plankton communities within the DJK and MPR.
Influencing factors of community assembly processes
In our study, the null model supported that the stochastic processes played a relatively more critical role than the deterministic processes (Fig. 7), which might be influenced by the connectivity of water bodies and the frequent hydrodynamic exchange within the DJK and the MRP. On one hand, the enhanced connectivity facilitated the homogenizing dispersal of eukaryotic plankton within the channel, aligning with findings from prior studies on groundwater bacteria91 and testate amoebae in reservoir systems92. The stochastic hydrodynamic disturbances can lead to the random aggregation of planktonic bacteria in hydrologically connected aquatic habitats93. Furthermore, the disordered hydraulic mixing effect within the DJK also contributed to a reduction in the spatial heterogeneity of environmental variables (Fig. S3), which may have attenuated the selective pressure exerted by deterministic processes. In ecosystems characterized by diminished environmental variability, stochastic processes are likely to supersede deterministic mechanisms94. Previous researches have elucidated that the biogeographical distribution of microorganisms is modulated by a complex array of factors. In addition to localized environmental conditions, historical and evolutionary determinants—encompassing geographic distance and dispersal history—also play a crucial role in shaping microbial spatial patterns95,96. Consequently, within interconnected habitats characterized by elevated hydrodynamic exchange, the assembly processes of eukaryotic plankton communities are likely to exhibit a greater propensity toward stochasticity.
Our results indicate that although stochastic processes predominantly govern the spatial distribution of eukaryotic plankton in both the DJK and the MRP, deterministic processes exert a more pronounced influence in the MRP compared to the DJK, accounting for 39.29% and 1.82%, respectively (Fig. 7b). For instance, the dominant influence of stochastic processes in community assembly observed in some lakes with low environmental gradients was similar to our study72. Conversely, in aquatic ecosystems where environmental gradients are more pronounced, deterministic processes have been shown to predominate in shaping community structure75,97. The previous study has identified that the MRP, in its efforts to optimize operational management by regulating flow velocity, discharge, and other hydrodynamic conditions, has been equipped with numerous hydraulic structures along its course, these interventions significantly influence the spatiotemporal distribution of nutrients within the water body, ultimately leading to the formation of diverse aquatic microhabitats53. Additional studies have revealed that more heterogeneous spaces, characterized by a multitude of microhabitat structures, can provide diverse survival conditions for bacteria, thereby enhancing the influence of deterministic processes on bacterial community assembly98. Thereby, we speculated that the increased environmental complexity along the MRP favors the predominance of deterministic processes in shaping the spatial patterns of eukaryotic plankton communities.
In addition, we found that the dominant effect on the eukaryotic plankton community assembly changed from primarily stochastic community assembly to homogeneous selection with increasing variations in TN, Chl.a and Spc (Fig. 8). The high concentration of nitrogen nutrient salts may affect the composition and diversity of bacterial communities99. Moreover, the difference in salinity and nutrient condition played a critical role in governing the composition, connectivity, and assembly process of prokaryotic communities100, the elevated TN and Spc concentrations in aquatic environments facilitate selective pressures that favor specific species, thereby diminishing the influence of stochastic processes on community assembly. The previous study has demonstrated that high concentrations of chlorophyll represent increased primary productivity, leading to a rise in organic carbon resources and intensified microbial competition, which in turn promotes deterministic processes101. In our study, we found that the concentrations of Chl.a in the water of the MRP in Hebei section are higher than those in Henan section, while the concentrations of Spc and TN are lower than those in Henan section (Fig. 5). The significant correlation between βNTI values and variations in TN, Spc and Chl.a further suggests that these are primary factors mediating the balance between stochastic and deterministic assembly processes in the eukaryotic plankton communities(Fig. 8). Collectively, these findings underscore the critical link between environmental variables and community assembly processes, which may, in turn, influence diversity and community stability.
