Abstract
This study aims to refine a radiomics-based diagnostic approach for detecting neonatal respiratory distress syndrome (NRDS) and examines the influence of rib suppression on the diagnostic precision of radiomics models using neonatal chest X-ray (CXR) images. A total of 138 CXR images were collected in this study. The data was partitioned into training and validation subsets based on chronological order. We applied rib suppression to the CXR images and extracted and analyzed radiomic features from lung regions both before and after rib suppression. This approach was designed to identify NRDS, develop radiomics models, and assess the impact of rib suppression on model performance. To establish these radiomics models, six machine learning models were utilized in the study. The performance was evaluated using the area under the receiver operating characteristic curve (AUC). On the validation set, the models demonstrated significant improvements after rib suppression. Specifically, the Gradient Boosting Machine (GBM) achieved an AUC of 0.781 post-suppression compared to 0.556 pre-suppression. Notably, Linear Discriminant Analysis (LDA) and Logistic Regression (LR) performed particularly well when combining features from both scenarios, achieving AUCs of 0.762 and 0.756. The results indicate the feasibility of developing radiomics models for diagnosing NRDS and highlight the enhancement in model performance due to rib suppression. This study provides a promising new method for the imaging diagnosis and prognosis evaluation of neonatal respiratory distress syndrome, showcasing the potential of radiomics in pediatric imaging.
Similar content being viewed by others
Introduction
Premature delivery poses a significant global health concern. Infants weighing less than 1500 g at birth constitute over 3.2% of births in China, with this proportion showing recent increases1. Lung dysplasia disorders, such as bronchopulmonary dysplasia and neonatal respiratory distress syndrome (NRDS), are prevalent among premature infants. NRDS, a leading critical condition among newborns globally, presents a substantial threat to infant survival. It is defined as a deficiency in pulmonary surfactant (PS), resulting in the rapid onset of dyspnea, cyanosis, and respiratory failure within hours of birth2. It is commonly known as hyaline membrane disease (HMD) due to the presence of hyaline membranes in the lungs3,4. Compared to Europe and America, China exhibits a lower NRDS incidence, approximately 7%. Bedside X-ray radiography has traditionally been considered the preferred method for diagnosing NRDS, given the high radiation dose associated with computed tomography (CT) scans and the impracticality of conducting bedside investigations.
Radiomics is a rapidly developing field in clinical research. In recent years, radiomics has increasingly been used in the diagnosis and prognosis based on medical imaging, enabling the extraction and analysis of quantitative features from medical images5,6. Currently, many radiomics studies have been able to help radiologists delve deeper into the hidden information within images, which can assist in the clinical diagnosis, treatment, and prediction of prognosis7,8,9,10.
Some researchers have proposed using radiomic features combined with contrastive learning to detect pneumonia in CXR images, demonstrating good performance and interpretability on the RSNA pneumonia detection challenge dataset7. By analyzing patients’ CXR images, some scholars have found that radiomic features have high accuracy in identifying COVID-19, which has positive implications for supporting clinical decision-making for physicians8,11,12. Researchers have developed a scoring system using radiomics, which analyzes and calculates the radiomic score (RadScore) of patients’ CXR images, the clinical score (TBScore), and the radiological score (RLscore). By examining the correlation between the changes in these three scores, the system aims to enable quantitative monitoring of the treatment response to tuberculosis (TB)9.
However, owing to the two-dimensional (2D) nature of X-rays, chest radiographs often exhibit overlapping images of ribs, soft tissues (such as bronchi, pulmonary arteries, pulmonary veins, etc.), and artifacts. These factors may impede the radiologist’s accuracy in lesion observation and disease diagnosis10,13,14. Deep learning techniques have demonstrated promising advancements in medical image processing. Convolutional neural network (CNN) models, such as MPR-CNN, DudeNet, and CCNN, have been employed in medical image preprocessing to improve imaging quality15,16,17,18.
