Abstract
This study investigates the association between mRNA levels of genes involved in 7-methylguanosine (m7G) metabolism and the prognosis of herpes zoster. By analyzing the transcriptional profiles of m7G-related genes in herpes zoster from the GSE242252 dataset, it was found that NSUN2, AGO2, and SNUPN were differentially expressed between herpes zoster and normal controls (p < 0.05). AGO2 and SNUPN were negatively correlated with multiple immune cell infiltrations, while NSUN2 was positively correlated with immature B cell infiltration. A nomogram model based on NSUN2, AGO2, and SNUPN was constructed and showed good predictive ability, validated through clinical impact curve analysis (CICA), calibration curve, and decision curve analysis (DCA). The results suggest that a nomogram based on NSUN2, AGO2, and SNUPN can predict the risk of herpes zoster, and the relationship between these genes and immune infiltration may influence the prognosis of herpes zoster.
Trial registration: This study was approved by the Institutional Review Board of the Fujian Medical University Union Hospital (No 2023QH003).
Similar content being viewed by others
Introduction
Varicella-zoster virus (VZV) reactivates in the body and causes herpes zoster (HZ), which is more common in the elderly. The clinical manifestations of HZ are diverse. An adverse event, known as HZ, has been reported in some people following COVID-19 vaccination1. As the number of people vaccinated against COVID-19 has increased during the pandemic, so has the incidence of HZ2. At the same time, with the aging of the population, it may lead to increased incidence. Immunosuppression and the use of specific drugs, including methotrexate, thiopurine, or corticosteroids, may induce VZV reactivation3. Therefore, HZ is also getting attention. In China, the incidence of HZ for all age groups is 4.28/1000 person-years, of which the incidence of HZ in people over 60 years old is 11.69 per 1,000 person-years4. This is similar to the 12.1 per 1,000 person-years reported for people over 70 years in the United States5. The manifestations of VZV infection vary from individual to individual, and the susceptibility and pathogenesis are not fully understood.
As a kind of RNA modification, N7-methylguanosine (m7G) is widely distributed in various biological RNAs and even in viral transcripts6. The m7G modification has been shown to play a significant role in the development of many human diseases by participating in various RNA metabolic processes and has attracted great attention in recent years7. Abnormally expressed levels of m7G-related genes have been found in many tumors and viral diseases8,9,10. Cluster analysis based on m7G-related genes found that COVID-19 patients with higher immune infiltration also had less symptoms8. Cell-mediated immunity (CMI) decline is thought to precede HZ onset. The CMI was negatively correlated with both the incidence and severity of HZ11. The immune function of HZ patients based on m7G modification level is also worth exploring.
Many studies have demonstrated the relationship between m7G modification and the pathogenesis of many viral infections. However, the specific mechanism of m7G modification in herpes zoster disease has not been clarified. In this study, we evaluated the m7G-related genes in patients with HZ. In addition, individuals were classified into disease subtypes by the consensus clustering method. Moreover, a nomogram model which based on three m7G genes was established for herpes zoster risk prediction.
Materials and methods
The RNA-seq of patients with herpes zoster and m7G-related genes
The GSE242252 dataset containing transcript profiles of herpes zoster patients was downloaded from GEO. The transcriptomic data presented in this study were derived from peripheral blood samples collected from patients diagnosed with herpes zoster. The blood samples were processed to isolate total RNA, which was then used to generate transcriptomic profiles. The transcriptome data were normalized before further analysis. The GSE242252 dataset included 49 herpes zoster and 31 healthy controls. According to literature searches and the GSEA database (http://www.gsea-msigdb.org/gsea/login.jsp), 24 m7G-related genes have been identified. All analyses were statistically analyzed by R software (version 4.3.1).
Classification of herpes zoster subtypes
This study classified herpes zoster patients’ subtypes based on different methods including m7G clustering, gene clustering, and m7G score clustering. In m7G clustering and gene clustering, we used R package “ConsensusClusterPlus” for grouping herpes zoster patients. The m7G clustering is to classify herpes zoster patients into two groups by the expression level of m7G-related genes. Then the gene clustering is according to the expression levels of differential expression genes (DEGs) detected between m7G-clusters. Principal component analysis (PCA) algorithm was used in the m7G score clustering. We also visualized the grouping of individual patients across three disease subtypes.
