Abstract
Toxoplasma gondii, a zoonotic protozoan parasite, affects up to one-third of the global population. It can be transmitted through consumption of raw or undercooked meat, vertical transmission, or oocysts from contaminated water, soil, or food. However, there are no reports on the molecular prevalence of T. gondii in environmental sources like soil, water, and vegetables in India. This study aimed to detect and analyze T. gondii in samples from Chandigarh city, India. A total of 100 each soil & water and 500 vegetable samples were collected and analyzed using conventional PCR assay, real-time PCR assay targeting the glycerol-3-phosphate dehydrogenase (B1) gene and real-time LAMP assay targeting both B1 and Toxoplasma gondii outer wall protein (TgOWP) genes. Results showed that 15% of water, 9% of soil, and 6.4% of vegetable samples were positive for T. gondii. Real-time PCR assay and real-time LAMP assay detected slightly higher positivity rates in water and vegetable samples. Phylogenetic analysis revealed that the T. gondii isolates clustered with those from other regions such as Iran, India, Mexico, and those found in cats, pigs, and humans. This study is the first report of T. gondii contamination in environmental sources and fresh produce in India. The findings highlight the potential risk of human infection from contaminated water, soil, and vegetables in the region.
Similar content being viewed by others
Introduction
Toxoplasma gondii (T. gondii) is a zoonotic, obligate intracellular protozoan parasite that affects nearly one-third of the global population, including most warm-blooded vertebrates1. It can be transmitted through three primary routes: ingestion of raw or undercooked meat from infected animals, vertical transmission from mother to fetus, and ingestion of oocysts via contaminated water, soil, or food1. In India, an estimated 56,737 to 176,882 children are at significant risk of congenital toxoplasmosis annually2.
As a major zoonotic pathogen and a notable cause of food- and waterborne diseases, T. gondii has garnered attention from international health organizations such as the World Health Organization, which emphasizes the need for precise epidemiological data on this parasite1. Humans and other susceptible animals are exposed to T. gondii through water contaminated with oocysts from feline feces3. These oocysts are highly resilient to various inactivation processes, including chemical treatments. Recent outbreaks of toxoplasmosis linked to water contaminated with oocysts have highlighted the need for increased awareness of safe water practices and effective remediation strategies. Waterborne outbreaks are more frequently reported in low- and middle-income countries4. The NHANES (National Health and Nutrition Examination Survey) survey (1999–2004; 2009–2010) in the USA also found that individuals consuming untreated well or tap water had a significantly higher risk of seropositivity for T. gondii compared to those consuming treated water5.
Soil contamination with T. gondii oocysts primarily results from felids, particularly young cats, which excrete millions of oocysts6. These oocysts pose a risk to humans, rodents, birds, and other intermediate hosts when soil becomes contaminated7. The prevalence of T. gondii in soil varies widely, ranging from 0% in the USA to approximately 50% in northeastern France8.
While the transmission of T. gondii via fresh produce has not been extensively studied in India, the absence of standardized techniques for detecting T. gondii DNA in vegetables and other fresh foods has hindered surveillance efforts. To date, only two outbreaks of toxoplasmosis linked to fresh produce have been reported globally9,10. Despite advancements in detection methods, the role of oocysts in T. gondii epidemiology has been underestimated due to the lack of standardized tests for environmental samples11. Further research into the presence of oocysts in various environmental matrices is essential for accurately assessing the risks posed to humans and animals through contaminated produce.
The significance of T. gondii as a zoonotic, foodborne, and waterborne pathogen underscores the critical need for comprehensive epidemiological data to identify infection sources, manage the disease, and mitigate its impacts on public health1. Unfortunately, only a few countries have conducted regular surveys to track T. gondii in humans, and even fewer have investigated its presence in animals or environmental sources12.
In India, there are currently no reports documenting the prevalence of T. gondii in soil, water, or vegetables. However, global studies increasingly recognize environmental transmission as a significant route of human infection13. While it was previously believed that contaminated meat was the primary transmission route, the high prevalence of T. gondii infection among vegetarians (e.g., Jains in India) suggests that environmental sources are also major contributors14.
