Abstract
Lung adenocarcinoma (LUAD), the most common type of lung cancer, is a leading cause of cancer-related mortality. NT5E, an ecto-5’-nucleotidase enzyme, has been implicated in cancer progression, particularly in efferocytosis. Despite its potential involvement, the prognostic significance of NT5E and relationship with immune cell infiltration in LUAD have not been extensively explored. In this study, we performed a comprehensive analysis to elucidate the expression patterns of NT5E and its prognostic implications in LUAD using data from diverse public databases. Multiple computational algorithms, including CIBERSORT, ESTIMATE, and xCell, were employed to assess the correlation between NT5E expression and immune cell infiltration. We found that NT5E was significantly overexpressed at both the mRNA and protein levels in LUAD tissues. Elevated NT5E expression was significantly linked to multiple clinicopathological factors, including metastasis and pathological stage, and served as a strong predictor of poor prognosis in LUAD patients. Gene Set Enrichment Analysis (GSEA) indicated that NT5E plays a crucial role in regulating immune responses, as evidenced by differential gene expression associated with NT5E levels. A strong positive correlation was observed between NT5E expression and the presence of immune cells, including dendritic cells, macrophages, and CD4+ T cells, as well as the expression of various immune cell markers, suggesting that NT5E may influence the prognosis of LUAD patients by regulating immune cell infiltration. Additionally, drug sensitivity analysis highlights the potential of selumetinib and PD318088, both MEK1/2 inhibitors, to target NT5E in LUAD treatment, suggesting their use as single agents or in combination with other therapies. Collectively, these findings establish NT5E as a promising prognostic biomarker and therapeutic target in LUAD, particularly in the context of immune cell infiltration.
Similar content being viewed by others
Introduction
Lung cancer remains the leading cause of cancer-related deaths, exceeding the combined mortality from colorectal, breast, and prostate cancers each year1. The majority of patients are diagnosed at a late stage, either with locally advanced or metastatic disease2. LUAD, the most prevalent histological subtype, accounts for about 50% of all lung cancer cases. For Stage I or II LUAD, the standard treatment typically includes surgical tumor removal, along with adjuvant therapy. However, as the disease advances to Stage III or IV, the approach shifts to chemotherapy or radiotherapy3. Regrettably, traditional chemotherapy drugs are hampered by common limitations such as non-specific targeting, low bioavailability, and the emergence of drug resistance, which restrict their effectiveness in treating cancer4. Nanomedicine delivery systems, with their unique properties, encapsulate therapeutic agents to prevent degradation, allowing for the precise delivery of anticancer drugs to tumor sites while minimizing damage to surrounding tissues, which makes nanomedicine a promising field to explore in lung cancer therapy5. Molecular targeted therapy has emerged as a vital strategy for LUAD treatment, with key targets including the epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (c-Met), and anaplastic lymphoma kinase (ALK)6. Tyrosine kinase inhibitors like gefitinib and erlotinib have been developed to target EGFR mutations7. However, acquired drug resistance, often due to epigenetic changes and tumor heterogeneity, is a significant barrier to the efficacy of LUAD treatments. Photothermal therapy is a promising minimally invasive treatment that has shown positive outcomes in tumor treatment, which combines photothermal agents with local laser irradiation to convert light energy into heat energy for treating tumors. The synergy between nanotechnology and photothermal therapy in tumor treatment shows promising prospects8. Immunotherapy strategies, such as checkpoint inhibitors, are safe and effective and may offer alternative treatment options for LUAD9. For instance, by acting as an inhibitor of the cystine/glutamate antiporter, erastin can enhance the responsiveness to anti-PD-L1 therapy, thereby effectively slowing the advance of lung cancer10. In some cases, a combination of multiple therapies or strategies with immunotherapy may be preferred. Despite advancements in therapeutic approaches, the prognosis for LUAD patients remains poor, with a five-year survival rate averaging below 20%1,2. Therefore, the discovery of novel biomarkers with high specificity and sensitivity is crucial for the accurate diagnosis, personalized treatment, and precise prognostic prediction of LUAD.
Efferocytosis, the pivotal process through which phagocytes remove apoptotic cells, is crucial for maintaining tissue homeostasis in healthy conditions and for restoring it after disease11,12. Ecto-5’-nucleotidase (NT5E), also known as CD73, is recognized for its diverse biological functions, especially its ability to convert extracellular purine 5’-mononucleotides into bioavailable nucleosides13. Previous studies have shown that NT5E plays a key role in modulating macrophage immune responses during efferocytosis. It converts AMP from apoptotic cells into adenosine, which reduces various pro-inflammatory cytokines, with NT5Eexpression on either the macrophage or the apoptotic cell being sufficient to mediate TNF suppression14. NT5E may exert a regulatory influence on cancer development and progression by participating in the efferocytosis process.
Increasing evidences have shown that NT5Eexpression is a major regulator of the invasive and metastatic potential of cancer cells15,16. High levels of NT5Ehave been found across various malignancies, including but not limited to breast, pancreatic, hepatic, ovarian, and gastric cancers13,17,18,19,20. The link between high NT5E levels and cancer progression is becoming clearer, possibly through adenosine-induced immunosuppressive and pro-angiogenic signaling cascades20,21. Furthermore, NT5E is not only found in cancer cells but also in various immune cells. These includes regulatory T cells, myeloid-derived suppressor cells, dendritic cells, and natural killer (NK) cells15,22,23. Notably, NK cells expressing NT5Ecan suppress T cell activity, possibly by increasing anti-inflammatory cytokines such as interleukin-10 (IL-10) and transforming growth factor-β (TGF-β)23. This complex interaction between NT5E and the immune microenvironment highlights the enzyme’s complex role in cancer’s immune response. Yet, the specific role of NT5E in the metastatic cascade of lung adenocarcinoma and its interaction with immune cell infiltration in the tumor microenvironment is not fully understood.
Considering the complex relationship between NT5E, efferocytosis, and tumorigenesis, the exact role and clinical relevance of NT5E in the LUAD pathogenesis and prognosis have not been thoroughly investigated. This study uses a comprehensive bioinformatics approach to examine NT5E’s potential role in LUAD metastasis and immune cell infiltration, as well as to elucidate its molecular mechanisms. Our analysis reveals a significant increase of NT5E in LUAD tissues compared to non-tumoral samples. Importantly, NT5E levels rise as the tumor progression and are strongly linked to axillary lymph node metastasis. Furthermore, high NT5E expression is inversely associated with the survival outcomes of LUAD patients. We also uncover a robust correlation between NT5E expression and the presence of immune cell infiltrates, including dendritic cells, neutrophils, CD4 + T cells, and macrophages, in LUAD. Notably, NT5E seems to affect LUAD patient prognosis, partly by its influence on immune cell infiltration. Furthermore, our analysis indicates that selumetinib and PD318088 are particularly promising as NT5E-targeting therapeutic agents for LUAD. These findings emphasize the pivotal role of NT5E in oncogenesis and suggest its critical involvement in the orchestration of the immune microenvironment, indicating its potential as a novel therapeutic target in LUAD.