Functional enrichment analysis and immune cell infiltration analysis
The DEGs between herpes zoster patients with the m7G disease subtype were analyzed for KEGG12 and GO analysis. Infiltration analysis of many types of immune cells in patients with herpes zoster was performed using the ssGSEA method8.
The nomogram model
By comparing the residuals, AUC values and reverse cumulative distribution of residuals of different machine learning methods, disease characteristic genes of herpes zoster were obtained after determining better machine learning methods. Then, a nomogram model for risk prediction of herpes zoster were constructed using disease characteristic genes. The validity of this model was also further verified using clinical impact curve analysis (CICA), calibration curve, and decision curve analysis (DCA).
qRT-PCR
We utilized peripheral blood from patients with herpes zoster to extract total RNA, which was then used to detect the relative expression levels of the relevant genes. The qRT–PCR was measured with the ABI 7900HT system with SYBR Select Master Mix (4472908, Thermo Fisher). The sequences of the primers are as follows: AGO forward, 5’- CGTCCTCTGGGACGACAATC-3’ and reverse, 5’- ATGGCTTCCTTCAGCACTGT − 3’; SNUPN forward, 5’- ACGTTC TGGATGTGATGTGCT-3’ and reverse, 5’- CATCACACAGGCTTTCGGGA-3’. NSUN2 forward, 5’-CCTGAAGATGACCCATTATTTCCA-3’ and reverse, 5’-GCACATTCCGCAACTCCTTA-3’. This study was performed according to the relevant medical ethics regulations and approved by the Human Research Ethics Committee of Fujian Medical University Union Hospital (Fuzhou, China). This study was carried out in accordance with the Helsinki Declaration. All participants gave written informed consent prior to collection of the specimens.
Results
Expression of m7G genes in herpes zoster patients
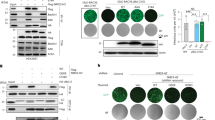
In the present study, 49 herpes zosters and 31 healthy controls were enrolled. Firstly, we identified the differential genes between the herpes zoster group and the healthy control group. The heat map in Fig. 1A shows the differential genes in each sample. Then we analyzed the m7G genes expression between herpes zoster and healthy controls. The level of NSUN2 was significantly up-regulated in herpes zoster. However, the expression of AGO2 and SNUPN were significantly down-regulated in herpes zoster (p < 0.05) (Fig. 1B). we generated heat maps of the expression of these genes in herpes zoster and healthy controls (Fig. 1C). The qRT-PCR analysis was conducted to compare the gene expression profiles of three genes (AGO2, SNUPN, and NSUN2) across 15 patients with herpes zoster and 15 healthy controls. As depicted in Fig. 1D, the relative expression levels of AGO2 and SNUPN were significantly downregulated in the herpes zoster patient group compared to the healthy controls. Conversely, the expression of NSUN2 was found to be upregulated in the patient cohort.
The expression and clustering of m7G genes. (A) Heat map of DEGs between herpes zoster and healthy controls. (B) The expression of m7G-related genes in herpes zoster and healthy controls. (C) Heat map of m7G genes between herpes zoster and healthy controls. (D) Using qRT-PCR to validate the relative expression levels of AGO2, SNUPN, and NSUN2. (E) The best consensus matrix. (F) AGO2 was differentially expressed in m7G cluster of herpes zoster patients. (G) Heat map of the expression of AGO2, NSUN2, and SNUPN. (H) The m7G cluster classification results are good.
