Abstract
Grass pea (Lathyrus sativus L.) stands out as an excellent choice for sustainable agriculture, thanks to its favorable agronomic characteristics, including a robust root system that penetrates deeply into the soil and its resilience against various biotic and abiotic stressors. In this study, dry-matter yield and seed yield of 16 grass pea genotypes were evaluated in rain-fed conditions at “Gachsaran”, “Mehran”, “Kuhdasht”, and “Shirvan-Chardavol” locations in Iran for three consecutive years. The experimental field trials were carried out using a randomized complete block design, and each experimental setup was replicated three times. The descriptive statistics showed a mean value of 4.030 (ton/ha) and 1.530 (ton/ha), with phenotypic coefficients of 54.77 and 61.56 for dry-matter yield and seed yield, respectively. The projection of geographical, climatic, and edaphic variables into yield measurements depicted remarkable divergence among the four studied environments. Elevation exerts a greater influence on both dry matter and seed yields in the Mehran location as compared to other environments. The climatic factors of rainfall and relative humidity played an important role in “Gachsaran” and “Shirvan-Chardavol”, respectively. While for seed yield, the temperature-related attributes were more significant in the “Mehran” location. Low broad-sense heritability was observed, and the R2 for genotype-environment interaction showed the existence of GEI for dry-matter yield (0.126) and seed yield (0.223). Both AMMI1 and AMMI2 could recognize unstable genotypes from other ones, and both AMMI’s identified genotypes G10 and G3 as high-yielding and stable genotypes. BLUP-based stability indices revealed G10 and G13 as superior genotypes for seed yield and dry-matter yield, respectively. Three and two mega-environments were identified using a GGE biplot for dry-matter yield and seed yield. For dry-matter-identified mega-environments, the G1, G13, and G2, and for seed-yield-recognized mega-environments, the G10 and G15 can be introduced. “Mehran” and “Gachsaran” out of the studied locations possessed diverse distributions considering dry-matter yield and seed yield and for further GE interaction studies, it is better to establish adaptability trials in these locations. The study concludes that for the promotion of sustainable agriculture in rain-fed regions, taking into account the influence of environmental factors, cultivation of the identified grass pea genotypes holds promise.
Similar content being viewed by others
Introduction
Lathyrus sativus L., commonly referred to as grass pea, is a leguminous plant known by various names, including white pea, chickling pea, Indian pea, cicerchia, white vetch, chickling vetch, and blue sweet pea. The production of this crop has been firmly established in several regions of Iran, serving as a fundamental source of food for both humans and animals. It is used for both feeding livestock and as a grain1. The grass pea is a resilient plant that may thrive in desert settings due to its ability to adapt to both cold and hot seasons with short growth cycles. The advantageous characteristics of this crop, such as increased yield, high protein content, ability to fix nitrogen, and tolerance to drought, salinity, and waterlogging, highlight its importance in promoting crop rotation, improving soil quality, and addressing issues related to weeds, pests, and diseases2. Grass pea cultivation spans across 1.5 million hectares, primarily in South Asia and Sub-Saharan Africa. However, its growth is declining in Mediterranean regions. The increasing global demand for legume protein highlights the potential of grass pea in cropping systems in marginal environments, such as those found in Mediterranean regions. The rusticity of grass pea makes it particularly suitable for these conditions3,4. Grass pea cultivation has been successfully implemented and expanded throughout Iran. Significant grass pea cultivation regions are situated in the Northwestern and Southern provinces of Iran. Owing to the extensive geographical range of production sites, there is considerable variation in edaphoclimatic conditions among different locations. The presence of genotypes × environment interactions (GEI) in crops like grass pea results in varied phenotypic manifestations of features due to the variability of environments5. The presence of GEI poses significant challenges to the enhancement of grass pea breeding. It hinders the identification of better genotypes with high yields and stability, as well as the prescription of genotypes suitable for specific agro-climatic zones6. Therefore, to mitigate the negative effects of Genotype-Environment Interaction (GEI) on the outcomes of breeding programs, breeders and researchers engaged in these initiatives must attain a thorough comprehension of GEI and precisely assess its magnitudes.
Various techniques have been documented in the literature to investigate adaptability and stability in multi-environment trials. A frequently employed approach for investigating genotype stability is the Additive Main Effects and Multiplicative Interaction (AMMI) method, as outlined by Gauch7. The AMMI model, along with ANOVA, generates AMMI1 and AMMI2 biplots. The AMMI1 biplot illustrates the genotypic means and their correlation with the first principal component analysis (PCA), while the AMMI2 biplot depicts the associations between genotypes and the first two PCAs8. An alternative method for assessing stability is by the use of genotype + genotype-by-environment (GGE) biplot analysis, as described by Yan et al.9. The GGE biplot offers a visual depiction of G × E effects, enabling the detection of trends, correlations, and outliers. This facilitates the interpretation and discussion of data. The AMMI and GGE models are well recognized as two prominent stability approaches, particularly when two principal components (PCs) hold major importance. Currently, plant breeding programs widely employ stability analysis methodologies that rely on mixed models (REML/BLUP)10,11. The REML approach is utilized to assess the variance components and genetic factors, while BLUP is considered the optimal selection procedure12. Within the framework of mixed models, certain stability parameters, such as the harmonic mean of the genotypic values (HMGV), the relative performance of genotypic values (RPGV), and the harmonic mean of the relative performance of the genotypic values (HMRPGV), can be computed to assess both yield stability and adaptability simultaneously13.