Materials and methods
Integrative analysis of NT5E expression and clinical outcomes
Transcriptional profiles and clinical data from cancerous tissues within the TCGA pan-cancer consortium and from normal human tissues within the Genotype-Tissue Expression (GTEx) project were sourced from the UCSC Xena platform (http://xenabrowser.net/)24. These expression profiles were standardized to transcripts per kilobase million (TPM) for subsequent analysis, including differential gene expression, tumor mutational burden (TMB), microsatellite instability (MSI), and the construction of receiver operating characteristic (ROC) curves. The Cox proportional hazards regression model was performed for univariate and multivariate survival analysis. A list of cancer types studied is provided in Supplementary Table 1.
Incorporating expression and survival analysis of NT5E
The “Expression DIY” module of GEPIA (Gene Expression Profiling Interactive Analysis) was used to contrast NT5Eexpression between LUAD and normal adjacent lung tissues, which serves as a database for exploring correlations between gene expression and cancer patient survival, leveraging data from both TCGA and GTEx25. This analysis incorporated matched GTEx normal samples and TCGA-LUAD data, applying a log2 transformation of transcripts per million (TPM + 1) for a logarithmic scale. GEPIA was also used to perform expression analysis and survival analysis of NT5E. Moreover, the TISIDB (Tumor-immune System Interactions Database), an integrated repository for the study of tumor-immune system interactions26, was also utilized to evaluate the correlation between NT5E gene expression levels and overall survival outcomes across various cancers.
Exploring NT5E expression patterns and clinicopathological correlations in LUAD patients
The UALCAN (University of Alabama at Birmingham Cancer Data Analysis) platform facilitates an extensive analysis of transcriptome data sourced from TCGA and the MET500 dataset27. In this study, UALCAN was utilized to examine the expression patterns of NT5E and to elucidate its correlation with a spectrum of clinicopathological parameters in lung cancer, such as sex, stage, nodal metastasis status, age, TP53 mutations in lung cancer.
Structure, localization, and expression analysis of the NT5E protein
The 3D structure of the NT5E protein was retrieved from the AlphaFold Protein Structure Database, a resource developed by DeepMind and EMBL-EBI28. To elucidate the subcellular localization of NT5E in cancerous cells, we utilized immunofluorescence staining on the human cancer cell line A-431, with images sourced from the Human Protein Atlas (HPA)29. HPA also provided data on NT5E protein expression in normal and lung cancer tissues.
Analysis of NT5E expression and its correlation with immune cell infiltration
To assess the expression of NT5Ein various malignancies, the “Diff Exp” module in TIMER (Tumor Immune Estimation Resource) was used, which is an interactive online platform that facilitates a comprehensive examination of immune cell infiltration across various cancer types30. Specifically, the correlation between NT5E expression and the infiltration of immune cells in lung adenocarcinoma (LUAD) was scrutinized. TIMER’s “Gene” module was then employed to explore the association between NT5E expression levels and the presence of distinct immune cell populations, including B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells, leveraging data from TCGA. Additionally, the platform’s “Correlation” module was applied to discern the relationships between NT5E expression and various gene markers of immune cells, such as CD8+ T cells, T cells (general), B cells, monocytes, TAMs, M1 macrophages, M2 macrophages, neutrophils, NK cells, DCs, Th1 cells, Th2 cells, Tfh cells, Th17 cells, Tregs, and exhausted T cells. This study’s statistical analysis involved the use of purity-correlated partial Spearman’s correlation to evaluate the strength of the associations, with statistical significance determined accordingly.
Survival outcomes of NT5E expression in different immune cell subsets of LUAD
To assess the prognostic relevance of NT5Ein LUAD, we utilized the Kaplan-Meier Plotter, an online repository that amalgamates mRNA expression profiles with clinical outcomes for over 513 Lung adenocarcinoma patients31. This analysis stratified patient cohorts based on NT5E expression with the “median” and “auto select best cutoff” parameters, differentiating between high and low expression groups. The impact of NT5E expression on overall survival (OS), relapse-free survival (RFS) was evaluated. Hazard ratios (HRs), accompanied by 95% confidence intervals (95% CIs) and log-rank P-values, were calculated to quantify the statistical association between NT5E expression and survival outcomes. Additionally, Kaplan-Meier analysis was applied to explore the relationship between NT5E expression levels and overall survival in various immune cell subsets with different infiltration levels in LUAD.
Identification and analysis of NT5E-interacting genes and proteins
Employing the GeneMANIA database, we constructed an interaction network for the NT5E gene32, a web-based tool that facilitates the prediction and visualization of gene functional linkages. This approach offers a comprehensive view of NT5E’s role within the broader genomic context, enhancing our grasp of its biological relevance. In parallel, we harnessed the STRING database to delineate a protein-protein interaction (PPI) network for NT5E33. This online platform specializes in the analysis of known and predicted interactions, offering insights into NT5E’s molecular associations in LUAD.
Gene ontology and pathway enrichment analysis of NT5E-associated DEGs
To further investigate the genes associated with variations in NT5E expression and their functions, we divided samples based on the median expression level of NT5E in the TCGA-LUAD cohort. The differential gene expression analysis was performed using the “limma” R package26. A total of 384 differentially expressed genes (DEGs) were identified with the statistical thresholds of fold change > 1.5 and false discovery rate (FDR) < 0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)34 pathway analyses were then performed to dissect the biological roles of NT5E in the context of lung cancer. GO analysis adeptly identifies the biological processes (BPs) associated with NT5E. GSEA complemented these findings by exploring the underlying mechanisms potentially influenced by NT5E. The execution of GO and KEGG enrichment analyses and visualization was performed utilizing the gseKEGG, gseGO and gseaplot functions of the R packages “enrichplot” and “clusterProfiler”35.
CIBERSORT algorithm-based correlation analysis between NT5E expression and immune cell infiltration
To clarify how NT5Egene expression relates to immune cell subsets, we used CIBERSORT to analyze immune cell composition in TCGA-LUAD patients36. CIBERSORT quantifies the relative proportions of tumor-infiltrating immune cells utilizing a curated leukocyte gene signature matrix comprising 547 genes representative of 22 distinct human immune cell subpopulations. Statistical significance was determined using a threshold P-value of less than 0.05 to identify immune cell populations affected by NT5E expression in LUAD patients.