Based on this background, the objective of this study is to develop and validate machine learning models based on radiomic features to distinguish between CXR images of NRDS patients and non-NRDS patients. We removed the rib images from the CXR images using a deep learning approach and used them as input for the machine learning model, comparing the performance of the model before and after rib suppression.
Methods
This study was reviewed and approved by the Institutional Review Board of Shandong Provincial Maternal and Child Health Care Hospital affiliated to Qingdao University (No.2024-001). The methods conducted in this study adhered to relevant guidelines and regulations. Informed consent was obtained from all subjects and/or their legal guardians after the study’s purpose, procedures, risks, and benefits were thoroughly explained.
Study design
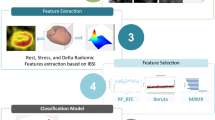
Our study introduces six machine learning models to develop radiomics models that encompass radiomic features extracted from neonatal chest X-rays both before and after rib suppression. We conducted a thorough evaluation and validation of each model. The study workflow is illustrated in Fig. 1.
Patients
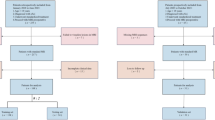
A total of 138 inpatients (including 101 neonates diagnosed with NRDS) with CXR imaging data obtained within hours after birth at the Institutional Review Board of Shandong Provincial Maternal and Child Health Care Hospital affiliated to Qingdao University between January 2022 and March 2023 were included in this study. The patients were divided into training (n = 96) and validation sets (n = 42) based on admission time. The details of the data are shown in Table 1.
Participants meeting the following criteria were extracted from patient’s medical records: (a) gestational age ≤ 37 weeks; (b) manifestation of progressive irregular respiration, dyspnea, cyanosis, and respiratory failure within a few hours of birth; (c) evidence of reduced lung transparency on X-ray, characterized by uniformly distributed fine granular shadows, with severe cases presenting a “white lung” phenomenon. All neonates meeting these criteria underwent CXR imaging within hours of birth. Participants were excluded if they had: (a) severe congenital malformations; (b) genetic metabolic diseases or chromosomal disorders; (c) liver, kidney, or thyroid diseases in either the preterm infant or their mother; (d) discontinued treatment or incomplete case data. Refer to Fig. 2 for the flowchart depicting the study population selection process.
Chest x-ray imaging acquisition
Bedside X-ray radiography is a standard procedure in the NICU, particularly when NRDS is suspected. The data utilized in this study were acquired using the uDR 370i (United Imaging Healthcare Co., Ltd, Shanghai, China) digital radiography system. Imaging was conducted in the supine anteroposterior position, with a fixed source to image receiver distance (SID) of 100 cm. Neonatal exposure parameters included a tube voltage of 65 kV, an exposure of 2.8mAs, and the use of a lead apron for radiation protection. Following parameter selection, adjust the grid size and initiate exposure. Post-exposure, verify image correctness and standardization; if not met, repeat exposure.
Bone suppression
The release of the image database with and without lung cancer nodules proposed by the Japanese Society of Radiological Technology (JSRT) opened this way for many research groups around the world19. Chest Diagnostic System Research Group (Budapest, Hungary) provided the bone shadow eliminated (BSE) version of the JSRT database (BSE-JSRT)20. BSE-JSRT database contains 247 images of the JSRT dataset, where shadows of ribs and clavicles were thoroughly removed from CXR images of the JSRT database by the special algorithms. Our analysis involved using CXR images and BSIs from this dataset, along with partially enhanced imaging data, for training and validating the rib suppression model. Then, we applied rib suppression to the neonatal chest X-rays collected in our study using models trained on public datasets. As for the self-built data utilized in our study, since it did not include BSIs, we assessed the rectal inhibition effect of this portion of the data through evaluation by two senior radiologists.
Initially, we employed publicly available code to construct the U-Net for our research21. This architecture, known for its “encoder-decoder” design, facilitates the extraction of contextual information from input data, enhancing output accuracy. Subsequently, we modified the model’s structure accordingly. The revised model incorporates a self-attention22 to learn spatial and feature information, diverging from the traditional U-Net framework reliant on cross-entropy loss. Detailed descriptions of the model, including its architecture and parameters, have been provided in the supplementary materials.