In this study, METs were employed to assess a cohort of recently developed grass pea lines, focusing on the examination of forage and seed yield, as well as adaptability in the rain-fed conditions of Iran. Additionally, the integration of geographical characters and climatic attributes into the yield data aims to elucidate environmental factors influencing grass pea yield.
Materials and methods
Plant materials, trial sites, and measured traits
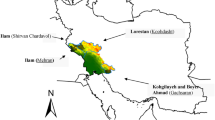
The field study was conducted at four locations, Gachsaran, Mehran, Kuhdasht, and Shirvan-Chardavol—over three consecutive years (2018–2019, 2019–2020, and 2020–2021). These locations, situated in three geographically diverse provinces of Iran (Fig. 1), were categorized as semi-warm regions based on climatic attributes. Monthly precipitation as well as soil physio-chemical properties of studied regions has been presented in Supplmentary Tables S1 and S2. The evaluation encompassed agro-morphological traits of 16 grass pea genotypes in 12 rain-fed environmental conditions. The plant materials used (Table 1) comprised seven grass pea inbred lines originating from Greece, Hungary, Nepal, Morocco, and Bangladesh, along with seven inbred lines of unknown origin. Additionally, one grass pea line from ICARDA (International Center for Agricultural Research in Dry Areas) and one inbred line (serving as a control) from Iran were included in the study. This germplasm was sourced from the gene bank section of DARI (Dryland Agriculture Research Institute) in Iran following the institute’s protocol for plant material collection and use. Prior to initiating the study, all necessary permissions for the collection and use of plant materials were obtained in compliance with institutional, national, and international guidelines and legislation. The research team worked closely with local authorities and communities to ensure sustainable collection methods that minimize impacts on natural habitats and promote the conservation of biodiversity. Documentation related to collection permits, Material Transfer Agreements (MTAs), and ethical approvals are maintained by the DARI (Dryland Agriculture Research Institute) and are available upon request.
In each environment (year × location), the research employed a randomized complete block design, incorporating three replications. Individual plots were comprised of four rows, each measuring 4.5 m in length, and spaced 25 cm apart. A consistent seeding rate of 150 seeds per m2 was maintained across all environments. Field practices, including weed control, were executed manually during crop growth and development. The dry matter yield at the 50% flowering stage and seed yield at physiological maturity (kg/ha) were calculated by extrapolating the yields obtained from the individual plots to hectares.
Additive multiplicative mean interaction
Firstly, the data was tested for normality distribution by the Anderson–Darling test and checked for outliers, then the Levene’s test was used for homogeneity of variance test to confirm the homogeneity of individual error mean squares. Subsequently, AMMI analysis was illustrated through biplot graphs, encompassing the AMMI1 biplot where the abscissa represents the average grain yield data of genotypes and average data of environments, while the ordinate represents the effects of interaction (IPCA1). The AMMI2 biplot portrays estimates of IPCA1 and IPCA2 on the abscissa and ordinate, respectively. The AMMI analysis was conducted using the R software with the “performs_ammi” function from the metan package14. The AMMI model for analyzing yield data in grass pea is expressed by the following equation:
Here, yij represents the yield response of the ith genotype in the jth environment, m is the grand mean, \({\alpha }_{i}\) and \({\tau }_{j}\) represent the ith genotypic effect and the jth environment effect, respectively. The term \(\sum_{k=1}^{p}{\lambda }_{k}{\alpha }_{ik}{t}_{jk}+{\rho }_{ij}+{\varepsilon }_{ij}\) models the multiplicative genotype × environment interaction effect, where \({\lambda }_{k}\) stands for the singular value for the kth interaction principal component axis (IPCA), \({\alpha }_{ik}\) is defined as the ith genotype eigenvector for axis k, tjk is the jth environment eigenvector for axis k, \({\rho }_{ij}\) stands for the residual not explained by the IPCAs used in the model, and \({\varepsilon }_{ij}\) is regarded as the error relevant to the model15.
Best linear unbiased predictor (BLUP)
The significance of each effect for the studied traits was assessed using the likelihood ratio test (LRT), employing a two-tailed chi-square test with one degree of freedom. So, for each environment, The traits were initially incorporated into a linear mixed-effect model, wherein environment and environment × genotype were treated as random effects, and genotype was considered a fixed effect, following the approach outlined by Olivoto et al.15. The standard linear mixed model, as defined by Piepho16, was computed using the function gamem_met from the metan package.
In this equation, y represents a response variable vector, b is a vector denoting fixed effects, u is a vector indicating random effects. The matrices X and Z consist of 0s and 1s, with X linking y to b and Z linking y to u. Additionally, e is a vector representing random errors.
After MLM analysis, it is assumed that genotype and G × E are random effects15 to predict genetic parameters using argument “genpar” in function gamem_met. By using “blup_indexes” from package “metan”, BLUP-based stability indices12 involved estimating HMGV (to deduce both yield and stability), RPGV (to examine mean yield and genotypic adaptability), and HMRPGV (to assess stability, adaptability, and yield concurrently).
where n is the number of environments (n = 6); GVij is the genetic value of ith genotype in jth environment where \({GV}_{ij}={\mu }_{j}+{g}_{i}+{ge}_{ij}\), µj is the average of jth environment, gi is the BLUP value of ith genotype, and geij is the BLUP value of the interaction between ith genotype and jth environment; Mj is the mean grain yield in the jth environment.