The PAM algorithm and the lever of NSUN2, AGO2, and SNUPN were performed to obtain the best consensus matrix. Then patients were best divided into 2 groups (Fig. 1E). So, we classify 49 herpes zoster patients into two groups, m7G-cluster A and B. The cluster B was characterized by a lower expression level of AGO2 (Fig. 1F). Then a specific transcription profile was obtained according to AGO2, NSUN2, and SNUPN between m7G clusters (Fig. 1G). The divide between m7G clusters was also clear (Fig. 1H).
m7G clustering and immune infiltration analysis
Due to the increasing number of herpes zoster patients, a deeper understanding of the immunological mechanisms involved in herpes zoster has become increasingly important. The results showed that the infiltration of activated CD4 + T cell and activated CD8 + T cell in m7G-cluster B was significantly higher than that in m7G-cluster A (Fig. 2A). We also further analyzed the correlation between NSUN2, AGO2, and SNUPN genes and immune infiltrating cells (Fig. 2B). The expression of AGO2 was negatively correlated with activated CD4 T cell and CD8 T cell. NSUN2 was positively correlated with immature B cell.
Analysis of m7G patterns for immune infiltration and functional enrichment. (A) The expression of activated CD4 + T cell and CD8 + T cell were higher in m7G cluster B than in cluster A. (B) The correlation between immune cells and NSUN2, AGO2, or SNUPN. The GO (C) and KEGG (D) analysis of DEGs which were obtained from m7G clusters A and B.
To further investigate the differences between the two subtypes, we counted the number of differentially expressed genes (DEGs). The total number of DEGs is 301. After that, the 301 genes were enriched by GO (Supplementary Table 1) and KEGG (Supplementary Table 2) analyses. GO analysis results are shown in Fig. 2C. Biological Process is mainly enriched in oxidative phosphorylation (GO:0006119), mitochondrial ATP synthesis coupled electron transport (GO:0042775), ATP synthesis coupled electron transport (GO:0042773), respiratory electron transport chain (GO:0022904), mitochondrial respiratory chain complex assembly (GO:0033108), and aerobic electron transport chain (GO:0019646). Cellular Component is mainly concentrated in the mitochondrial inner membrane (GO:0005743) or mitochondrial protein-containing complex (GO:0098798). Molecular Function is mainly concentrated in the structural constituent of ribosome (GO:0003735). KEGG results indicated that enrichment was concentrated in neurological diseases and viral infection-related diseases. It mainly included Pathways of neurodegeneration - multiple diseases, Amyotrophic lateral sclerosis, Alzheimer disease, Huntington’s disease, Prion disease, Coronavirus disease, and Viral life cycle - HIV-1(Fig. 2D).
Gene clustering and m7G score clustering
The patients of herpes zoster were re-divided into 2 groups according to the expression of 301 DEGs (Fig. 3A). The 301 DEGs will be elaborated upon in Supplementary Table 3. We refer to these two groups of patients as gene cluster A and gene cluster A, respectively. Heat maps show all gene expression characteristics of patients in both groups (Fig. 3B). The expression of AGO2 was significantly higher in gene cluster A than in B (Fig. 3C). The infiltration abundance of activated B cells, activated CD4 + T cell, and activated CD8 + T cell in gene cluster A was significantly lower than in cluster B (Fig. 3D). These results are consistent with previous findings that AGO2 was negatively correlated with activated CD4 + T cell, and activated CD8 + T cell.
(A) Patients were categorized into two gene clusters (A and B) based on 301 DEGs expression. (B) Heat map of gene expression from herpes zoster patients in gene clusters A and B. (C) AGO2 expression is higher in cluster A than in cluster B. (D) Lower infiltration of activated B cells, CD4 + T cells, and CD8 + T cells in cluster A versus B. The m7G-score distribution of patients showed that gene cluster B or m7G cluster B had lower scores than cluster A (E, F). (G) Patients divided into high and low m7G-score groups show mostly consistent positioning in subtypes.
The concept of m7G-score has also been developed, based on a comprehensive assessment of the expression of m7G genes in herpes zoster patients. Then the m7G-score was assigned to each patient based on the analysis. Compared to gene/m7G cluster A, gene/m7G cluster B had a lower m7G score (Fig. 3E and F).
According to the m7G score of each herpes zoster patient, we divided them into high and low m7G score groups in turn. In addition, we compared the positioning of everyone in three subtypes. The result shows that the group coincidence is well (Fig. 3G).