The dataset was partitioned into a training set and a validation set at a 7:3 ratio. Metrics such as cross-entropy loss, structural similarity index measure (SSIM) and mean absolute error (MAE) were employed to evaluate the efficacy of the rib suppression model and pinpoint disparities between anticipated and observed outcomes. Preprocessing was conducted on the experimental image data, resizing it to 512 pixels by 512 pixels. The CNN model described above was implemented using the TensorFlow 2.3 framework and programmed in Python 3.6. All deep learning computations were executed on a system featuring an NVIDIA Quadro P2000 (with 5GB of graphics memory) and an Intel(R) Xeon(R) CPU E5-1620 v4.
Segmentation methods
Following radiological grading and diagnostic standards for NRDS, the region of interest (ROI) in this study was delineated as the area encompassing the thoracic cavity, including lung parenchyma, cardiac structures, and diaphragmatic borders. Manual delineation was employed to segment the ROIs.
Images underwent resampling and normalization to mitigate variations stemming from equipment and scanning disparities. In the ROI delineation process, two junior radiologists utilized the RadCloud platform (Version 7.2; Huiying Medical Technology Co., Ltd., Beijing) to outline preterm neonatal CXR images23. Adhering to radiological diagnostic criteria for NRDS, they defined lung fields with smooth closed curves, encompassing cardiac and diaphragmatic boundaries. Subsequent to initial delineation, a senior radiologist reviewed the completed ROI, requesting revisions for non-compliant tracings and implementing necessary adjustments. Thirty days later, 30 images were randomly selected for ROI redefinition by the same two radiologists. Senior radiologists validated all ROI segmentations and resolved any discrepancies through consensus.
Radiomics features extraction and selection
This study employed the RadCloud platform for feature extraction and screening. PyRadiomics (version 3.1.0, https://pyradiomics.readthedocs.io/), based on Python (version 3.7.0, https://www.python.org), was utilized to extract radiomics features from medical images24. PyRadiomics standardized CXR images, involving resampling and gray-level discretization with a bin width of 25, while conforming to the Image Biomarker Standardization Initiative (IBSI, https://theibsi.github.io/)25. In our study, we extracted a total of 1316 image features from the ROI of each patient. The intraclass correlation coefficient (ICC) was used to evaluate intra- and inter-observer reproducibility. Radiomics features with good intra- and inter-observer reproducibility (ICC > 0.75) were selected for comparison between the two groups.
To enhance model reliability and reduce feature dimensionality, we initially applied the variance threshold method, setting the threshold at 0.8 to exclude features with variances less than 0.8. Next, SelectKBest univariate analysis was used to select radiomics characteristics with a significance level of P-value < 0.05 for further investigation. The best radiomics features were determined using LASSO regression on the filtered features. The LASSO approach was employed to create a penalty function that compressed the selected regression coefficients. L1 regularization served as the loss function for LASSO, with a 5-fold cross-validation error and a maximum iteration count of 1000. Finally, the most effective radiomics features were chosen for modeling26,27,28.
Development and validation of models
In this study, we utilized six machine learning models to develop and validate predictive models: Gradient Boosting Machine (GBM), Linear Discriminant Analysis (LDA), Logistic Regression (LR), Multilayer Perceptron (MLP) Classifier, Random Forest (RF), and Extreme Gradient Boosting (XGBoost). To achieve discrimination of NRDS in neonatal CXR, these six models were trained on radiomics features extracted from images both before and after rib suppression. A range of validation techniques were employed to evaluate the performance of these models and identify the most effective one. Specifically, we compared the performance of each machine learning model across three groups: A (After rib suppression), B (Before rib suppression), and C (Combined A and B). ML analysis was performed using the Python software Scikit-learn (version 1.0.2, https://scikit-learn.org). The discriminative performance of the developed radiomics models was assessed using a comprehensive set of evaluation metrics, including receiver operating characteristic (ROC) curves, area under the curve (AUC), sensitivity, specificity, precision, and F1-Score29. These metrics collectively provided a thorough evaluation of the models’ ability to accurately distinguish NRDS cases from controls.