GGE biplot analysis
Model 1, which relies on singular value decomposition (SVD) of tester-centered data derived from the first two principal components, was chosen for generating diverse biplots. This model is applicable to datasets where all testers share the same unit, as exemplified by a genotype-by-environment table for a single trait such as seed yield in this investigation. The model is represented by the equation: Yij−u−βj = gi1 e1j + gi2 e2j + εij. In this equation, Yij signifies the expected seed yield of genotype (entry) i in location (environment) j; µ is the overall mean of the observations; βj is the main effect of location j; g1i and e1j are the primary scores for the ith genotype in the jth environment, respectively; gi2 and e2j are the secondary scores for the ith genotype in the jth environment, respectively; and εij is the residual not explained by primary and secondary effects9. Subsequently, the GGE biplot model constructs a biplot by dispersing gi1 and gi2 for genotypes and e1j and e2j for environments, through singular value decomposition. This is in accordance with the equation Yij−μ−βj = λ1ξi1η1j + λ2ξi2η2j + εij. In this equation, λ1 and λ2 are the largest eigenvalues of the first and second principal components, PC1 and PC2, respectively; ξi1 and ξi2 are the eigenvalues of the ith genotype for PC1 and PC2, respectively; and η1j and η2j are the eigenvalues of the jth environment for PC1 and PC2, respectively9. The GGE model is fitted using the ‘gge’ function in the ‘metan’ package. Subsequently, the ‘plot()’ function is employed to generate a biplot, with the type of biplot determined by the ‘type’ argument (type = 3 for which-won-where, and type = 10 for the relationship among environments).
Geographical, climatic, and edaphic data applying
The ordination method was used to find the effect of important environmental variables on grass pea dry-matter yield as well as seed yield in four regions across 3 years17. In each of the 12 environments, the climatic factors, including minimum and maximum temperature, absolute temperature, relative humidity, and rainfall were acquired by synoptic meteorological stations. The usual tests to check the quality of the data, including randomness test (run test), homogeneity test, outlier data check (Grubb’s test) and data normality (Kolmogrove-Smirnov) were investigated. Edaphic parameters, including sand percentage, clay percentage and silt percentage, and soil organic matter (Organic Carbon) accompanied with topographic parameters comprising elevation, longitude and latitude were investigated in four locations. Detrended correspondence analysis (DCA) showed that for all variables, the resulting gradient length is suitable for linear ordination (Linear response) by PCA (principal component analysis) method. Then, all collected values were converted to standard values and afterward incorporated into both dry-yield and seed yield data analysis using CANOCO software version 4.518.
Results
Discrimination among test environments
The descriptive statistics for the DY and SY data of 16 grass pea genotypes during 3 consecutive years in 4 locations showed the existence of considerable variation among genotypes (Table 2). Accordingly, the grand means for DY and SY were 4.030 (ton/ha) and 1.530 (ton/ha) with phenotypic coefficients of 54.770 and 61.560, respectively. In the present study, the minimum value of DY was seen in E7 (Mehran in the first year) for G6, while its maximum value belonged to G13 in E2 (Gachsaran in the second year). Vice versa for DY, the maximum value of SY was found for G10 in E7 (Mehran in the first year) and the minimum value of SY was detected for G16 in E12 (Shirvan and Chardavol in the third year) (Table 2). PCA analysis based on geographical, climatic, and edaphic data revealed the first two PCs explained 95.7% of the total variation. The distribution of studied environments in a bi-plot span showed variability among studied locations regarding DY (Fig. 2A) and SY (Fig. 2B) of grass pea. Among studied locations, “Shirvan Chardavol” possessed maximum content of organic carbon (OC) in its soil texture.
As shown in Fig. 2, the OC content had a negative correlation with temperature and a positive correlation with rainfall. Interestingly, among studied locations, “Mehran” more than other locations has been influenced by elevation for both DY and SY (Fig. 2).
Regarding the DY of grass pea, the climatic factors of rainfall and relative humidity played such an important role in “Gachsaran” and “Shirnan-Chardavol”, respectively (Fig. 2A), while temperature factors were affective in the “Koohdasht” location. Bi-plot view for SY variable represented highest value of OC for “Shirvan-Chardavol” location (Fig. 2B). Among climatic factors influencing SY of grass pea, the temperature-related attributes were more significant in the “Mehran” location.
Additive main effects and multiplicative interaction analysis
Both DY and SY data underwent combined analysis of variance (ANOVA) and AMMI analysis, following confirmation of homogeneity of error variance through Levene’s test (p > 0.01). The mean squares obtained from the combined ANOVA indicated significant variations (p < 0.01) for environments, genotypes, and genotype-environment interaction (GEI) in the case of both DY and SY (Table 2). The AMMI analysis also revealed significant variations (p < 0.01) among the studied genotypes, environments, and GEI (Table 2). Further, the analysis demonstrated that GEI was significantly explained by the first three principal components (PCs). Specifically, the first PC contributed 33.2% and 39.6% to the total GEI of DY and SY, respectively, while the second and third PCs contributed 19.7% and 15.7% for DY and 23.1% and 16.8% for SY, respectively.