Establish a prediction model for herpes zoster
Subsequently, we used machine learning methods to screen out disease characteristic genes associated with herpes zoster. First, we applied two machine models, random forest (RF) model, and support vector machine (SVM) model, for comparison. By comparing residual, reverse cumulative distribution of residual and AUC values, we finally selected the RF model for screening herpes zoster-associated genes (Fig. 4A-C). The error of the RF model is minimal when the option Trees are 21(Fig. 4D).
According to the RF model, we selected genes with importance scores greater than 10, including NSUN2, SNUPN, and AGO2 (Fig. 5A). Next, we used these three genes to construct a nomogram for risk prediction of herpes zoster (Fig. 5B). The nomogram presented here is a graphical tool designed to estimate the risk of developing herpes zoster based on the expression levels of three key genes: AGO2, NSUN2, and SNUPN. In interpreting the nomogram for herpes zoster risk, the first step is to locate the patient’s AGO2 expression score on the AGO2 axis and note the corresponding points. Next, repeat the same process for NSUN2 and SNUPN on their respective axes, recording the points for each gene’s expression. Then, sum the points obtained from all three genes to calculate the total score. Finally, find the total score on the Risk scale to determine the estimated risk of herpes zoster for the patient. The validity of the model is also verified. Firstly, the model is well-calibrated (Fig. 5C). Second, the model has a better net benefit in DCA (Fig. 5D). The predicted high-risk group of herpes zoster had a good coincidence with the actual high-risk group (Fig. 5E).
Establish and validate the nomogram model. (A) Key genes identified by RF model. Genes with importance scores > 10, such as NSUN2, SNUPN, and AGO2, are highlighted. (B) Nomogram for herpes zoster risk prediction using the selected genes. (C) Model calibration curve, confirming the model’s predictive accuracy. (D) DCA showing the model’s net benefit in risk assessment. (E) Correlation between predicted and actual high-risk groups, indicating good agreement.
Discussion
Both primary chickenpox and reactivated HZ occur in susceptible individuals. In immunocompromised individuals, pregnant women, and healthy adults, it can cause serious illness and even be fatal. VZV enters the innervated dermatome from the axon when reactivated, leading to HZ. By identifying the genetic basis for differences in disease susceptibility among individuals, it is helpful to understand how gene-environment interactions affect infection outcomes.
We found that three genes were differentially expressed in patients with HZ, namely NSUN2, AGO2, and SNUPN. Based on m7G clusters, it can distinguish disease subgroups well. SNUPN and AGO2 were significantly downregulated in this study. At present, many studies have found that AGO2 is closely related to viral replication and translation. Viruses such as Hepatitis C Virus and Seneca Virus A are affected by AGO2 during replication and translation13,14. Enterovirus 71 replication and translation are stimulated by AGO2, which binds to its internal ribosome entry site15. AGO2 not only colocates with HBcAg and HBsAg, but also affects the production of HBV DNA and HBsAg16. An antiviral RNA interference pathway mediated by RDE-1/AGO2 is associated with endoplasmic reticulum homeostasis17. Whether AGO2 is also related to the replication of VZV is worth further investigation. In previous studies, CD4 + T cells decreased, CD8 + T cells increased, Th1/Th2 ratio decreased and Th17/Treg ratios increased in herpes zoster18. However, in this study, the AGO2 gene was also found to be negatively correlated with many immune genes, including CD8 + T cells and CD4 + T cells. The changing trend of T cells is affected by many factors, so the specific mechanism needs to be verified experimentally. Furthermore, T cells lacking AGO2 tend to induce more lineage-specific cytokines, induce an increase in IFN-γ, and show no particular predisposition to Th1 and Th2 differentiation19.
A majority of the enrichment was found in viral infection-related and neurological diseases, according to the KEGG results. Although the specific signaling mechanism is not directly stated, this suggests that m7G may have a significant role in VZV infection.