Drug sensitivity analysis and identification of NT5E-targeting agents
To elucidate the correlation between NT5Eexpression levels and drug sensitivity, we utilized the IC50 values of small molecules across various cell lines sourced from the Cancer Therapeutics Response Portal version 2 (CTRP2)37. Concurrently, we procured gene expression sequencing data for the same cell lines from the Cancer Cell Line Encyclopedia (CCLE) database. Alternatively, these data can be directly downloaded from the R package “oncoPredict” available at the following link: https://osf.io/c6tfx/. Subsequently, we performed a Pearson correlation analysis to determine the relationship between NT5E mRNA expression levels and drug IC50 values. To find drugs that might target NT5E-expressing cancer cells, we used the Biomarker Exploration for Solid Tumors (BEST) database (https://rookieutopia.hiplot.com.cn/app_direct/BEST/) to identify candidate agents that could be effective for high NT5E expression utilizing data from TCGA-LUAD and GEO cohorts. A negative correlation means that gene expression is suppressed indicating sensitivity to that drug, and vice versa.
Western blot assay
The NT5E antibody (catalog number D221879) was procured from Sangon Biotech. Cells were detached from culture plates and lysed to isolate whole-cell protein extracts. The protein lysates were prepared using RIPA Buffer (Cell Signaling Technologies, USA) supplemented with a protease and phosphatase inhibitor cocktail (Roche, Switzerland). Protein concentrations were ascertained utilizing the BCA Protein Quantification Kit (Vazyme, China). Equal volumes of cell extracts containing denatured proteins were subjected to SDS-polyacrylamide gel electrophoresis on 12% gels, followed by transfer to a NC membrane. The membranes were incubated with primary antibodies for 1 h. Subsequent to primary antibody binding, secondary antibodies were applied. Chemiluminescence detection was performed using the ECL Western blotting detection reagents (Pierce, USA).
RNA extraction and qPCR analysis
Total RNA was isolated from samples using TRIzol reagent (Invitrogen, USA) and converted into complementary DNA (cDNA) with a reverse transcription kit (Vazyme Biotech, China). Quantitative real-time PCR (qPCR) was performed using a qPCR kit from the same manufacturer. The relative quantification of gene expression was achieved by applying the 2^-ΔΔCt method. The qPCR reaction mixture comprised 12.5 µL of Maxima SYBR Green qPCR Master Mix, 0.3 µM each of forward and reverse primers, and up to 500 ng of cDNA, with the volume adjusted to 25 µL with nuclease-free water. The reactions were carried out in a 96-well plate with an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The qPCR program concluded with a melt curve analysis consisting of 95 °C for 15 s, 60 °C for 60 s, and a final step at 95 °C for 15 s. Gene expression levels were normalized to a housekeeping gene, RPL13A, to ensure the accuracy of relative quantification. The primer sequences for NT5E were as follows: Forward Primer 5’-GCCTGGGAGCTTACGATTTTG-3’, Reverse Primer 5’-TAGTGCCCTGGTACTGGTCG-3’. The housekeeping gene RPL13A was amplified using the following primers: Forward Primer 5’-GCCATCGTGGCTAAACAGGTA-3’, Reverse Primer 5’-GTTGGTGTTCATCCGCTTGC-3’.
Statistical analysis and data visualization
Statistical computations and graphical illustrations within this study were executed using the R platform (www.R-project.org/), unless otherwise noted. To evaluate the significance of the findings, a range of tests was employed, including the Student’s t-test for parametric data, the Pearson’s correlation coefficient for linear relationships, the Spearman’s correlation coefficient for non-linear associations, and the Wilcoxon rank-sum test for non-parametric comparisons. The results of Kaplan-Meier Plotter, GEPIA are presented alongside hazard ratios (HR) and associated P-values, which were ascertained through log-rank testing. Spearman’s rank correlation was performed to evaluate the statistical relationships between NT5E expression and other factors, such as immune cell infiltration levels, immune regulator genes, TMB, and MSI, with statistical significance determined at the conventional threshold. A heatmap illustrating the correlation matrix of NT5E with genes implicated in efferocytosis was crafted using the R packages “ggplot2” and “linkET”, with Pearson’s correlation coefficient. The UCSC genome browser was used to visualize ATAC-seq and ChIP-seq data obtained from the Cistrome database (http://cistrome.org/db). The thresholds for statistical significance were defined as * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
Results
Aberrant expression of NT5E in pan-cancer landscape
To reveal the expression pattern of NT5E across cancers, we analyzed NT5EmRNA levels in both normal and cancerous tissues using the Tumor Immune Estimation Resource (TIMER) database30. Notably, higher NT5E mRNA levels were observed in tumor tissues as compared to normal tissues in lung adenocarcinoma (LUAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), and thyroid carcinoma (THCA) (P < 0.001) (Fig. 1A). Conversely, NT5E expression was found to be significantly downregulated in breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), liver hepatocellular carcinoma (LIHC), prostate adenocarcinoma (PRAD), uterine corpus endometrial carcinoma (UCEC) (P < 0.001) (Fig. 1A; Supplementary Fig. 1). These findings highlight the complex and variable expression of NT5E across different cancer types.
Pan-cancer analysis identifiesNT5Eas a potential prognostic biomarker in LUAD. (A) The expression level of NT5E between tumor and normal tissues in each type of cancer based on the TIMER database. Red-shaded and blue-shaded areas indicate significantly increased or decreased NT5E expression in the tumor tissue, respectively. * P < 0.05, ** P < 0.01, *** P < 0.001. (B) Forest plots representing the hazard ratio (HR) and 95% confidential interval (CI) of NT5E using a Cox proportional hazard regression model across 33 human cancer types in TCGA pan-cancer dataset. P-values below 0.05 were considered statistical significance and highlighted in red font. (C) Correlations between NT5E expression and tumor mutation burden (TMB) in pan-cancer. * P < 0.05, ** P < 0.01. (D) Correlations between NT5E expression and microsatellite instability (MSI) in pan-cancer. * P < 0.05, ** P < 0.01. (E) Diagnostic ROC curves of NT5E for predicting the 5-year survival in the TCGA-LUAD dataset. AUC, area under the curve.
NT5E is a promising candidate biomarker for LUAD
We then performed studies to confirm the prognostic significance of NT5E expression in a wide range of cancers from TCGA database. Univariate Cox regression analysis revealed that high NT5E expression was associated with worse prognostic outcomes in a variety of cancers, and the overexpression of NT5E could strongly predict short overall survival (OS) in LUAD, HNSC, MESO, PAAD, STAD, and UVM (P < 0.01), and CESC (P < 0.05), which underscored the potential of NT5E as a cancer prognostic biomarker (Fig. 1B; Supplementary Fig. 2).