The AMMI bi-plot analysis functions as an informative tool for evaluating the stability of genotypes across various field test conditions. In particular, the AMMI1 bi-plot depicts genotypes on the ordinate and the abscissa, illustrating the additive main effect of genotypes and the effects of the interaction between genotype and environment, as shown in Figs. 3 and 4. In Fig. 3A, the abscissa represents the yield means of the examined grass pea genotypes, while the ordinate reflects the scores of the Interaction Principal Component Axis 1 (IPCA1) for each grass pea genotype. The origin, situated at the intersection of both axes, symbolizes the average yield and zero effects of genotype and environment interaction. In this study, dry yield (DY) means on the right side of the abscissa axis were greater than those on the left side (Fig. 3A). Deviations from the origin along the ordinate axis indicated environmental influences on DY. Genotypes positioned farther away from the abscissa axis suggested greater environmental effects on dry yield means, indicating less stability (Fig. 3A).
Consequently, G13, G14, G11, G12, G2, G5, G10, and G15 exhibited DY above the average yield of all grass pea genotypes (Fig. 3A). Regarding stability, G15, G5, G10, and G9 were relatively close to the abscissa on the bi-plot, suggesting stability in the order of G15 = G5 > G5 > G9. Considering both DY means and stability, grass pea genotypes G10 and G15 exhibited higher DY than the mean yield and lower interaction with the environment, leading to their selection as the best-performing genotypes (Fig. 3A). About SY (Fig. 4A), genotypes including G10, G12, G1, G13, G7, and G3 possessed SY higher than the grand mean. From a point view of stability, G8, G5, G3, G13, G7, and G4 with relatively close distance to bi-plot origin could be calculated as SY stable genotypes (Fig. 4A).
As illustrated in Fig. 4A, G13, G7, G3 with either SY higher than mean yield and lower values of genotype-environment interaction can be recorded as high-yielding and stable genotypes.
In contrast to AMMI1, the AMMI2 bi-plot (Figs. 3B and 4B) employs an IPCA1 and IPCA2 score-based approach to depict the influences of genotype and environment interaction on genotype ranking. In this way, genotypes closer to the starting point of the AMMI2 bi-plot are less affected by interactions between genotype and environment, which means they are more stable across environments. Regarding DY in Fig. 3B, G10 exhibited the shortest distance from the bi-plot origin, indicating the highest adaptability among all tested genotypes. Following G10, genotypes G13 and G14 demonstrated high stability (Fig. 3B). Genotypes like G1, G2, G4, G16, G7, G11, G12, and G16, which interacted positively or negatively with environments, were positioned farther from the bi-plot origin, suggesting their unstable DY performance across different environments (Fig. 3B). For SY in Fig. 4B, G3 had the shortest distance from the bi-plot origin, followed by G4, and G2, suggesting higher stability for these genotypes. However, some genotypes like G10, G9, G1, and G15 exhibited instability across the studied environments in terms of SY.
MLM-BLUP
In this study, mixed linear model through calculation of likelihood ratio test (LRT) was implemented to test variation sources (Table 3). Results showed that the genotype effect is significant for DY and non-significant for SY. The genotype × environment interaction was also significant for both DY and SY of grass pea (Table 3). Both DY and SY showed low estimates of broad-sense heritability, as observed in the calculated statistics through REML (Table 3). Additionally, the genotypic accuracy of selection (AS) (Table 3), reflecting the correlation between predicted and observed values, was 0.808 for DY and 0.642 for SY. For both DY and SY, the genotypic CV (6.170% and 5.990%) was found to be less than half of the residual CV (22.30%, 31.200%). The coefficient of determination for genotype-environment interaction (GEIr2) also showed the existence of GEI for DY (0.126) and SY (0.223).
Multi-environment trials, as the last step of most plant breeding programs, need to be applied with a robust and concise statistical method to increase their ability to identify the correct genotypes as well as mega-environments. Hence, statistical methods with more predictive ability, like BLUP were introduced. Comparison of predicted DY performance using BLUP method (Fig. 5A) revealed that G5, G3, G2, G1, G12, G10, G15, G11, G14, and G13 have predicted means higher than the grand mean. In return, G9, G7, G3, G12, G13, G1, and G10 have predicted SY (Fig. 5B) more than the total mean. In general, G13 and G10 emerged as standout genotypes with the highest predicted means among the tested genotypes for DY and SY, respectively (Fig. 5A,B). Subsequently, BLUP-based stability indices, including HMGV, RPGV, and HMRPGV (Table 4), were computed using BLUP-derived values for both DY and SY.
This was undertaken to assess which method serves as a superior choice for selecting genotypes that are both stable and high-yielding. Genotypes G13, G14, and G15 were recognized as highly stable and productive in terms of dry matter yield, as indicated by stability parameters including HMGV, RPGV, and HMRPGV, as presented in Table 4. In the other hand, genotypes G1, G10, and G13 were assigned the first three ranks of BLUP-based stability parameters for seed yield (Table 4) and identified as stable and high seed yielding.
GGE bi-plot analysis
Singular value decomposition (SVD) unveiled the initial two principal components (PCs) associated with dry matter yield (DY), elucidating 39.52% and 17.08% of the total genotype + genotype × environment (G + G × E) variation. Similarly, these components were related to seed yield (SY), accounting for 39.09% and 22.20% of the total variation. The relationships between environments were illustrated in Fig. 6. A smaller angle between two environments signifies a higher correlation between them, indicating co-occurrence in genotype ranking or a high level of repeatability. On the contrary, a wider angle between two environments indicates a lower correlation or conflicts with genotypic performance (Habtegebriel, 2022). Categorizing test environments according to DY data (Fig. 6A) showed that E9 and E10 formed a group (Group I) with smaller angles between them, indicating similar performance and genotypic ranks among the grass pea genotypes. Similarly, comparable patterns were observed for E3, E2, E7, E11, E1, E8, E12, and E14 (Group II), as well as for E5 and E6 (Group III). Regarding SY trait (Fig. 6B), E1, E8, E2, E6, E7, E5, E12 and E11 with close angles made group I and possessed same genotype ranking, while E9 and E10 located in Group II. In this study, E4 out of the test environments had a different genotype ranking for SY trait compared with other environments.