Conclusion
The present study revealed spatial heterogeneity of eukaryotic plankton communities. The similarity in community composition of DJK is significantly higher than that of MRP, exhibiting distance-decay patterns. Environmental heterogeneity exhibits significant differences between DJK and MRP, and it significantly influences community composition and alpha diversity. Additionally, the assembly processes of eukaryotic plankton in both DJK and MRP are predominantly influenced by stochastic processes. However, in comparison to DJK, deterministic processes have a more pronounced impact on MRP. The variations in TN, Chl.a, and Spc have led to a transition in the assembly of eukaryotic phytoplankton communities in MRP from a stochastic process to a deterministic process. These findings provide the basis for further insights into the important role of eukaryotic plankton communities in maintaining ecological function and water quality stability in a long-distance water diversion channel and its water source.
Data availability
All sequencing data are available through the NCBI Sequence Read Archive under the accession number PRJNA1177785.
References
Seymour, J. R., Amin, S. A., Raina, J. B. & Stocker, R. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2 (7), 17065 (2017).
Buysschaert, B. et al. Flow cytometric fingerprinting to assess the microbial community response to changing water quality and additives. Environ. Science: Water Res. Technol. 5 (10), 1672–1682 (2019).
Jiang, T. et al. Microbial diversity characteristics and the influence of environmental factors in a large drinking-water source. Sci. Total Environ. 769 (3), 144698 (2021).
Zhang, L. et al. Untangling microbiota diversity and assembly patterns in the World’s Largest Water Diversion Canal. Water Res. 204, 117617 (2021).
Niu, A. et al. Impact of water quality on the microbial diversity in the surface water along the three Gorge Reservoir (TGR), China. Ecotoxicol. Environ. Saf. 181, 412–418 (2019).
Saingam, P., Li, B. & Yan, T. Fecal indicator bacteria, direct pathogen detection, and microbial community analysis provide different microbiological water quality assessment of a tropical urban marine estuary. Water Res. 185, 116280 (2020).
Bianchi, F. et al. Can plankton communities be considered as bio-indicators of water quality in the lagoon of Venice? Mar. Pollut. Bull. 46 (8), 964–971 (2003).
Mai, S., He, Y., Li, W. & Zhao, T. Effects of environmental factors on vertical distribution of the eukaryotic plankton community in early summer in Danjiangkou Reservoir, China. Front. Ecol. Evol. 11, 1324932 (2023).
Wurzbacher, C. M., Bärlocher, F. & Grossart, H. P. Fungi in lake ecosystems. Aquat. Microb. Ecol. 59 (2), 125–149 (2010).
Nagano, Y. & Nagahama, T. Fungal diversity in deep-sea extreme environments. Rev. High. Press. Sci. Technol. 20 (4), 321–329 (2010).
Sun, Z., Li, G., Cheng, W., Li, G. & Zhou, Q. Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir. Sci. Rep. 4 (1), 6966 (2014).
Piwosz, K. et al. Bacterial and eukaryotic small-subunit amplicon data do not provide a quantitative picture of microbial communities, but they are reliable in the context of ecological interpretations. mSphere 5 (2), e00052–e00020 (2020).
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C. & Martiny, J. B. H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10 (7), 497–506 (2012).
Vellend, M. Conceptual synthesis in Community Ecology. Q. Rev. Biol. 85 (2), 183–206 (2010).
Stegen, J. C. et al. Quantifying community assembly processes and identifying features that impose them. ISME J. 7 (11), 2069–2079 (2013).
Yan, Q. Y. et al. Nearly a decade-long repeatable seasonal diversity patterns of bacterioplankton communities in the eutrophic Lake Donghu (Wuhan, China). Mol. Ecol. 26 (14), 3839–3850 (2017).
Danczak, R. E., Johnston, M. D., Kenah, C., Slattery, M. & Wilkins, M. J. Microbial community cohesion mediates community turnover in unperturbed aquifers. mSystems 3 (4), e00066–e00018 (2019).