To further assess the predictive capacity of NT5Ein immune checkpoint inhibitor (ICI) therapy outcomes, we examined its expression in relation to tumor mutation burden (TMB) and microsatellite instability (MSI), recognized predictive biomarkers in immunotherapy38,39. We found that there is a positive correlation between NT5E expression and TMB in ACC and SKCM, contrasting with a negative correlation observed in LUAD, BRCA, HNSC, LIHC, PCPG, and TGCT (Fig. 1C). Additionally, NT5E expression showed a positive association with MSI in COAD, TGCT, and UCEC, and a negative association in LUAD, DLBCL, HNSC, and PRAD (Fig. 1D). These correlations imply that NT5E may predict ICI therapy efficacy in specific cancers.
Our extensive pan-cancer analysis revealed a significant correlation between NT5E expression and various malignancies, with a particularly robust association in LUAD. Evidence from multiple studies indicates that increased NT5E expression in LUAD patients is a risk factor for poor overall survival and is inversely correlated with TMB and MSI (Fig. 1A and D; Supplementary Fig. 3). The prognostic implications of NT5E in LUAD was also supported by receiver operating characteristic (ROC) analysis, which was based on a 5-year survival time point (Fig. 1E). These connections position NT5E as a candidate for prognostic assessment and a potential immunotherapy target in LUAD.
NT5E expression closely relates to clinical progression and metastasis in LUAD
To further confirm the expression pattern of NT5E in LUAD, we performed an integrative mRNA expression analysis comparing LUAD with normal lung tissues. This analysis utilized data from the TCGA-LUAD and GTEx datasets, accessed via multiple online databases, including UCSC Xena (Fig. 2A), GEPIA (Fig. 2B), UALCAN (Fig. 2C), and BEST (Fig. 2D)24,25,27. The results consistently showed higher mRNA expression of NT5E in LUAD tissues than in normal lung tissues. This significant upregulation of NT5E was also evident in a cohort of 58 paired tumor and normal samples (Fig. 2E). Furthermore, our analysis confirmed a consistent NT5E expression pattern in LUAD patients across GEO cohorts, reinforcing the reliability of our findings (Fig. 2F).
Utilizing the UALCAN platform, we next analyzed NT5E mRNA expression in LUAD patient subgroups categorized by various clinical parameters. Analysis by sex showed significant upregulation of NT5E in lung cancer samples from both males and females compared to normal lung tissues (Fig. 2G). Analysis by tumor stage indicated a stage-dependent increase in NT5E expression, with elevated levels in LUAD patients from stage 1 to stage 4 (Fig. 2H). Additionally, higher NT5E expression was associated with nodal metastasis status, with increased levels in patients classified as N0, N1, N2, and N3 (Fig. 2I). Notably, NT5E upregulation was observed in LUAD patients with both TP53-mutant and TP53 wild-type genotypes when compared to normal controls (Fig. 2J). A general inverse relationship between promoter methylation and mRNA expression levels was observed, suggesting regulatory implications (Fig. 2G and J).
mRNA expression analysis ofNT5Ein normal lung tissues and LUAD tissues. (A) Differential expression of NT5E between tumors and normal samples using combined data from TCGA-LUAD and GTEx datasets. (B) Increased expression of NT5E in LUAD compared to normal tissues in the GEPIA database. (C)NT5E expression in LUAD was examined by using the UALCAN database. (D) Analysis of NT5E expression in LUAD and adjacent normal tissues in the TCGA-LUAD database. (E) Statistical analyses of NT5E expression in 58 pairs of LUAD tissues and adjacent normal tissues in the TCGA database. (F) Statistical analyses of NT5E expression in normal lung tissues and LUAD tissues in Biomarker Exploration of Solid Tumors (BEST) database. (G–J) Box plots represent the mRNA expression and promoter methylation levels of the NT5E gene across various LUAD patient cohorts, categorized by clinical parameters, utilizing data from the UALCAN portal. The analysis was stratified by sex (G), cancer stage (H), presence of metastasis (I), and TP53 mutation status (J). Lymph node metastasis was classified according to the number of involved axillary lymph nodes: N0 for no metastasis, N1 for 1 to 3 nodes, N2 for 4 to 9 nodes, and N3 for 10 or more nodes. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
Next, we extended our investigation to understand the protein structure, subcellular localization, and expression of NT5E, utilizing AlphaFold2 and the HPA databases28,29. Our findings showed that NT5E is primarily located in the plasma membrane, with additional presence in the nucleoplasm and cytosol (Fig. 3A and B). Consistent with the mRNA expression data, NT5E protein levels were significantly higher in lung cancer tissues than in normal tissues (Fig. 3C). These findings indicate a robust correlation between aberrant NT5E expression and the clinical progression and metastasis of LUAD.
To further validate our analytical findings, we conducted Western blot and qPCR experiments to assess the gene expression of NT5E in both normal lung fibroblast cell lines (MRC-9, IMR-90, WI-38) and lung cancer cell lines (PC-9, A549, HCC44, NCI-H1437, NCI-H1568, NCI-H838). The results revealed that NT5E expression, at both the protein and mRNA levels, was significantly higher in cancer cells compared to normal cells, with in vitro experimental outcomes corroborating our database analysis (Fig. 3D and E; Supplementary Fig. 5). Furthermore, utilizing epigenomic data from the Cistrome DB database (http://cistrome.org/db), we analyzed and compared ATAC-seq and H3K27ac ChIP-seq data between normal (IMR-90) and LUAD cancer cells (PC-9, A549, NCI-H524). Our findings indicated that lung cancer cells displayed greater chromatin accessibility near the NT5E promoter and a higher enrichment of H3K27ac marks (Fig. 3F). The integration of multi-omics data with our experimental results suggests that NT5E should play a pivotal role in the development of LUAD.
The structure, subcellular localization, protein expression, chromatin accessibility and H3K27ac ofNT5E. (A) The predicted 3D structure of human NT5E protein taken from the AlphaFold Protein Structure Database. (B) Subcellular localization of NT5E in A-431 cancer cell lines obtained from the Human Protein Atlas (HPA) database. Scale bar = 10 μm. (C) Immunohistochemistry (IHC) staining of NT5E protein expression in normal lung tissues (left panel) and lung cancer tissues (right panel) determined by the HPA database (antibody HPA017357). Scale bar = 100 μm. (D) Western blotting showing the expression levels of the NT5E protein in three normal lung fibroblast cell lines (MRC-9, IMR-90, WI-38) and six lung cancer cell lines (PC-9, A549, HCC44, NCI-H1437, NCI-H1568, NCI-H838). (E) The qPCR analysis of relative NT5E expression levels normalized to the housekeeping gene RPL13A in both normal and cancer cell lines as indicated in (D). (F) The UCSC genome browser tracks showing normalized ATAC-Seq and ChIP-Seq signals sourced from the Cistrome Database at the NT5E gene locus in normal lung fibroblast cell lines (IMR-90) and lung cancer cell lines (PC-9, A549, NCI-H524). The red shaded area indicates the transcription start site and promoter regions of NT5E.