The polygon view of the GGE bi-plot (Fig. 7) indicates the which-won-where patterns for both DY and SY attributes of 16 grass pea genotypes. The dashed lines, positioned perpendicular to the borders of the polygons, partition the bi-plots into six distinct regions.
Each region, encompassing various environments, constitutes a group of environments. The vertices of the polygon signify the genotypes demonstrating the most favorable performance within their respective groups of environments. Accordingly, for DY character (Fig. 7A), the genotypes G1, G13, G2, G4, G16, and G6 were winners, while for the SY character (Fig. 7B), the genotypes G10, G6, G11, G15, and G13 were winners.
Discussion
Field crops such as wheat and barley have been reported as well-suited for rain-fed conditions, especially when rotated with other crops such as beans and oil plants in crop rotation. The concern revolves around limitations encountered during autumn planting of beans and oil crops in rain-fed regions characterized by a cold climate. Meanwhile, crop rotation through supplying soil nutrients, improving soil structure, as well as mitigating soil erosion is mandatory to achieve sustainable agriculture. Hence, extending the variety of crops in rain-fed regions and moving onto sustainable agriculture is being highlighted in numerous reports19,20. In this regard, annual forage legumes such as Lathyrus sativus L. could be effectively utilized in cereal crop rotation21. The grass pea is a robust crop that demonstrates resilience to stress conditions and holds significant nutritional value for human consumption through seed usage as well as for animals through feeding. Therefore, breeding grass pea genotypes with remarkable performance for dry-matter yield and seed yield in rain-fed regions of Iran has become important. Yields, being a multifaceted trait, display variability influenced by both genotypes (G) and environments (E). The complexity is heightened by the challenges of genotype × environment interaction (GEI)22. The existence of notable GEI effects indicates distinct expressions of phenotypes for genotypes in diverse environments23. Similarly, the present study achieved large variations for DY and SY traits (Table 2), and revealed notably GEI effects (p < 0.01) on these traits in the METs of three consecutive years (Tables 2 and 3). The analysis of variance (ANOVA) in the present study aligns with findings reported by Ahmadi et al.5 and Vaezi et al.6, indicating a consistent observation. Specifically, the analysis indicates that significant environmental (E) effects contribute to a proportion of the total effects considerably higher than both genetic (G) and genotype-environment interaction (GEI) effects across all traits. This implies that the variations in phenotypic expressions of the traits are primarily driven by environmental factors, with genotypes playing a role in causing phenotypic differences, albeit to a lesser extent than environmental factors. In line with our ANOVA outputs, PCA analysis using climatic, geographical and edaphic variables manifested variation among four test locations and also effectiveness of each environment from specific environmental factors. In this regard, Rahman et al.24 also showed that environmental variables could drive plant species composition, distribution, and yield. While rainfall is acknowledged as the most limiting factor in rain-fed regions25, it’s important to note that the plant’s requirements for biomass and seed production extend beyond the need for water alone. Numerous factors within the soil matrix and the microclimate surrounding the plant may influence the production of sugar, the plant’s ability to cope with stresses, and overall plant maintenance. Among these factors, soil organic carbon (OC) emerges as one of the most crucial, influenced by various elements and playing a significant role in the efficiency of water consumption and nutrient utilization26. In this research, the soil OC content of the “Shirvan-Chrdavol” location (with the lowest mean temperature) was the highest. This finding highlights that, while augmented rainfall and diminished temperatures can contribute to an increase in soil organic carbon (OC) according to Zhang et al.26, the influence of temperature appears to play a more substantial role than rainfall in determining soil OC content. Supporting this perspective, researchers such as Hartley et al.27 have demonstrated the significance of temperature compared to rainfall. The rationale behind this observation is that elevated temperatures enhance the activity of microorganisms, leading to the release of carbon from the soil source and its subsequent addition to the atmosphere26. Based on our findings, attributes such as rainfall, elevation, and temperature have the potential to differentiate the four rain-fed locations from one another. Similarly, Fikre et al.28 inspected effects of climatic, edaphic and altitudinal factors affecting grass pea seed yield through five locations during 2 years and emphasized the importance of rainfall as the most considerable factor. Our research revealed that rainfall significantly influenced the variation in dry yield, while temperature played a crucial role in the variation of seed yield between locations. In the studied area encompassing four locations, we observed a negative correlation between elevation and rainfall. This aligns with the findings of Javari29, who reported that, in most cases, precipitation tends to decrease with an increase in altitude, except in situations where the slopes are oriented towards rain-producing winds.