Result
Bone suppression
The results of rib suppression on adult CXR images by the model are depicted in Fig. 3. Throughout the training and validation process, we evaluated the network’s performance using metrics such as cross-entropy loss, MAE, and SSIM. Detailed results are provided in Table S1 in the Supplementary Material.
We aimed to apply a rib suppression model, developed using adult CXR images, to neonatal CXR images. Due to the absence of matching BSIs in neonatal CXRs, the rib suppression effect was assessed by two senior radiologists and two junior radiologists. They unanimously agreed that the BSIs generated by the model performed well and were beneficial for image interpretation and diagnosis. Figure 4 shows a comparison of neonatal CXR images before and after rib suppression.
Radiomics features extraction and selection
Leveraging PyRadiomics technology, 1,316 radiomics features were extracted from the CXR images, 1,036 of 1,316 radiomics features remained following ICC analysis (ICC > 0.75) within the Region of Interest (ROI). From these 1,036 features, 265 were selected using a variance threshold approach, with a threshold set at 0.8. The SelectKBest method was subsequently employed to further reduce this to 57 features. Finally, the LASSO technique was employed to identify the optimal set of 21 features for the model. The statistics from the LASSO analysis are illustrated in Fig. 5. Table S2 in the supplementary material provides a detailed breakdown of the number of features and their respective groupings at each stage of the feature extraction and selection process, offering an overview of how features were categorized and the final count of features selected for use in the model.
Results of machine learning models
After applying rib suppression, the performance of six machine learning models significantly improved, as quantified by the AUC metric. The AUC scores for GBM, LDA, LR, MLP, RF, and XGBoost were 0.781, 0.725, 0.738, 0.744, 0.759, and 0.769 respectively. In contrast, before rib suppression, the corresponding AUCs were notably lower at 0.556, 0.625, 0.625, 0.638, 0.647 and 0.544.
When considering combined features from both before and after rib suppression datasets, the AUCs for GBM, LDA, LR, MLP, RF, and XGBoost were 0.731, 0.762, 0.756, 0.762, 0.544, and 0.750, indicating that the integration of these features further enhanced model performance in some cases.
Additionally, Table 2 presents a comprehensive overview of the machine learning model’s AUCs across different settings. For the training set, all models achieved high performance metrics after rib suppression, with particularly outstanding results for RF and XGBoost, achieving near-perfect scores of 0.999 and 1.000, respectively, on index A. On the validation set, the models also showed substantial improvements after rib suppression, with GBM reaching an AUC of 0.781 compared to 0.556 before suppression. Notably, LDA and LR performed especially well when combining features from both scenarios, with AUCs of 0.762 and 0.756, respectively.
The ROC curves for validation sets of each model are illustrated in Fig. 6, providing visual confirmation of the models’ discriminatory power. Figure 7 presents radar maps that we created based on various performance metrics, including sensitivity, specificity, precision, and F1-Score, for each of the models. The specific values for these metrics are detailed in Table S3 of the supplementary materials.
Discussion
Neonatal respiratory distress syndrome, a prevalent pulmonary condition in premature infants, stands as a leading cause of respiratory insufficiency and neonatal mortality in NICUs30,31,32. Its incidence rises with decreasing childbearing age, reaching nearly 93% in very early births (< 28 weeks). Although the rates decrease in late, premature, and post-menstrual births, they remain elevated, ranging from 0.3 to 11%. NRDS gives rise to various complications, including persistent pulmonary arterial hypertension, dysphoria, pneumonia, and, in severe cases, neonatal demise. Precise early identification of NRDS and the development of robust artificial intelligence (AI) diagnostic models based on bedside X-ray radiography hold significant clinical value for managing NRDS complications and predicting adverse outcomes.