Logares, R. et al. Contrasting prevalence of selection and drift in the community structuring of bacteria and microbial eukaryotes. Environ. Microbiol. 20 (6), 2231–2240 (2018).
Isabwe, A. et al. Community Assembly mechanisms underlying the core and Random Bacterioplankton and Microeukaryotes in a river–Reservoir System. Water 11 (6), 1127 (2019).
Liu, K., Liu, Y., Wang, J., Yang, H. & Zhu, Y. Different community assembly mechanisms underlie similar biogeography of bacteria and microeukaryotes in tibetan lakes. FEMS Microbiol. Ecol. 96 (6), 1–12 (2020).
Wu, W., Yang, J., Li, H., Duan, X. & Liu, Y. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 11, 1377–1388 (2017).
Kong, J. et al. Patterns of relative and quantitative abundances of marine bacteria in surface waters of the subtropical Northwest Pacific Ocean estimated with high-throughput quantification sequencing. Front. Microbiol. 11, 599614 (2021).
Kong, J. et al. Diversity distribution and assembly mechanisms of planktonic and benthic microeukaryote communities in intertidal zones of Southeast Fujian, China. Front. Microbiol. 10, 2640 (2019).
Li, S. et al. Antibiotics in water and sediments of Danjiangkou Reservoir, China: spatiotemporal distribution and indicator screening. Environ. Pollut. 246, 435–442 (2019).
Han, B., Chen, S., Zhang, L. & Wang, Y. Assessment and management of pressure on water quality protection along the Middle Route of the South-to-North Water Diversion Project. Sustainability 11 (13), 3665 (2019).
Long, D. et al. South-to-north water diversion stabilizing Beijing’s groundwater levels. Nat. Commun. 11 (1), 3665–3675 (2020).
Wang, Y. et al. Climatic changes and anthropogenic activities driving the increase in nitrogen: evidence from the South-to-North water diversion project. Water 13 (18), 2517–2527 (2021).
Editorial Board of Monitoring and Analysis Methods of Water and Wastewater, Ministry of Environmental Protection of the People’s Republic of China. Monitoring and Analysis Methods of Water and Wastewater: Fourth Edition. Beijing: China Environmental Science press (2002).
Stoeck, T. et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19 (S1), 21–31 (2010).
Piredda, R. et al. Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean LTER site. FEMS Microbiol. Ecol. 93 (1), 1–13 (2017).
Vierna, J., Doña, J., Vizcaíno, A., Serrano, D. & Jovani, R. PCR cycles above routine numbers do not compromise high-throughput DNA barcoding results. Genome 60 (9), 868–873 (2017).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), 884–890 (2018).
Magoc, T., Salzberg, S. L. & FLASH Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963 (2011).
Zhang, C. et al. Diversity and assembly processes of microbial eukaryotic communities in Fildes Peninsula Lakes (West Antarctica). Biogeosciences 19 (18), 4639–4654 (2022).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 10 (10), 996–998 (2013).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 (16), 5261–5267 (2007).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (1), 590–596 (2013).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 (8), 1621–1624 (2012).
Clarke, K. R. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 18 (1), 117–143 (1993).
Teittinen, A. et al. Variation in stream diatom communities in relation to water quality and catchment variables in a boreal, urbanized region. Sci. Total Environ. 530–531, 279–289 (2015).
Murtagh, F. & Legendre, P. Ward’s hierarchical agglomerative clustering method: which algorithms implement ward’s criterion? J. Classif. 31 (3), 274–295 (2014).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), R60 (2011).
Quinn, G. P. & Keough, M. J. Experimental Design and Data Analysis for Biologists (Cambridge University Press, 2022).
Jolliffe, I. T. & Cadima, J. Principal component analysis: a review and recent developments. Philosophical Trans. Royal Soc. A: Math. Phys. Eng. Sci. 374 (2065), 20150202 (2016).