Previous studies have established some nomogram models associated with HZ, but most have been based on clinical indicators (age, VAS, site of involvement, etc.), in addition to focusing on predicting limb weakness or motor dysfunction after HZ20,21. However, no predictive models based on m7G-related genes have been found to predict the risk of HZ. The nomogram model constructed by the three m7G genes was also a good predictor of disease risk. The model has been verified by many parties and the prediction ability of the model is satisfactory. Unfortunately, there is no way to predict a specific disease phenotype. The manifestations of VZV infection vary from individual to individual, and the susceptibility and pathogenesis are not fully understood. It is worth noting that postherpetic neuralgia is the most common sequelae of HZ, and the incidence of this pain lasting more than 1 month is about 40.2%, which seriously affects the quality of life of patients22.
There are still some shortcomings in this study. Firstly, transcriptome-based data is a good way to find host genetic characteristics, but lack of clinical data, it is impossible to correlate transcriptome-based analysis with clinical characteristics. We hope to explore the correlation between the expression levels of m7G-related genes and various clinical parameters, such as disease severity, duration of symptoms, and frequency of outbreaks. Concurrently, the current study has identified certain m7G-related genes that show significant differential expression between herpes zoster patients and healthy controls. We aim to determine whether these genes can serve as biomarkers for disease progression or response to treatment. The recommendations for future research that would translate these findings into clinical practice include, of course, the need for larger patient cohorts and longitudinal studies to validate the predictive and prognostic value of the identified genes. Secondly, it is suggested to expand the sample size and multi-center clinical samples to continue to verify our conclusions. Finally, the mechanism of m7G modification in the pathogenesis of HZ needs to be further verified in vivo and in vitro trials.
In conclusion, disease risk models based on NSUN2, AGO2, and SNUPN can predict the onset of HZ. m7G-related genes can be used to classify HZ patients into disease subgroups. This will help further explore the relationship between m7G and other phenotypes of HZ. Moreover, these three m7G-related genes are closely related to immune infiltration, which may affect the prognosis of herpes zoster.
Data availability
All the data in this study can be obtained in the GSE242252 dataset at https://ftp.ncbi.nlm.nih.gov/geo/series/GSE242nnn/GSE242252/matrix/.
References
Shahrudin, M. S. & Mohamed-Yassin, M. S. Nik Mohd Nasir, N. M. herpes zoster following COVID-19 vaccine booster. Am. J. case Rep. 24, e938667. https://doi.org/10.12659/ajcr.938667 (2023).
Patil, A., Goldust, M. & Wollina, U. Herpes zoster: A review of clinical manifestations and management. Viruses 14, (2022). https://doi.org/10.3390/v14020192
Singer, D. et al. Incidence and risk of herpes zoster in patients with ulcerative colitis and Crohn’s disease in the USA. Gastroenterol. Rep. 11, goad016, (2023). https://doi.org/10.1093/gastro/goad016
Zhang, Z. et al. The incidence of herpes zoster in China: a meta-analysis and evidence quality assessment. Hum. Vaccines Immunother.. 19, 2228169. https://doi.org/10.1080/21645515.2023.2228169 (2023).
Singer, D. et al. Incidence and risk of herpes zoster in patients with ulcerative colitis and Crohn’s disease in the USA. Gastroenterol. Rep. 11 https://doi.org/10.1093/gastro/goad016 (2023).
Ramanathan, A., Robb, G. B. & Chan, S. H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 44, 7511–7526. (2016).
Pandolfini, L. et al. METTL1 promotes let-7 MicroRNA Processing via m7G methylation. Mol. Cell. 74, 1278–1290e1279. https://doi.org/10.1016/j.molcel.2019.03.040 (2019).
Lu, L. et al. The m7G modification level and Immune infiltration characteristics in patients with COVID-19. J. Multidiscipl. Healthc. 15, 2461–2472. https://doi.org/10.2147/jmdh.S385050 (2022).
Cruz, A. & Joseph, S. Interaction of the Influenza A Virus NS1 protein with the 5’-m7G-mRNA·eIF4E·eIF4G1 complex. Biochemistry 61, 1485–1494. https://doi.org/10.1021/acs.biochem.2c00019 (2022).