The AMMI model stands out as a powerful statistical tool extensively employed for the analysis of genotype-environment interaction (GEI) effects30. In the present study, the application of the AMMI model allowed the significant GEI effects to be decomposed into 11 multiplicative terms (Interaction Principal Component Axes—IPCAs), with the first three demonstrating significant effects (Table 2). This suggests that the interaction between the 16 grass pea genotypes and the four locations over three consecutive growing seasons was effectively captured by IPCA1 and IPCA2, accounting for 52.9% and 62.7% of the total variation for DY and SY, respectively. The present study identified two crucial IPCAs, which prove adequate for discerning superior genotypes. Gauch and Zobel31 validated the adequate accuracy of projecting the AMMI model using the first two significant IPCAs. These significant IPCAs form the basis for conducting further stability analysis through the AMMI approach32. The AMMI model generates AMMI1 and AMMI2 bi-plots for evaluating significant multiplicative interactions of agronomic traits among genotypes and environments in multi-environment field trials. The AMMI1 bi-plot visually communicates information about the yield potential and stability performance of the assessed genotypes33.
The AMMI1 bi-plot employs the ordinate and abscissa axes to represent environmental effects and mean yields, respectively15. According to Esan et al.33 and Habtegebriel34, genotypes positioned to the right of the bi-plot origin along the abscissa axis exhibit higher yields, while those on the left side demonstrate lower yields. Additionally, the impact of environments is more pronounced as genotypes move farther away from the abscissa axis. Their outcomes align with our current study, indicating that G10 and G15 are considered superior in terms of both high dry yields (DY) and stability. Similarly, G13, G7, and G3 have been identified as superior genotypes with respect to high seed yields and stability. Furthermore, following the methodology outlined by Hossain et al.35, Ljubicic et al.36, and Habtegebriel34, the AMMI2 bi-plot in our study indicates that deviations from the bi-plot origin signify the magnitudes of genotype-environment interaction (GEI) effects. To elaborate, the greater the distance from the origin, the more pronounced the influence of GEI effects, and vice versa. Our evaluation, consistent with the approach taken by Hossain et al.35, Ljubicic et al.36, and Habtegebriel34, reveals G10 > G13 > G14 as widely adapted genotypes with high environmental stability in terms of dry yield (DY) performance. Similarly, G3 > G4 > G2 are identified as superior, stable genotypes for seed yield (SY). For further evaluation of grass pea genotypes and also for doing comparable analyses, the BLUP values related to each genotype in each environment were calculated. Our results showed that G13 and G10 had the highest predicted means among the tested genotypes for DY and SY, respectively. Parallel to previous reports37, the high-yielding genotypes identified by BLUP may not necessarily be winners in each environment. Here in, BLUP predicted means over environments resulted in similar output for just DY. Subsequently, the ranking of grass pea genotypes based on BLUP-based stability indices (Table 4) underscored the robustness of BLUP in effectively screening highly adaptable genotypes. The outcomes of BLUP analysis for the dry yield (DY) character, specifically for genotypes G13, G14, and G15, exhibited some degree of overlap with the AMMI analysis. Notably, genotypes identified by AMMI were also selected by BLUP-based indices. In contrast to the AMMI analysis, the BLUP-based indices identified genotypes G1, G10, and G13 as highly adaptable across the studied test environments for seed yield (SY) character.
The GGE bi-plot analysis stands as a reliable statistical tool for assessing test environments and genotypes, as well as offering genotype recommendations for specific environments9. As outlined by Yan and Tinker38, GGE bi-plot analysis utilizes principal component analysis (PCA) to break down the varied responses of genotypes across multiple environments. Subsequently, it projects the genotypic main effects (PC1) and genotype-environment interaction effects (PC2) onto the abscissa and ordinate axes of the bi-plot, respectively. This approach allows for a visual and straightforward assessment of genotype performances and the identification of mega-environments, as emphasized by Zhang et al.39. The illustration of six mega-environments, defined by winner genotypes positioned farthest from the bi-plot origin, is depicted in Fig. 7. Regarding GGE bi-plot analysis of DY, only the genotypes G1, G13, and G2 out of the winner genotypes determined the mega-environments I (Mehran), II (Koohdasht and Gachsaran), and III (Shirvan Chardavol), respectively. For SY, GGE bi-plot analysis showed presence of two mega-environments including I (Mehran, Koohdasht and Shirvan Chardavol) and II (Gachsaran) that were in correspondence with G10 and G15, respectively. In essence, the genotypes identified in this manner are recommended for locations encompassed within each corresponding mega-environment. Paralleled with previous reports in grass pea6, the Mehran region is distinct from other mega-environments based on DY, and so the Mehran location contributed most to the GE interaction. Similarly, Gachsaran out of other mega-environments possessed a diverse distribution considering SY. Hence, for further studies about GE interaction of DY and SY, it is better to establish adaptability trials in these locations.
Conclusion
The 3-years study of 16 grass pea genotypes in four rain-fed Iranian locales revealed their performance, adaptability, and stability. Dry yield (DY) and seed yield (SY) varied greatly among genotypes, highlighting the role of genotype-environment interaction (GEI). PCA analysis showed that geographic, climatic, and edaphic factors affected grass pea yield. Notably, soil organic carbon (OC) concentration was a key driver, with higher OC in certain places, emphasizing the complex link between soil qualities and plant performance. At different sites, rainfall, temperature, and elevation affected grass pea yield. Genotype differences and performance stability were shown by statistical tools like the Additive Main Effects and Multiplicative Interaction (AMMI) method. G10 and G15 were excellent for DY and SY, yielding high yields and being stable across environments. AMMI analysis, backed by BLUP, highlighted genetic adaptability, giving a solid foundation for breeding program genotype selection. GGE bi-plot analysis also helped identify mega-environments, providing a viable framework for genotype recommendation. The results showed that G1, G13, and G2 performed well in different mega-environments, sharpening the need for customized genotype recommendations. Grass pea breeding programs benefit from the study’s deep understanding of genotype-environment interactions. The found high-yielding and stable genotypes and environmental parameters affecting grass pea yield can guide future breeding efforts to improve this important crop’s resilience and productivity in rain-fed settings.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Unander, D. Grass Pea. Lathyrus sativus L. Promoting the conservation and use of underutilized and neglected crops, vol. 18. Econ. Bot. 56, 105 (2002).