Increased expression of NT5E in LUAD patients predicted a poor prognosis
The investigation of NT5E gene expression in LUAD has delineated its association with disease progression and metastasis. Using data from GEPIA, Kaplan-Meier plotter, and the BEST databases, we found that LUAD patients with higher NT5E expression levels were correlated with worse overall survival (OS) and relapse-free survival (RFS) (Fig. 4A and C). Especially, the BEST database provided strong evidence that high NT5E expression was associated with a poor prognosis in the GSE3141 and GSE26939 cohorts (Fig. 4C).
To further explore the prognostic value and underlying mechanisms of NT5E expression, we performed a Cox regression analysis using the TCGA-LUAD dataset. This analysis revealed that in stage IV LUAD patients, increased NT5E expression was significantly linked to diminished overall survival (Fig. 4D). The correlation between NT5E expression and poor OS was also confirmed in patients with AJCC stage T3 and N1 LUAD, where a significant association was observed (Fig. 4D). Altogether, these results highlight the importance of NT5E expression as a prognostic factor in lung adenocarcinoma.
Survival analyses evaluating the prognostic value of NT5Ein lung adenocarcinoma. (A) The overall survival analysis of NT5E in LUAD patients was examined by using the GEPIA database. (B) Overall survival (OS) and relapse free survival (RFS) of NT5E in lung adenocarcinoma from the Kaplan-Meier plotter database. (C) Overall survival analysis of NT5E expression using TCGA-LUAD and GSE-LUAD datasets from the web application BEST. (D) Forest plot depicting the correlations between NT5E expression and clinicopathological parameters in LUAD and patients.
Identification of NT5E-interacting genes and proteins
To understand how NT5Einteracts with other molecules in LUAD, we employed GeneMania32 to construct a gene-gene interaction network encompassing NT5E and its neighboring genes. The analysis identified a cohort of 20 genes that demonstrated significant correlation with NT5E, including PCDH7, HIF1A, CFAP20, and RNF6 (Fig. 5A). Further exploration of the protein-protein interaction (PPI) landscape of NT5Ewas performed using the STRING database33, which revealed a network with 32 edges and 11 nodes, prominently featuring UPP1, UPP2, NMNAT1, and NMNAT2 (Fig. 5B). Additionally, using the TCGA-LUAD database, we looked into how NT5E relates to genes involved in efferocytosis. A positive and significant correlation was observed between NT5E and key efferocytosis-related genes (Supplementary Table 2), including AXL, ATG7, ITGB5, and MFGE8 in LUAD (Fig. 5C; Supplementary Fig. 4). Herein, we provide an extensive characterization of the molecular context surrounding NT5E, elucidating its putative involvement in the pathophysiology of lung adenocarcinoma.
NT5E-Interacting genes and proteins. (A) A gene-gene interaction network for NT5E was constructed using the GeneMANIA database. (B) The PPI network of NT5E was generated using STRING database. (C) A heatmap shows the correlations between NT5E and efferocytosis-related genes based on TCGA-LUAD dataset. The Mantel test was used to analyze the interaction influences of NT5E on efferocytosis-related genes. Genes with absolute Pearson correlation coefficients below 0.5 relative to other genes were omitted from the heatmap plot.
Function and pathway enrichment of NT5E-associated DEGs in LUAD
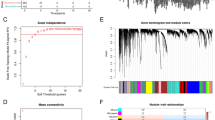
To investigate the molecular impact of NT5Ein LUAD, we performed a comprehensive differential gene expression analysis. Utilizing the “limma”26 R package, we identified 384 differentially expressed genes (DEGs) with stringent criteria (fold change > 1.5, FDR < 0.05) (Supplementary Table 3), grouped by median NT5E expression levels. The expression profiles of the top 15 upregulated and downregulated DEGs were depicted in a heatmap for visual correlation (Fig. 6A). To assess the clinical relevance of these DEGs, we employed the Gene Set Variation Analysis (GSVA)40 to calculate GSVA scores for the top 15 upregulated DEGs, including EREG, MET, IL37, and FOXQ1. Notably, Kaplan-Meier survival analysis of the TCGA-LUAD cohort demonstrated a significant association between high GSVA scores and shorter overall survival, indicating a possible prognostic value for these genes (Fig. 6B).
We gained further insight into the biological implications of NT5E-associated DEGs was obtained through GO term annotation and KEGG pathway analysis using GSEA41. The GO analysis highlighted enrichment of immune-related pathways, particularly those involved in antigen processing and presentation, cellular response to type II interferon, and T cell-mediated cytotoxicity and immunity (Fig. 6C; Supplementary Table 4). Similarly, the KEGG analysis revealed an enrichment of immune functional gene sets in lung cancer, including pathways related to autoimmune thyroid disease, inflammatory bowel disease, cytokine-cytokine receptor interactions, Th17 cell differentiation, antigen processing and presentation, NK cell-mediated cytotoxicity, chemokine signaling, and Th1 and Th2 cell differentiation (Fig. 6D; Supplementary Table 5). Together, the results from GO and KEGG analyses highlight the key role of NT5E in modulating the immune response in LUAD, and provide a foundation for further investigation into the immunotherapeutic potential of targeting NT5E in lung cancer treatment.
Biological function and pathway enrichment analysis of DEGs between low and high NT5E groups in TCGA-LUAD patients. (A) The heatmap illustrates the top 15 up-regulated and top 15 down-regulated DEGs stratified by high and low NT5E expression levels in the TCGA-LUAD dataset. (B) Overall survival analysis based on the gene set variance analysis (GSVA) score of the top 15 up-regulated DEGs in (A). (C) Top 10 enrichment terms in BP categories of the DEGs in TCGA-LUAD. (D) Top 10 KEGG enrichment pathways of the DEGs in TCGA-LUAD.
NT5E expression associated with diverse immune cell infiltration in LUAD
Our analysis revealed a significant correlation between NT5E expression and the infiltration of various immune cell types in LUAD. Specifically, higher NT5E levels were linked to increased dendritic cells, neutrophils, CD4 + T cells, and macrophages, yet there was no significant link with B cell infiltration, as shown by the TIMER database (Fig. 7A). To further delineate the influence of NT5E on the tumor microenvironment (TME) in LUAD, we utilized multiple algorithms, including CIBERSORT, TIMER, and xCell to estimate the correlation between NT5Eexpression and immune cell infiltration, drawing on data from TCGA and GEO datasets36. We found positive correlation with resting dendritic cells, M2 macrophages, monocytes, CD4 + memory resting T cells, and resting mast cells. In contrast, NT5E expression was negatively correlated with plasma cells, follicular helper T cells, M0 macrophages, resting NK cells, CD8 + T cells, and memory B cells in LUAD (Fig. 7B and C). These results suggest that NT5E plays a complex role in modulating the immune landscape within LUAD, with both stimulatory and inhibitory effects on different immune cell subsets. The observed correlations provide valuable insights into the potential mechanisms by which NT5E may influence the TME and contribute to the lung cancer progression.