Blanchet, F. G., Legendre, P. & Borcard, D. Forward selection of explanatory variables. Ecology 89 (9), 2623–2632 (2008).
Habagil, M., Martiny, J. B. H. & Martiny, A. C. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 4 (7), 1183–1195 (2019).
Tai, X. et al. Pollution gradients altered the bacterial community composition and stochastic process of rural polluted ponds. Microorganisms 8 (2), 311 (2020).
Hijmans, R. J. geosphere: spherical trigonometry. R package version 1.5–10 (2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis 2nd edn (Springer, 2016).
Stegen, J. C., Lin, X., Konopka, A. E. & Fredrickson, J. K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6 (9), 1653–1664 (2012).
Stegen, J. C. et al. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6, 370 (2015).
Chen, H. et al. Spatiotemporal variation of water quality in the main canal of Middle Route of South-to North Water Diversion Project based on principal component and cluster analysis. J. Yangtze River Sci. Res. Inst. 39 (07), 36–44 (2022).
Zhang, C. M. et al. Spatiotemporal pattern of phytoplankton community structure and its determining factors in the channel of the middle route of South-to-North Water Diversion Project. J. Lake Sci. 33 (03), 675–686 (2021).
Fang, R. Y. A survey and development of methods for determination of permanganate index in surface water. China Resour. Compr. Utilization. 36 (05), 76–78 (2018).
Fan, A. X. et al. The characteristics and cause analysis of oxygen consumption substances for the waterbody in the main channel of the Middle Route of South-to North Water Diversion Project. Acta Sci. Circum. 40 (03), 871–879 (2020).
Wu, L. D., Zhou, J. Z., Deng, Y. & He, Z. L. PCR-DGGE Fingerprinting Analysis of Plankton Communities and its relationship to Lake Trophic Status. Int. Rev. Hydrobiol. 94 (5), 528–541 (2010).
Zheng, B. H. et al. Structural Characteristics and Driving Factors of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir, China. Water 12(12), 3499–3514 (2020).
Zhang, C., Li, X. J., Deng, Y., Zhou, J. Z. & Wu, L. D. Assembly processes of eukaryotic Plankton communities in the World’s largest drinking Water Diversion Project. Sci. Total Environ. 884, 163665 (2023).
Held, A. A. Rozella and Rozellopsis: Naked Endoparasitic Fungi which Dress-up as their hosts. Mycologia 73 (4), 451–515 (1981).
Zhang, M. et al. Feedback regulation between aquatic microorganisms and the bloom-forming cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 85 (21), 01362–01319 (2019).
Yan, M. et al. Community compositions of phytoplankton and eukaryotes during the mixing periods of a drinking water reservoir: Dynamics and interactions. Int. J. Environ. Res. Public Health. 17 (4), 1128 (2020).
Reynolds, C. S., Huszar, V., Kruk, C., Naselli-Flores, L. & Melo, S. Towards a functional classification of the freshwater phytoplankton. J. Plankton Res. 24 (5), 417–428 (2002).
Zhou, G. J., Kuang, Q. J., Hu, Z. Y. & Cai, Q. H. Assessment of algal diversity and water blooms prevention in four tributaries of Three Gorges Reservoir, China. China Environ. Sci. 03, 337–341 (2006).
Li, N. X. et al. Effect of local watershed landscapes on the nitrogen and phosphorus concentrations in the waterbodies of reservoir bays. Sci. Total Environ. 716, 137132 (2020).
Dang, C. Y. et al. Rare biosphere regulates the planktonic and sedimentary bacteria by disparate ecological processes in a large source water reservoir. Water Res. 216, 118296 (2022).
Li, F. P., Gao, Y., Zhang, H. P., Xiao, Y. H. & Chen, L. Simulation experiment on the effect of flow velocity on phytoplankton growth and composition. J. Lake Sci. 27 (01), 44–49 (2015).