X-ray, a form of radioactive imaging, poses heightened sensitivity to neonates, particularly premature infants. Despite this, its distinct advantages in lung structure assessment, coupled with significantly lower radiation doses compared to CT scans, maintain bedside CXR as a crucial and versatile imaging modality for neonatal lung evaluation10. However, interpreting neonatal CXR images demands specialized training, and the diagnostic yield is constrained, and prone to subjective interpretation. Imaging remains a rapidly evolving field in clinical research, increasingly integral in medical imaging diagnosis and prognosis. It aids radiologists in extracting quantitative features from images, offering novel avenues for imaging diagnostics and prognostics5,6. This methodology has also found application in numerous studies concerning neonatal and fetal imaging. Prayer et al. utilized radiomics techniques to extract microstructural and morphometric fetal lung characteristics from MRI, facilitating the analysis of expected lung capacity to predict postnatal abnormalities and optimize treatment for future lung development issues33. Standard ultrasound images effectively capture fetal lung texture, and their fusion with AI offers a novel approach to predicting prenatal respiratory diseases by analyzing image flow characteristics34.
Currently, limited imaging research is conducted on neonatal CXR images. This limitation arises from the scarcity of CXR data for neonates, especially premature infants. Additionally, the challenges posed by neonatal photography, including the infants’ smaller body size and tendency to move or cry during imaging, result in suboptimal image quality. Incorrect positioning and increasing artifact cases further complicate the standardization of image quality, hindering effective statistical analysis. Image heterogeneity can influence the diagnosis made by imaging physicians, leading to potential subjective bias. Extracting synthesized images of pertinent areas of interest for diagnostic classification can mitigate observer bias to some degree. Hence, establishing a reliable model for assembling image characteristics is crucial for enhancing the accuracy of human intelligence diagnosis of NRDS.
The overlap of ribs and lungs in neonatal chest imaging is the most prominent structural overlap in this region and occurs relatively uniformly. We posit that suppressing rib images notably enhances image stability and data reliability. Although dual-energy subtraction radiography can provide bone and soft tissue images separately, reducing diagnostic inaccuracies, the dual-exposure method increases radiation exposure. Additionally, this technique necessitates specialized, costly equipment, limiting its availability to only a few hospitals35.
In recent years, AI has seen widespread application in medical imaging research, demonstrating notable efficacy36,37,38,39. Building upon the Attention U-Net model, we trained a rib suppression model using an adult CXR dataset and successfully applied it to perform rib suppression on neonatal chest X-rays. The specific results are shown in Figs. 3 and 4. Moreover, the ROC demonstrated superior performance of the geometric model trained on rib-suppressed data compared to the primary dataset. Notably, CXR images exhibited enhanced geometric features post-rib suppression, resulting in improved diagnostic model performance (Figs. 6 and 7), with heightened sensitivity and heterogeneity.
This study has several limitations. Our data originate from a single center, and multi-center data are needed to validate the generalizability of the machine learning model; additionally, the sample size of this study is small, and a larger sample size is necessary to help improve the stability of the model. These limitations highlight the necessity for further research. In the future, we will continue to address these issues to enhance the stability and generalizability of our research findings.
Conclusion
The neonatal BSI-based model has the potential to clinically aid radiologists in diagnosing NRDS in premature infants, thereby expediting therapeutic development and prognostic assessments to mitigate NRDS-related complications. Currently, clinical diagnosis and management of NRDS heavily rely on clinical expertise. Our research indicates that a combined model integrating radiological features and clinical indicators offers promising adjunctive diagnostic utility, aiding primary neonatal practitioners in NRDS assessment. Further applications may involve assessing NRDS treatment efficacy in premature births and predicting adverse outcomes.
Data availability
The dataset utilized for rib suppression analysis is publicly accessible via the following URL: https://www.kaggle.com/datasets/hmchuong/xray-bone-shadow-supression/data. Additional data associated with this study can be obtained from the corresponding author upon reasonable request.
References
Lee, A. C. et al. National and regional estimates of term and preterm babies born small for gestational age in 138 low-income and middle-income countries in 2010. Lancet Global Health. 1, e26–e36 (2013).