This study aims to detect the prevalence of T. gondii in vegetables from open markets and environmental samples, such as water and soil, from the Chandigarh region using molecular techniques. Accurate detection of T. gondii in these sources is critical for mitigating the risk of toxoplasmosis in human populations. As the first study in India to investigate the prevalence of T. gondii in environmental samples, it represents a significant step toward understanding the parasite’s transmission dynamics and informing public health interventions.
Results
Toxoplasma gondii DNA was detected in water collected from tube wells, ponds, tap water collected from government schools, houses, canteens, gurudwaras, public parks, hospitals etc. The parasite was detected in water collected from different areas of Chandigarh which includes Dhanas, Kaimwala (all villages), sector 15, sector 42, sector 45, sector 35, Bapu Dham Colony, Industrial area, sector 12 (Table 1).
Soil samples that tested positive were gathered from various locations throughout the city. The positive samples were collected from urban slums, industrial areas, hospital, public toilet area, kitchen garden, organic farm and public park and all were confirmed to contain Toxoplasma gondii using molecular methods (Table 2).
In vegetable samples, thirty-three samples were found positive by real-time LAMP against both the genes for Toxoplasma gondii. Coriander, Coriandrum sativum (n = 5) and methi, Trigonella foenum-graecum (n = 5) were found to be highly contaminated with T. gondii followed by radish, Raphanus sativus (n = 3), cabbage, Brassica oleracea (n = 3), Beetroot & greens, Beta vulgaris (n = 2), spinach, Spinacia oleracea (n = 2), green chili, Capsicum annuum (n = 2), sprinkle onion, Allium cepa (n = 2), mint, Mentha (n = 1), lettuce, Lactuca sativa (n = 1), swiss chard, Beta vulgaris (n = 1), parsley, Petroselinum crispum (n = 1), arugula, Eruca sativa (n = 1), rape, Brassica napus (n = 1) and tomatoes, Solanum lycopersicum (n = 1). The detailed results of the positive vegetable samples of different types with molecular techniques and from different areas are given in Table 3.
Conventional PCR assay
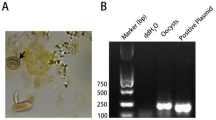
A total of 100 water samples were analysed out of which 15 (15%) were found positive for Toxoplasma gondii by conventional PCR assay. Nine out of 100 samples of soil were found to be positive for Toxoplasma gondii. All the samples showed band for 18s housekeeping gene. Of the 500 vegetable samples, 32 (6.4%) were positive for T. gondii. All the positive samples showed a band at 193 bp corresponding to B1 gene of parasite (Supplementary Fig. 1).
Real- time PCR assay
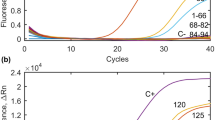
SYBR Green based Real-Time PCR assay was performed in all the water, soil and vegetables samples for B1 gene of Toxoplasma gondii. Eighteen out of 100 water samples were positive of T. gondii (18%). In case of soil samples, out of 100 analysed, 9 were found positive for the parasite. In vegetable samples, thirty-three (33, 6.6%) samples were found positive out of 500 analysed with real-time PCR assay. The samples with Ct value < 30 was considered positive whereas the samples with Ct value > 30 were considered as negative. Melt curve analysis was also performed in which the samples with fluorescence more than threshold were considered positive and below threshold were considered negative (Supplementary Fig. 2).
Real-time LAMP assay
Real-Time LAMP detected T. gondii DNA in 18 samples out of 100 (18%) analysed water samples. Both the B1 gene targeted LAMP and TgOWP gene targeted LAMP was detected same number of positive samples. Water samples collected from tube wells, ponds, tap water collected from government schools, houses, canteens, gurudwaras, public parks, hospitals etc. were found to be contaminated with T. gondii oocysts (Supplementary Fig. 3).
Overall, the positivity rate of Toxoplasma gondii in water were 15% with conventional PCR assay and 18% with real-time PCR assay & real-time LAMP assay, soil 9% and vegetables 6.4% with conventional PCR assay and 6.6% with real-time PCR assay & real-time LAMP assay (Fig. 1).