Morgan-Kiss, R. M., Lizotte, M. P., Kong, W. & Priscu, J. C. Photoadaptation to the polar night by phytoplankton in a permanently ice-covered Antarctic lake. Limnol. Oceanogr. 61 (1), 3–13 (2016).
Izaguirre, I., Allende, L. & Marinone, M. C. Comparative study of the planktonic communities of three lakes of contrasting trophic status at Hope Bay (Antarctic Peninsula). Exp. Neurol. 25 (9), 1079–1097 (2003).
Gleason, F. H. et al. Ecological potentials of species of Rozella (Cryptomycota). Fungal Ecol. 5 (6), 651–656 (2012).
Carrara, F., Altermatt, F., Rodriguez-Iturbe, I. & Rinaldo, A. Dendritic connectivity controls biodiversity patterns in experimental metacommunities. Proc. Natl. Acad. Sci. 109 (15), 5761–5766 (2012).
Yuan, Y. X. et al. Environmental variables influencing phytoplankton communities in hydrologically connected aquatic habitats in the Lake Xingkai basin. Ecol. Ind. 91, 307–314 (2018).
Bai, C. R. et al. Geographic patterns of bacterioplankton among lakes of the middle and lower reaches of the Yangtze River Basin, China. Appl. Environ. Microbiol. 86 (6), e02423–e02419 (2020).
Bai, C. R. et al. Spatial patterns and community assembly mechanisms of bacterioplankton in large shallow lakes: a case study of Lake Taihu. J. Lake Sci. 35 (4), 1433–1442 (2023).
Liu, L., Yang, J., Yu, Z. & Wilkinson, D. M. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9 (9), 2068–2077 (2015).
Li, H. et al. Contrasting patterns of diversity of abundant and rare bacterioplankton in freshwater lakes along an elevation gradient. Limnol. Oceanogr. 62 (4), 1570–1585 (2017).
Liao, J. et al. Similar community assembly mechanisms underlie similar biogeography of rare and abundant bacteria in lakes on Yungui Plateau, China. Limnol. Oceanogr. 62 (2), 723–735 (2017).
Liu, Z., Zhang, Y., Yu, Z. & Yang, J. Spatial and Seasonal Bacterioplankton Community Dynamics in the Main Channel of the Middle Route of South-to-North Water Diversion Project. Res. Microbiol. 169 (2), 101–109 (2018).
Kunlasak, K., Powtongsook, S. & Limsuwan, C. Relationships of dissolved oxygen with Chlorophyll-a and Phytoplankton Composition in Tilapia Ponds. Int. J. Geosci. 4 (5), 46–53 (2013).
Yan, X. et al. Hydrologic and physicochemical factors co-drive seasonal changes of phytoplankton during dynamic water diversion processes in the Danjiangkou Reservoir. J. Lake Sci. 33 (5), 1350–1363 (2021).
Fang, L. J., Liu, D. F., Yang, Z. J. & Tian, Z. B. Effects of water temperature on the phytoplankton community structure. Environ. Sci. Technol. 37 (S2), 45–50 (2014).
Da, W. Y. et al. Long-term variation of phytoplankton community and driving factors in Qiandaohu Reservoir, Southeast China. J. Lake Sci. 31 (05), 1320–1333 (2019).
Ren, H. P. et al. Spatial distribution and variation of suspended solids in the Main Channel of the Middle Route of the South-to North Water Diversion Project and relationships with phytoplankton community. Resour. Environ. Yangtze Basin. 31 (11), 2473–2480 (2022).
Wang, C. et al. Annual variation pattern of phytoplankton community at the downstream of Xijiang River. Acta Ecol. Sin. 33 (14), 4398–4408 (2013).
Cudowski, A., Pietryczuk, A. & Hauschild, T. Aquatic fungi in relation to the physical and chemical parameters of water quality in the Augustów Canal. Fungal Ecol. 13, 193–204 (2015).