NT5E expression correlates with immune infiltration levels in lung adenocarcinoma. (A) The expression of NT5E exhibits a significant positive correlation with tumor purity and the infiltration levels of various immune cell types, as elucidated by the TIMER database. (B) Utilizing the CIBERSORT algorithm, a substantial correlation is observed between NT5E expression and the immune cell infiltration in LUAD. (C) The heatmap showed the infiltration levels of various immune cell subtypes associated with NT5E expression evaluated by multiple algorithms in LUAD samples from TCGA and GEO datasets. (D) Scatterplots depict the significant correlations between NT5E expression levels and the expression of immune checkpoint genes CD274 (PD-1), PDCD1G2 (PD-L2), HAVCR2 (TIM-3, CD366), SIGLEC15 (CD33L3), CTLA4 (CD152), TIGIT (CD226), HLA-A, HLA-B, TNFSF4, CD80, CD70 and CD40 in LUAD patients from TCGA-LUAD dataset.
Correlation between NT5E expression and immune markers
To address the immunomodulatory role of NT5E in LUAD, we utilized the TIMER database to assess the correlation between NT5E expression and various immune markers. A panel of genes (Table 1) was employed to represent various immune cell types, including B cells, T cells, and their subsets, as well as myeloid cells such as tumor-associated macrophages (TAMs) and dendritic cells. Aware of the impact of tumor purity on immune infiltration analysis, we adjusted for this variable and observed significant associations between NT5E expression and the majority of immune markers across different immune cell types in LUAD (Table 1).
Broadening our investigation to functional T cell subsets, we examined the correlation between NT5E expression and markers for Th1, Th2, regulatory T cells (Tregs), and other T cell states (Table 2). After adjusting for tumor purity, we found that NT5E expression significantly correlated with 24 of 31 T cell markers in LUAD, post-tumor purity adjustment, highlighting extensive interactions between NT5E and the adaptive immune response (Table 2).
Diving deeper into the immunological landscape, we investigated the relationship between NT5E expression and immune checkpoint genes (ICGs) within the TCGA-LUAD cohort, and found that NT5E expression significantly correlated with the expression of multiple ICGs, including CD274 (PD-1), HLA-A, HLA-B, PDCD1G2 (PD-L2), TIGIT (CD226), CTLA4 (CD152), and CD80 (Fig. 7D; Supplementary Table 6). These correlations suggest that NT5E not only modulates immune cell infiltration but also influences the expression of key regulators of immune response, possibly involving NT5E in immune evasion strategies within the lung cancer microenvironment.
LUAD prognosis is influenced by different immune cell infiltration and NT5E expression
Considering the known link between NT5E expression and immune cell infiltration, and its connection to poor prognosis in LUAD, we aimed to find out if NT5E expression affects LUAD prognosis by altering the immune microenvironment. We conducted a stratified prognostic analysis based on NT5Eexpression levels in relation to various immune cell subgroups of LUAD using the Kaplan-Meier Plotter database31. We discovered that LUAD patients with elevated NT5E expression and diminished infiltration of B cells, CD4 + T cells, and natural killer T cells exhibited a significantly poorer prognosis (Fig. 8A, B). Conversely, high NT5E expression coupled with increased infiltration of macrophages, regulatory T cells, and Th2 cells was also linked to a worsened prognosis (Fig. 8A and B). These findings underscore the complex interplay between NT5E expression and the immune landscape, suggesting that NT5E may modulate immune cell dynamics to influence clinical outcomes. However, we observed no correlation with prognosis in patients with varying levels of CD8 + T cells and type 1 helper T cells (Fig. 8A, B). Notably, high NT5E expression consistently predicted a poor prognosis, regardless of the infiltration levels of these cell types, suggesting that NT5E expression might impact LUAD outcomes independently of CD8 + T cells and type 1 helper T cells infiltration levels. Overall, these results indicate that NT5E may impact LUAD prognosis partly due to different infiltration level of various immune cells, highlighting the intricate relationship between NT5E expression and the immune microenvironment of LUAD.
Overall survival analysis relating NT5E expression to immune cell subgroups in lung adenocarcinoma. (A) The forest plot delineates the prognostic significance of NT5E expression levels in various immune cell subgroups of patients with LUAD. (B) The Kaplan-Meier plotter was utilized to estimate the association between NT5E expression and overall survival (OS) across different immune cell subgroups in LUAD patients.
Potential NT5E-targeting drugs in lung adenocarcinoma
The 50% inhibitory concentration (IC50) is a key measure in pharmacology, indicating the concentration of a drug necessary to inhibit the growth of tumor cells by half. This value is widely recognized as an indicator of a drug’s efficacy, with a lower IC50 meaning the tumor cells are more sensitive to the drug. For drug sensitivity analysis with the CTRP v2 database37, we gathered IC50 values for 545 drugs across various cancer cell lines, complemented by transcriptome sequencing data sourced from the CCLE database42. Subsequently, the expression levels of the NT5E gene within each cell line were correlated with the respective IC50 values for each drug, aligning them by cell line identity. A Pearson correlation analysis was performed to explore the relationship between gene expression levels and drug IC50 values, thereby determining the correlation between the NT5E gene and each drug’s effectiveness. Furthermore, leveraging the BEST database, we performed a correlation analysis between NT5E gene expression and drug sensitivity, utilizing gene expression profiles from the TCGA-LUAD and GEO datasets. This comprehensive approach provides a robust framework for understanding the interplay between gene expression and drug response in lung cancer treatment.
A negative correlation between a gene’s expression and the IC50 of a drug suggests that increased expression of the gene correlates with increased drug sensitivity. In our study, we observed a positive correlation between NT5E gene expression and the IC50 values of belinostat, vincristine, and VX-680, indicating a potential resistance to these agents in tumor cells with a higher NT5E expression level (Fig. 9A). This finding is particularly relevant given the distinct drug and tumor cell profiles assessed in the CTRP and BEST databases, which may account for the observed variations in results of resistant drugs (Fig. 9A and B; Supplementary Tables 7 and 8). Conversely, we found a significant negative correlation between NT5E expression and the IC50 values of selumetinib, PD318088, the combination of selumetinib and PLX-4032 (8:1 mol/mol), and the combination of selumetinib and MK-2206 (8:1 mol/mol). This inverse relationship was consistently observed across cancer cell lines, TCGA-LUAD, and GEO cohorts with lung cancer, suggesting that higher NT5E expression is associated with increased sensitivity to these drugs (Fig. 9). These results indicate that drugs like selumetinib and PD318088 (Fig. 9C and D), which are MEK1/2 inhibitors and show negative correlations with NT5E expression, could be effective therapeutic options for targeting NT5E in LUAD.