Voltas, J. et al. Genotype by environment interaction and adaptation in barley breeding: Basic concepts and methods of analysis. Barley Sci. Recent Adv. from\nMol. Biol. to Agron. Yield Q. (2002).
Vaz Patto, M. C. et al. Lathyrus improvement for resistance against biotic and abiotic stresses: From classical breeding to marker assisted selection. Euphytica https://doi.org/10.1007/s10681-006-3607-2 (2006).
Lambein, F., Travella, S., Kuo, Y. H., Van Montagu, M. & Heijde, M. Grass pea (Lathyrus sativus L.): Orphan crop, nutraceutical or just plain food?. Planta 250, 821–838. https://doi.org/10.1007/s00425-018-03084-0 (2019).
Ahmadi, J. et al. Non-parametric measures for yield stability in grass pea (Lathyrus sativus L.) advanced lines in semi warm regions. J. Agric. Sci. Technol. 17, 1825–1838 (2015).
Vaezi, B. et al. Graphical analysis of forage yield stability under high and low potential circumstances in 16 grass pea (Lathyrus sativus L.) genotype. Acta Agricult. Slov. https://doi.org/10.14720/aas.2023.119.1.2227 (2023).
Gauch, H. G. Model selection and validation for yield trials with interaction. Biometrics 44(3), 705. https://doi.org/10.2307/2531585 (1988).
Bocianowski, J., Warzecha, T., Nowosad, K. & Bathelt, R. Genotype by environment interaction using AMMI model and estimation of additive and epistasis gene effects for 1000-kernel weight in spring barley (Hordeum vulgare L.). J. Appl. Genet. 60(2), 127–135. https://doi.org/10.1007/s13353-019-00490-2 (2019).
Yan, W. & Kang, M. S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists (2002).
Torres Filho, J. et al. Genotype by environment interaction in green cowpea analyzed via mixed models. Rev. Caatinga 30, 687–697. https://doi.org/10.1590/1983-21252017v30n317rc (2017).
de Sousa, A. M. C. B., Silva, V. B., Lopes, Â. C. A., Gomes, R. L. F. & Carvalho, L. C. B. Prediction of grain yield, adaptability, anstability in landrace varieties of lima bea(Phaseolus lunatus L.). Crop Breed. Appl. Biotechnol. 20, 450. https://doi.org/10.1590/1984-70332020v20n1a15 (2020).
de Resende, M. D. V. Software Selegen-REML/BLUP: A useful tool for plant breeding. Crop Breed. Appl. Biotechnol. 16(4), 330–339. https://doi.org/10.1590/1984-70332016v16n4a49 (2016).
Resende, M. D. V. de. SELEGEN-REML/BLUP: Sistema Estatístico e Seleção Genética Computadorizada via Modelos Lineares Mistos. Embrapa Florestas. (2007).
Olivoto, T. & Lúcio, A. D. C. metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 11(6), 783–789 (2020).
Olivoto, T. et al. Mean performance and stability in multi-environment trials I: Combining features of AMMI and BLUP techniques. Agron. J. 111(6), 2949–2960. https://doi.org/10.2134/agronj2019.03.0220 (2019).
Piepho, H.-P. Best Linear Unbiased Prediction (BLUP) for regional yield trials: A comparison to additive main effects and multiplicative interaction (AMMI) analysis. Theoret. Appl. Genet. 89(5), 647–654. https://doi.org/10.1007/BF00222462 (1994).
Maleki, H. H., Khoshro, H. H., Kanouni, H., Shobeiri, S. S. & Ashour, B. M. Identifying dryland-resilient chickpea genotypes for autumn sowing, with a focus on multi-trait stability parameters and biochemical enzyme activity. BMC Plant Biol. https://doi.org/10.1186/s12870-024-05463-0 (2024).
ter Braak, C. J. F. & Šmilauer, P. CANOCO Reference manual and canodraw for windows user’s guide: Software for canonical community ordination (Version 4.5). Sect. Permut. Methods. Microcomput. Power, Ithaca, New York (2002) citeulike-article-id:7231853.
Ates, S., Feindel, D., El Moneim, A. & Ryan, J. Annual forage legumes in dryland agricultural systems of the West Asia and North Africa Regions: Research achievements and future perspective. Grass Forage Sci. 69(1), 17–31. https://doi.org/10.1111/gfs.12074 (2014).
Rubiales, D. & Mikic, A. Introduction: Legumes in sustainable agriculture. Crit. Rev. Plant Sci. 34(1–3), 2–3. https://doi.org/10.1080/07352689.2014.897896 (2014).
Gonçalves, Letice, Rubiales, D., Bronze, M. R. & Vaz Patto, M. C. Grass Pea (Lathyrus sativus L.)—A sustainable and resilient answer to climate challenges. Agronomy 12(6), 1324. https://doi.org/10.3390/agronomy12061324 (2022).