Reich, M. et al. Impacts of a reduction in seawater pH mimicking ocean acidification on the structure and diversity of mycoplankton communities. Aquat. Microb. Ecol. 79 (3), 221–233 (2017).
Wang, Y., Bai, Y., Liao, K., Zhao, C. & Qu, J. Impact of environmental gradients on the abundance and diversity of planktonic fungi across coastal habitats of contrasting trophic status. Sci. Total Environ. 683, 822–833 (2019).
Bahram, M., Harend, H. & Tedersoo, L. Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME J. 10 (5), 1259–1269 (2016).
Li, C. et al. The ecology of the plastisphere: microbial composition, function, assembly, and network in the freshwater and seawater ecosystems. Water Res. 202, 117428 (2021).
Zhang, W., Zhang, X., Chen, G., Wang, X. & Li, Y. Nitrogen rather than phosphorus driving the biogeographic patterns of abundant bacterial taxa in a eutrophic plateau lake. Sci. Total Environ. 806, 150947 (2022).
Clark, D. R., Underwood, G. J. C., McGenity, T. J. & Dumbrell, A. J. What drives study-dependent differences in distance-decay relationships of microbial communities? Glob. Ecol. Biogeogr. 30 (4), 811–825 (2021).
Beaton, E. D. et al. Local and regional diversity reveals dispersal limitation and drift as drivers for groundwater bacterial communities from a fractured granite formation. Frontiers in Microbiology 7, (2016). (1933).
Wang, W. et al. Seven-year dynamics of testate amoeba communities driven more by stochastic than deterministic processes in two subtropical reservoirs. Water Res. 185, 116221 (2020).
Mo, Y. et al. Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J. 12 (9), 2198–2210 (2018).
Wang, J. et al. Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J. 7 (7), 1310–1321 (2013).
Zhou, J. & Ning, D. Stochastic Community Assembly: does it Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 81 (4), 2–17 (2017).
Martiny, J. B. H. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4 (2), 102–112 (2006).
Logares, R. et al. Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J. 7 (5), 937–948 (2013).
Jiao, C. et al. Disentangling the seasonal co-occurrence patterns and ecological stochasticity of planktonic and benthic bacterial communities within multiple lakes. Sci. Total Environ. 740, 140010 (2020).
Chen, Z., Wang, H., Xia, S. & Zhang, P. Illumina MiSeq sequencing and network analysis the distribution and co-occurrence of bacterioplankton in Danjiangkou Reservoir, China. Arch. Microbiol. 202 (4), 859–873 (2020).
Zhou, N., Liu, H., Zhao, X. & Zhang, H. Salinity and nutrient condition as key factors drive the assembly of sediment prokaryotic communities. Int. Biodeterior. Biodegrad. 193, 105466 (2024).
Maria, S., Anderson, R., Dupont, C. L. & Ellis, E. E. Interplay between stochastic and deterministic processes in the maintenance of alternative community states in Verrucomicrobia-dominated shallow lakes. FEMS Microbiol. Ecol. 93 (7), 1–11 (2017).
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2022YFC3203905), Key Research and Development Projects of Hubei Province (2024BCB064), Finance Project of the Ministry of Water Resources (102126222020060009026) and Wuhan Knowledge Innovation Special Basic Research Project (2022020801010171; 2023020201020299).
Author information
Authors and Affiliations
Contributions
Liming Zhu: Conceptualization, Methodology, Writing-original draft. Lihui Feng; Daoxi Zhang and Qing Yang: Environmental factors analysis. Xi Zou: Supervision. Shan He and Wen Zhu: Sample collection. Fang Shi: Conceptualization, Writing-review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, L., Feng, L., Zhang, D. et al. Eukaryotic plankton community and assembly processes in a large-scale water diversion project in China. Sci Rep 15, 4365 (2025). https://doi.org/10.1038/s41598-025-87983-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87983-9