Drug sensitivity analysis and potential inhibitors targeting NT5E. (A) The correlation between drug sensitivity (IC50) and the NT5E gene expression in pan-cancer (left) and lung cancer (right) based on IC50 data from CTRP2 and transcriptomic data from CCLE database. The numbers in parentheses of labels indicate the arrangement of related drugs based on PCC Z-score. PCC, Pearson’s correlation coefficient. (B) Heatmap illustrates the correlations of NT5E gene expression with top 20 drugs across TCGA-LUAD and GEO cohorts from BEST database. Higher-ranked drugs indicate that high expression level of NT5E predict drug resistance and vice versa. (C and D) The correlations between NT5E gene expression and IC50 of selumetinib (C) and PD318088 (D) across TCGA-LUAD and GEO cohorts in (B). P-values less than 0.05 were considered statistically significant and less than 0.01 were considered highly significant.
Discussion
Lung cancer remains the leading malignant neoplasm, causing the highest number of cancer-related
deaths globally, with a significant incidence in both sexes4,43. Despite the progress in early detection and the development of various treatments including chemotherapy, radiotherapy, immunotherapy, and targeted therapies, LUAD is often not identified until it has reached an advanced stage. This late diagnosis is associated with a low five-year survival rate and a generally poor prognosis, primarily because of LUAD’s aggressive nature and the limited availability of effective treatment targets43,44,45. Consequently, there is an urgent need to map out the molecular pathways that drive LUAD metastasis and to identify robust prognostic biomarkers for this malignancy.
In this study, we utilized bioinformatics to analyze public databases, including TCGA, GTEx, TIMER, UALCAN and HPA, uncovering a significant upregulation of the enzyme NT5E at both the mRNA and protein levels in LUAD tissues compared to normal lung tissues. Survival analyses showed that patients with higher NT5E expression had a notably lower survival rate than those with lower expression levels. Furthermore, our findings revealed a significant association between increased NT5E expression and advanced clinical staging as well as an enhanced metastatic propensity. These results align with existing literature, suggesting that NT5Emay act as an oncogene, facilitating the malignant progression across a spectrum of cancers16,20. The results highlight NT5E’s potential as a novel independent prognostic biomarker for LUAD, paving the way for the development of innovative targeted and precision oncology strategies.
NT5E, a membrane-bound homodimer, converts extracellular AMP into adenosine, which is a key player in inflammation and hypoxia responses13. Studies have shown that NT5Eis upregulated in various human malignancies and is implicated in tumor immune evasion, making it a potential target for immune checkpoint modulation15,16. NT5E is also recognized as an immunosuppressive protein that plays a critical role in tumorigenesis, proliferation, metastasis, apoptosis evasion, and resistance to therapy15,16,18,19,46. Higher NT5Eexpression correlates with adverse prognosis, as demonstrated across studies on pancreatic19, gastric21, bladder17, and other cancers16,18, underscoring its potential as a therapeutic target. Our investigation reveals that in lung cancer tissues, both mRNA and protein levels of NT5E are significantly upregulated compared to normal lung tissues, implying a role for NT5E in LUAD proliferation. However, strategies to modulate NT5E expression need further investigation, as systemic reduction of NT5E could lead to iron deposition, which is a risk factor for tumor progression. Over the past decade, there has been a surge in the development of pulmonary drug delivery systems tailored for LUAD, with a variety of nanocarriers, such as nanoparticles, liposomes, and polymers, designed to selectively target the tumor with anticancer agents. Inhalation of NT5E-targeted drugs may represent a promising therapeutic approach for LUAD.
Understanding NT5E’s interaction network with other genes and proteins helps us to unravel its function in LUAD. Our GO and KEGG pathway analyses, which show an enrichment of immune-related pathways, substantiate NT5E’s role in immune modulation. Notably, the interplay between NT5E and immune cell infiltration in LUAD has not been previously explored. Our analyses indicate that NT5E is intricately involved in multiple pathways, particularly those pertinent to the immune system in LUAD. Moreover, we also report for the first time that high NT5E expression in LUAD is associated with increased infiltration of dendritic cells, neutrophils, CD4 + T cells, macrophages, and CD8 + T cells. A significant correlation between NT5E and a spectrum of immune cell markers is observed in LUAD indicating that NT5E could serve as a novel immune-related therapeutic target in lung adenocarcinoma.
NT5E plays a key role in LUAD patient survival by partially mediating immune cell infiltration, which shapes the tumor microenvironment. The robust positive correlation between NT5E expression and the infiltration of immune cells, particularly dendritic cells and CD8 + T cells, suggests that NT5E is crucial for mounting anti-tumor immune responses and could influence the effectiveness of immunotherapy. Furthermore, NT5E’s connection to the regulation of immune checkpoint genes such as PD-1, PD-L2, and CTLA-4 indicates its possible involvement in immune evasion, adding another layer to its role in the tumor microenvironment. However, the complex interactions of NT5E within the tumor-immune microenvironment require more in-depth research to fully understand.
Finally, our analysis of drug sensitivity data reveals a potential resistance to belinostat, vincristine, and VX-680 in LUAD cells with high NT5E expression. Conversely, a negative correlation between NT5E expression and the IC50 values of selumetinib and other combination therapies suggests that these drugs may be more effective in LUAD patients with higher NT5E levels. Here, we propose that selumetinib and PD318088, both of which are inhibitors of MEK1/2, whether employed in isolation or in synergistic combinations with other drugs, present as promising therapeutic strategies that target NT5E for the treatment of LUAD.
This study improves our understanding of how NT5E interacts with LUAD, but we recognize some limitations. Firstly, while NT5E is highly expressed in LUAD cells, its molecular basis and diverse roles in cancer development, spread, and immune system interaction need further exploration. Secondly, although we’ve identified a link between NT5E and immune infiltration in LUAD patients, the interpretation of immune profiles across different patient groups is still largely unexplored. Larger, more diverse studies are needed to confirm and build on our findings. Thirdly, the current analyses is mainly based on mRNA levels of NT5E; a more compelling argument could be constructed through protein-level assessments. Lastly, this study did not address the diagnostic and prognostic significance of NT5E in large and small cell lung adenocarcinomas, which could be valuable areas for future research. Overall, our research indicates that NT5E could be a promising prognostic and immunotherapeutic biomarker for LUAD. Additionally, our evidence supports a role of NT5E in modulating immune cell infiltration within the tumor microenvironment of LUAD. These insights could significantly enhance our understanding of NT5E’s role and its potential application in lung cancer prognosis and immunotherapy.