Wei, W. et al. Comprehensive pan-cancer analysis of N7-methylguanosine regulators: Expression features and potential implications in prognosis and immunotherapy. Front. Genet. 13, 1016797. https://doi.org/10.3389/fgene.2022.1016797 (2022).
Asada, H. VZV-specific cell-mediated immunity, but not humoral immunity, correlates inversely with the incidence of herpes zoster and the severity of skin symptoms and zoster-associated pain: the SHEZ study. Vaccine 37, 6776–6781. https://doi.org/10.1016/j.vaccine.2019.09.031 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d592. https://doi.org/10.1093/nar/gkac963 (2023).
Wu, X. et al. CRISPR/Cas9-Mediated knockout of the Dicer and Ago2 genes in BHK-21 cell promoted Seneca Virus A replication and enhanced autophagy. Front. Cell. Infect. Microbiol. 12, 865744. https://doi.org/10.3389/fcimb.2022.865744 (2022).
Wilson, J. A., Zhang, C., Huys, A. & Richardson, C. D. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J. Virol. 85, 2342–2350. https://doi.org/10.1128/jvi.02046-10 (2011).
Lin, J. Y., Brewer, G. & Li, M. L. HuR and Ago2 bind the internal ribosome entry site of enterovirus 71 and promote virus translation and replication. PloS One. 10, e0140291. https://doi.org/10.1371/journal.pone.0140291 (2015).
Hayes, C. N. et al. Hepatitis B virus-specific miRNAs and Argonaute2 play a role in the viral life cycle. PloS One. 7, e47490. https://doi.org/10.1371/journal.pone.0047490 (2012).
Efstathiou, S. et al. ER-associated RNA silencing promotes ER quality control. Nat. Cell Biol. 24, 1714–1725. https://doi.org/10.1038/s41556-022-01025-4 (2022).
Chen, W., Zhu, L., Shen, L. L., Si, S. Y. & Liu, J. L. T lymphocyte subsets Profile and Toll-Like receptors responses in patients with herpes zoster. J. pain Res. 16, 1581–1594. https://doi.org/10.2147/jpr.S405157 (2023).
Bronevetsky, Y. et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 210, 417–432. https://doi.org/10.1084/jem.20111717 (2013).
Li, S. J. & Feng, D. Risk factors and nomogram-based prediction of the risk of limb weakness in herpes zoster. Front. NeuroSci. 17 https://doi.org/10.3389/fnins.2023.1109927 (2023).
Tang, J., Tao, J., Luo, G., Zhu, J. & Yao, M. Analysis of risk factors and construction of a prediction model of motor dysfunction caused by limb herpes zoster. J. pain Res. 15, 367–375. https://doi.org/10.2147/jpr.S346564 (2022).
Zhu, Q. et al. Epidemiology of herpes zoster among adults aged 50 and above in Guangdong, China. Hum. Vaccines Immunother.. 11, 2113–2118. https://doi.org/10.1080/21645515.2015.1016672 (2015).
Acknowledgements
Not applicable.
Funding
This study was funded by Joint Funds for the innovation of science and Technology, Fujian province (Grant Number: 2023Y9160) and Startup Fund for scientific research, Fujian Medical University (Grant Number: 2023QH1025).
Author information
Authors and Affiliations
Contributions
Study design and concept (All), acquisition of data (Lingling Lu and Fangze Cai), data interpretation and analysis (Lingling Lu and Fangze Cai), drafting of the manuscript (Lingling Lu and Fangze Cai), critical revision of the manuscript (All). All authors contributed to the study supervision, read, and approved the final version of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This study was approved by the Ethics Committee of the Fujian Medical University Union Hospital and carried out in accordance with the Helsinki Declaration. All the data are obtained from the GEO databases so that informed consent can be guaranteed.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lu, L., Cai, F. & Luo, Y. m7G gene expression and disease risk model construction in patients with herpes zoster. Sci Rep 15, 4881 (2025). https://doi.org/10.1038/s41598-025-88664-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88664-3