Detection of Toxoplasma gondii DNA in water, soil and vegetable samples using three molecular techniques (Blue color: Conventional PCR, Red Color: Real-Time PCR, Green Color: Real-Time LAMP, n: Total number of samples analyzed for specific category, Numbers above the bars: Total no. of positive samples by technique).
Sanger sequencing and phylogenetic analysis
Sanger sequencing was performed for the 12 positive T. gondii isolates from water (n = 4), soil (n = 2) and vegetable samples (n = 6) for the B1 gene. The results for sequencing were obtained in FASTA files which were analysed using FinchTV software. The sequences were submitted to NCBI GenBank database and the accession number obtained were OQ448549, OQ448550, OQ448551, OQ448552. OQ448553. OQ448554 (vegetables samples) OP709775, OP709776 (soil samples) OQ448545, OQ448546, OQ448547, OQ448548 (water samples). MEGA software was used for phylogenetic proximity between the different T. gondii isolates (n = 12) obtained from the study with other non-typing B1 gene sequences (n = 45) available on the NCBI database. The phylogenetic analysis revealed that the T. gondii isolates from environment and vegetables from our study were clustering with other partial B1 gene sequences of cat, sheep, pig T. gondii isolates from Iran, Mexico, India and also with patient’s T. gondii isolate from North Indian region. Moreover, the isolates from current study also clustered with ME49 reference strain and RH reference strain of T. gondii (Fig. 2). The statistical analysis performed on the positivity rate of water samples using different molecular techniques from different water sources showed significance with p value < 0.0001 and F value 81.00.
Molecular phylogenetic analysis of different types of Toxoplasma gondii, using the B1 gene region by the Neighbour joining statistical method. The isolates within square box represents the T. gondii sequences from the present study (VD1, VD2, SL1, WT4, VD5, VD6, SL2, SL3, WT3, VD3, WT2, WT1, VD4), while the blue star denotes reference sequences (ME49, VEG, Reference sequence (RH)). The unsquared black star marked represent pigs and cats B1 gene sequences from India (PTH3, PTH2, PTH1, Cat1). Other represent similar B1 gene sequences from the GenBank. Percentages (bootstrap values) – Indicates confidence estimate in the branch.
Discussion
Toxoplasma gondii (T. gondii) is obligate protozoan with wide intermediate host range such as warm-blooded animals including humans, wild mammals etc. and is of significant veterinary importance15,16. It affects up to 30% of people worldwide. The infection is often contracted by consuming contaminated water or food with sporulated oocysts from cats infected with T. gondii, or by eating raw meat containing bradyzoites or tissue cysts17,18.
Chandigarh is near the Shivalik Range of the Himalayas (North India) and has an area of around 114 km2 and is situated on the border with Punjab and Haryana. Climate plays a vital role in the survival of Toxoplasma gondii oocysts in the environment. In regions with warm and moist climates, oocysts can survive for extended periods, increasing the risk of infection. Conversely, in colder and drier climates, oocysts may perish faster, leading to a lower prevalence of the parasite.
Hygiene practices can significantly impact the spread of T. gondii. Poor sanitation, improper waste disposal, and limited access to clean water increase the likelihood of environmental contamination with oocysts, leading to higher infection rates. At present, the Union Territory of Chandigarh has 23 villages which are divided into the sectoral and non-sectoral villages. There are around a million people residing in Chandigarh, and mostly residing in metropolitan areas. Although they are considered to be non-sectarian villages connected to the city, Kansal and Maloya have less developed infrastructure and socioeconomic status than the city proper and Rajiv slum colony is located in Sector 38 West. Chandigarh has seen an increase of slum settlements in recent years, particularly on the city’s outskirts where low-income families prefer to live owing to the availability of affordable homes19,20,21.