Data availability
All data supporting the findings of this study are available within the article and its supplementary files. Further inquiries can be directed to the corresponding author.
References
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74(1), 12–49 (2024).
The, L. Lung cancer treatment: 20 years of progress. Lancet 403(10445), 2663 (2024).
Mathieu, L. N. et al. FDA approval summary: Atezolizumab as adjuvant treatment following surgical resection and platinum-based chemotherapy for stage II to IIIA NSCLC. Clin. Cancer Res. 29(16), 2973–2978 (2023).
Megyesfalvi, Z. et al. Clinical insights into small cell lung cancer: Tumor heterogeneity, diagnosis, therapy, and future directions. CA Cancer J. Clin. 73(6), 620–652 (2023).
Ezhilarasan, D., Lakshmi, T. & Mallineni, S. K. Nano-based targeted drug delivery for lung cancer: Therapeutic avenues and challenges. Nanomedicine (Lond.) 17(24), 1855–1869 (2022).
Karuppasamy, R., Veerappapillai, S., Maiti, S., Shin, W. H. & Kihara, D. Current progress and future perspectives of polypharmacology: From the view of non-small cell lung cancer. Semin Cancer Biol. 68, 84–91 (2021).
Shah, M. P. & Neal, J. W. Targeting acquired and intrinsic resistance mechanisms in epidermal growth factor receptor mutant non-small-cell lung cancer. Drugs 82(6), 649–662 (2022).
Saleem, H. M. et al. Nanotechnology-empowered lung cancer therapy: From EMT role in cancer metastasis to application of nanoengineered structures for modulating growth and metastasis. Environ. Res. 232, 115942 (2023).
Reck, M., Remon, J. & Hellmann, M. D. First-line immunotherapy for non-small-cell lung cancer. J. Clin. Oncol. 40(6), 586–597 (2022).
Tang, B. et al. Macrophage xCT deficiency drives immune activation and boosts responses to immune checkpoint blockade in lung cancer. Cancer Lett. 554, 216021 (2023).
Doran, A. C., Yurdagul, A. Jr. & Tabas, I. Efferocytosis in health and disease. Nat. Rev. Immunol. 20(4), 254–267 (2020).
Adkar, S. S. & Leeper, N. J. Efferocytosis in atherosclerosis. Nat. Rev. Cardiol.(2024).
Gao, Z. W., Dong, K. & Zhang, H. Z. The roles of CD73 in cancer. Biomed. Res. Int. 2014, 460654 (2014).
Murphy, P. S. et al. CD73 regulates anti-inflammatory signaling between apoptotic cells and endotoxin-conditioned tissue macrophages. Cell. Death Differ. 24(3), 559–570 (2017).
Magagna, I. et al. CD73-mediated immunosuppression is linked to a specific fibroblast population that paves the way for new therapy in breast cancer. Cancers (Basel) 13(23) (2021).
Li, H. et al. CD73/NT5E is a potential biomarker for Cancer Prognosis and Immunotherapy for multiple types of cancers. Adv. Biol. (Weinh) 7(2), e2200263 (2023).
Koivisto, M. K. et al. Cell-type-specific CD73 expression is an independent prognostic factor in bladder cancer. Carcinogenesis 40(1), 84–92 (2019).
Chen, Y. H., Lu, H. I., Lo, C. M. & Li, S. H. CD73 promotes tumor progression in patients with esophageal squamous cell carcinoma. Cancers (Basel) 13(16) (2021).
Yu, X. et al. CD73 induces gemcitabine resistance in pancreatic ductal adenocarcinoma: A promising target with non-canonical mechanisms. Cancer Lett. 519, 289–303 (2021).
Wang, H. et al. NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. Br. J. Cancer 106(8), 1446–1452 (2012).
Yuan, J. et al. Current strategies for intratumoural immunotherapy - beyond immune checkpoint inhibition. Eur. J. Cancer 157, 493–510 (2021).
Alam, M. S. et al. CD73 is expressed by human regulatory T helper cells and suppresses proinflammatory cytokine production and Helicobacter felis-induced gastritis in mice. J. Infect. Dis. 199(4), 494–504 (2009).
Neo, S. Y. et al. CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J. Clin. Invest. 130(3), 1185–1198 (2020).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38(6), 675–678 (2020).
Li, C., Tang, Z., Zhang, W., Ye, Z. & Liu, F. GEPIA2021: Integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 49(W1), W242–W246 (2021).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47 (2015).
Chandrashekar, D. S. et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 25, 18–27 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596(7873), 583–589 (2021).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347(6220), 1260419 (2015).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48(W1), W509–W514 (2020).
Gyorffy, B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 181(3), 362–374 (2024).
Warde-Farley, D. et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Web Server issue), W214–220 (2010).
Szklarczyk, D. et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51(D1), D638–D646 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16(5), 284–287 (2012).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12(5), 453–457 (2015).
Rees, M. G. et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 12(2), 109–116 (2016).
Zhao, P., Li, L., Jiang, X. & Li, Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J. Hematol. Oncol. 12(1), 54 (2019).
Chan, T. A. et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 30(1), 44–56 (2019).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102(43), 15545–15550 (2005).
Nusinow, D. P. et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell 180(2), 387–402 (2020).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018).
Hirsch, F. R. et al. Lung cancer: Current therapies and new targeted treatments. Lancet 389(10066), 299–311 (2017).
Lahiri, A. et al. Lung cancer immunotherapy: Progress, pitfalls, and promises. Mol. Cancer 22(1), 40 (2023).
Yu, M. et al. CD73 on cancer-associated fibroblasts enhanced by the A(2B)-mediated feedforward circuit enforces an immune checkpoint. Nat. Commun. 11(1), 515 (2020).
Acknowledgements
We thank the staffs of the Core Facility for Cell Biology of SIBCB for technical support.
Funding
This work was supported by the Wuxi Municipal Health Commission Project (Grant No. Z202216).
Author information
Authors and Affiliations
Contributions
MZ and XW designed and conceived this project. LC, TQ, and BZ performed bioinformatics analysis and generated data. XW and MZ analyzed and interpreted data. LC and XW wrote the draft. MZ revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Our study protocols were approved by the Ethics Committee of the Affiliated Wuxi People’s Hospital of Nanjing Medical University. We confirmed that all experiments were performed in accordance with relevant guidelines and regulations. All the datasets were retrieved from the online databases, so it was confirmed that all written informed consent was obtained.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, L., Qi, T., Zhang, B. et al. NT5E (CD73) as a prognostic biomarker and therapeutic target associated with immune infiltration in lung adenocarcinoma. Sci Rep 15, 4340 (2025). https://doi.org/10.1038/s41598-025-88964-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88964-8