Pour-Aboughadareh, A., Khalili, M., Poczai, P. & Olivoto, T. Stability indices to deciphering the genotype-by-environment interaction (GEI) effect: An applicable review for use in plant breeding programs. Plants 11(3), 414. https://doi.org/10.3390/plants11030414 (2022).
da Silva, K. J. et al. Identification of mega-environments for grain sorghum in Brazil using GGE biplot methodology. Agron. J. 113(4), 3019–3030 (2021).
Rahman, I. U. et al. Environmental variables drive plant species composition and distribution in the moist temperate forests of Northwestern Himalaya, Pakistan. PLoS ONE 17(2), e0260687. https://doi.org/10.1371/journal.pone.0260687 (2022).
Beyer, M. et al. Rainfall characteristics and their implications for rain-fed agriculture: A case study in the Upper Zambezi River Basin. Hydrol. Sci. J. 61(2), 321–343. https://doi.org/10.1080/02626667.2014.983519 (2016).
Zhang, K., Dang, H., Zhang, Q. & Cheng, X. Soil carbon dynamics following land‐use change varied with temperature and precipitation gradients: Evidence from stable isotopes. Global Change Biol. 21(7), 2762–2772. https://doi.org/10.1111/gcb.12886 (2015).
Hartley, I. P., Hill, T. C., Chadburn, S. E. & Hugelius, G. Temperature effects on carbon storage are controlled by soil stabilisation capacities. Nat. Commun. https://doi.org/10.1038/s41467-021-27101-1 (2021).
Fikre, A., Negwo, T., Kuo, Y.-H., Lambein, F. & Ahmed, S. Climatic, edaphic and altitudinal factors affecting yield and toxicity of Lathyrus sativus grown at five locations in Ethiopia. Food Chem. Toxicol. 49(3), 623–630. https://doi.org/10.1016/j.fct.2010.06.055 (2011).
Javari, M. Assessment of temperature and elevation controls on spatial variability of rainfall in Iran. Atmosphere 8(3), 45. https://doi.org/10.3390/atmos8030045 (2017).
Rodrigues, P. C., Monteiro, A. & Lourenço, V. M. A robust AMMI model for the analysis of genotype-by-environment data. Bioinformatics 32(1), 58–66. https://doi.org/10.1093/bioinformatics/btv533 (2016).
Gauch, R. H. & Zobel, W. AMMI analysis of yield trials. In Genotype-by-Environment Interaction (eds Gauch, H. G. & Kang, M.) 85–122 (CRC Press, 1996). https://doi.org/10.1201/9781420049374.ch4.
Gauch, H. G. A simple protocol for AMMI analysis of yield trials. Crop Sci. 53(5), 1860–1869. https://doi.org/10.2135/cropsci2013.04.0241 (2013).
Esan, V. I., Oke, G. O., Ogunbode, T. O. & Obisesan, I. A. AMMI and GGE biplot analyses of Bambara groundnut [Vigna subterranea (L.) Verdc.] for agronomic performances under three environmental conditions. Front. Plant Sci. https://doi.org/10.3389/fpls.2022.997429 (2023).
Hailemariam Habtegebriel, M. Adaptability and stability for soybean yield by AMMI and GGE models in Ethiopia. Front. Plant Sci. https://doi.org/10.3389/fpls.2022.950992 (2022).
Amir Hossain, Md. et al. Integrating BLUP, AMMI, and GGE Models to explore GE interactions for adaptability and stability of winter lentils (Lens culinaris Medik.). Plants 12(11), 2079. https://doi.org/10.3390/plants12112079 (2023).
Ljubičić, N. et al. Multivariate interaction analysis of Zea mays L. genotypes growth productivity in different environmental conditions. Plants 12, 2165 (2023).
Daba, S. D., Kiszonas, A. M. & McGee, R. J. Selecting high-performing and stable pea genotypes in multi-environmental trial (MET): Applying AMMI, GGE-biplot, and BLUP procedures. Plants 12, 2165 (2023).
Tinker, N., Yan, W. & Tinker, N. A. Biplot analysis of multi-environment trial data : Principles and applications Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 6, (2015).
Zhang, P. P. et al. GGE biplot analysis of yield stability and test location representativeness in proso millet (Panicum miliaceum L.) genotypes. J. Integr. Agric. 15(6), 1218–1227. https://doi.org/10.1016/S2095-3119(15)61157-1 (2016).
Author information
Authors and Affiliations
Contributions
Hamid Hatami Maleki conceptualized the study, designed the research methodology, and played a leading role in drafting and revising the manuscript. Behrouz Vaezi contributed to the experimental design, data collection, and significantly to the analysis and interpretation of data. Askar Jozeyan and Amir Mirzaei were involved in conducting the experiments, data acquisition, and preliminary data analysis. Reza Darvishzadeh contributed to the literature review, theoretical framework, and critically revised the manuscript for important intellectual content. Shahryar Dashti assisted in data interpretation, statistical analysis, and played a pivotal role in manuscript revision. Mousa Arshad, Hossein Zeinalzadeh-Tabrizi, and Mojtaba Kordrostami contributed to the experimental work, data collection, and provided logistical support. All authors reviewed the manuscript, provided feedback, and approved the final version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Maleki, H.H., Vaezi, B., Jozeyan, A. et al. Grass pea dual purpose dry matter and seed yields in rainfed conditions across diverse environments. Sci Rep 15, 4960 (2025). https://doi.org/10.1038/s41598-025-89050-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89050-9