Reuter, S., Moser, C. & Baack, M. Respiratory distress in the Newborn. Pediatr. Rev. 35, 417–429 (2014).
Wu, X. et al. Efficacy and safety of surfactant administration via thin catheter in preterm infants with neonatal respiratory distress syndrome: A systematic review and meta-analysis. Pediatr. Pulmonol. 56, 3013–3025 (2021).
Zang, R. et al. Association between Vitamin D Level and neonatal respiratory distress syndrome: A systematic review and Meta-analysis. Front. Pediatr. 9, 803143 (2022).
Mayerhoefer, M. E. et al. Introduction to radiomics. J. Nucl. Med. 61, 488–495 (2020).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 278, 563–577 (2016).
Han, Y., Chen, C., Tewfik, A., Ding, Y. & Peng, Y. Pneumonia Detection On Chest X-Ray Using Radiomic Features And Contrastive Learning. in IEEE 18th International Symposium on Biomedical Imaging (ISBI) 247–251 https://doi.org/10.1109/ISBI48211.2021.9433853 (2021)
Prinzi, F., Militello, C., Scichilone, N., Gaglio, S. & Vitabile, S. Explainable machine-learning models for COVID-19 prognosis prediction using clinical, Laboratory and Radiomic features. IEEE Access. 11, 121492–121510 (2023).
Du Plessis, T., Rae, W. I. D., Ramkilawon, G., Martinson, N. A. & Sathekge, M. M. Quantitative chest X-ray radiomics for therapy response monitoring in patients with pulmonary tuberculosis. Diagnostics 13, 2842 (2023).
Bao, L. Y., Dao, X. Y. & Du, K. Progress in the application of lung ultrasound for the evaluation of neonates with respiratory distress syndrome. JMDH Volume. 17, 1–9 (2024).
Zhang, Y. et al. Radiomics-based detection of COVID-19 from chest X-ray using interpretable soft label-driven TSK fuzzy classifier. Diagnostics (Basel). 12, 2613 (2022).
Hu, Z., Yang, Z., Lafata, K. J., Yin, F. & Wang, C. A radiomics-boosted deep‐learning model for COVID‐19 and non‐COVID‐19 pneumonia classification using chest x‐ray images. Med. Phys. 49, 3213–3222 (2022).
Li, Z. et al. Encoding CT Anatomy Knowledge for Unpaired Chest X-ray Image Decomposition. Preprint at (2019). http://arxiv.org/abs/1909.12922
Liu, Y. et al. Bone suppression of lateral chest x-rays with imperfect and limited dual-energy subtraction images. Comput. Med. Imaging Graph. 105, 102186 (2023).
Jiang, X., Zhu, Y., Zheng, B. & Yang, D. Images denoising for COVID-19 chest X-ray based on multi-resolution parallel residual CNN. Mach. Vis. Appl. 32, 100 (2021).
Sahu, A., Rana, K. P. S. & Kumar, V. An application of deep dual convolutional neural network for enhanced medical image denoising. Med. Biol. Eng. Comput. 61, 991–1004 (2023).
Ren, G. et al. Deep learning-based bone suppression in chest radiographs using CT-derived features: A feasibility study. Quant. Imaging Med. Surg. 11, 4807–4819 (2021).
Cho, K. et al. Bone suppression on pediatric chest radiographs via a deep learning-based cascade model. Comput. Methods Progr. Biomed. 215, 106627 (2022).
Shiraishi, J. et al. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 174, 71–74 (2000).
Juhász, S., Horváth, Á., Nikházy, L., Horváth, G. & Horváth, Á. Segmentation of anatomical structures on chest radiographs. in XII Mediterranean Conference on Medical and Biological Engineering and Computing 2010 (eds. Bamidis, P. D. & Pallikarakis, N.) vol. 29, pp. 359–362 (Springer Berlin Heidelberg, 2010).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional networks for biomedical image segmentation. Preprint at (2015). http://arxiv.org/abs/1505.04597
Vaswani, A. et al. Attention is All you Need.
Yayuan, G. et al. RadCloud—An artificial intelligence-based research platform integrating machine learning-based radiomics, deep learning, and data management. JoAIMS 2, 97–102 (2021).
Van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017).
Zwanenburg, A. et al. The image Biomarker Standardization Initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338 (2020).
Li, Z. et al. The feasibility of MRI-based radiomics model in presurgical evaluation of tumor budding in locally advanced rectal cancer. Abdom. Radiol. 47, 56–65 (2022).
Karalar, H., Kapucu, C. & Gürüler, H. Predicting students at risk of academic failure using ensemble model during pandemic in a distance learning system. Int. J. Educ. Technol. High. Educ. 18, 63 (2021).
Dai, H. et al. Considerable effects of imaging sequences, feature extraction, feature selection, and classifiers on radiomics-based prediction of microvascular invasion in hepatocellular carcinoma using magnetic resonance imaging. Quant. Imaging Med. Surg. 11, 1836–1853 (2021).
Parmar, C., Grossmann, P., Bussink, J., Lambin, P. & Aerts, H. J. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 5, 13087 (2015).
Sweet, D. G. et al. European consensus guidelines on the management of respiratory distress syndrome: 2022 update. Neonatology 120, 3–23 (2023).
Rocha, G., Rodrigues, M. & Guimarães, H. Respiratory distress syndrome of the preterm neonate—placenta and necropsy as witnesses. J. Maternal-Fetal Neonatal Med. 24, 148–151 (2011).
Donda, K. et al. Trends in epidemiology and outcomes of respiratory distress syndrome in the United States. Pediatr. Pulmonol. 54, 405–414 (2019).
Prayer, F. et al. Fetal MRI radiomics: Non-invasive and reproducible quantification of human lung maturity. Eur. Radiol. 33, 4205–4213 (2023).
Du, Y. et al. Ultrasound-based radiomics technology in fetal lung texture analysis prediction of neonatal respiratory morbidity. Sci. Rep. 12, 12747 (2022).
Yamazaki, A., Koshida, A., Tanaka, T., Seki, M. & Ishida, T. Development of Artificial Intelligence-based dual-energy subtraction for chest radiography. Appl. Sci. 13, 7220 (2023).
Gichoya, J. W. et al. AI recognition of patient race in medical imaging: A modelling study. Lancet Digit. Health. 4, e406–e414 (2022).
Rajpurkar, P. & Lungren, M. P. The current and future state of AI interpretation of medical images. N. Engl. J. Med. 388, 1981–1990 (2023).
Sun, L. et al. Few-shot medical image segmentation using a global correlation network with discriminative embedding. Preprint at (2020). http://arxiv.org/abs/2012.05440
Zhang, J., Wu, J., Zhou, X. S., Shi, F. & Shen, D. Recent advancements in artificial intelligence for breast cancer: Image augmentation, segmentation, diagnosis, and prognosis approaches. Sem. Cancer Biol. 96, 11–25 (2023).
Funding
This study was supported by Shandong Provincial Natural Science Foundation (No. ZR2024MH223), the Chinese Association for Maternal and Child Health Studies (No. 2023CAMCHS003A17) and Shandong Provincial Medical and Health Science and Technology Project (No. 202425021067).
Author information
Authors and Affiliations
Contributions
Yudong Wang: writing original draft preparation, formal analysis. Zongjin Qu: formal analysis, review and editing. Zhengjun Dai: software, validation. Yuhong Li: methodology. Yanyan Liu & Wei Wang: data curation. Lianxiang Xiao: conceptualization, methodology, funding acquisition. Yi Zhang: conceptualization, project administration, review and editing, funding acquisition. All authors reviewed the manuscript. Funding
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Qu, Z., Dai, Z. et al. Rib suppression-based radiomics for diagnosis of neonatal respiratory distress syndrome in chest X-rays. Sci Rep 15, 4416 (2025). https://doi.org/10.1038/s41598-025-88982-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88982-6