Oocysts of T. gondii have been identified in water samples across various regions globally such as France, Poland, Germany, Russia, Bulgaria, Scotland, Turkey, Brazil, and Ecuador7. The current study represents the detection of T. gondii detection in environmental water samples collected from the Chandigarh region using highly sensitive molecular techniques for first time from India according to literature review. In the current study, Conventional PCR for B1 gene of T. gondii detected 15% samples positive for the parasite whereas real-time PCR and real-time LAMP for B1 and TgOWP detected 18% samples positive for T. gondii. The findings from current study were similar to other studies such as Pinto-Durate et al., 20,222 which found T. gondii in 9.1% of water samples collected from Quindio River basin, Colombia21. Hernandez-Cortazar et al., 2017 found 5.4% of the drinking water samples positive for T. gondii from an endemic region in Southern Mexico22. Adamska. 2018 detected T. gondii in 19.4% of water samples collected from natural surface water bodies i.e., lakes, rivers and ponds in Poland20. Lass et al., 2022 investigated a total of 214 water samples taken from wastewater treatment plants, slaughterhouse and rivers in China. They detected T. gondii type I in 4 samples (1.9%) via real-time PCR and multilocus genotyping23. There may be a number of reasons of this the variable incidence (9–58%) of T. gondii DNA in water samples across various studies world-wide. Firstly, the source of water in these studies are different and differs from natural environmental sources like rivers, lakes, ponds; to sewage and treated drinking water. These different water sources have diverse environmental, and climatic conditions, as well as range of temperature, pH, and concentration of disinfectants which influence the diverse makeup of host populations. These variable factors which are crucial to the survival of T. gondii oocysts cause differences in potential of water sources being source of T. gondii.
In the present study, most of the positive samples for Toxoplasma were from the village areas and urban slums of Chandigarh city, India. This data would suggest that not all treatments are efficient or that treated water may get polluted, which is especially common in nations with poor water supply infrastructure. As a result, in addition to preventing the contamination of water that has been kept (in tanks, cisterns, and other containers), it is also essential to take into account the efficiency of treatments and handling of treated water for prevention of exposed population from waterborne toxoplasmosis.
This is the first study from India to our knowledge, using molecular techniques to estimate the degree of soil contamination with T. gondii at the Chandigarh city, India. It is difficult to detect low oocysts concentration in soil due to lack of techniques with sufficient sensitivity. Molecular methods such as conventional PCR, real-time PCR and real-time LAMP were used in the current study which are highly sensitive and specific and time saving tests. It was seen that 9% of soil samples were positive for T. gondii by all the three molecular techniques used. T. gondii DNA was found in soil samples collected from urban slum areas, hospitals, public parks, organic farms, kitchen gardens, industrial area etc. Similar to the findings of our study, Ahmed et al., 2019 conducted a study in soil samples from Baghdad and Kut al-imara cities by targeting B1 gene of Toxoplasma gondii. The results showed highest prevalence in gardens, schools and backyards of homes in Baghdad city (21.42%, 17.4%, 16.66%) and in the Kut al-imara city is (6%, 5.5%, 4%) respectively24. Other studies like Pinto-Durate et al. detected 28% positivity rate with nested PCR targeting B1 gene for T. gondii in soil samples collected from Quindio River basin, Colombia21. Haghparast-kenari et al., 2020 found Toxoplasma gondii DNA in 78.1% of environmental soil samples in Mazandaran Province, Northern Iran by nested PCR of RE gene25.
Soil is one of the environmental sources of T. gondii oocysts and these can remain infective for months to years. Understanding the spread of T. gondii soil contamination is important for determining the risk factors of toxoplasmosis in intermediate hosts as well as in humans26. Routes for soil contamination with T. gondii can be many. Cats, including kittens play in the soil residential and educational settings as seen in our study. Cats typically bury their waste in the primary dwelling area, while they occasionally leave smell trails along the outside of their territory27,28. Children are considerably more likely to get toxoplasmosis since they may not be aware of adequate hygiene precautions so to stop the spread of T. gondii29.
In the present study, Toxoplasma gondii was also detected with molecular methods in green leafy vegetables for the first time in India. Positivity rate for T. gondii was found to be 6.4% in vegetable samples with conventional PCR for B1 whereas real-time PCR and real-time LAMP for B1 and TgOWP detected the parasite in 6.6% of samples and thus prevalence of T. gondii in vegetables correlates with other studies. Berrouch et al., 2021 from Marrakech, Morocco revealed an overall contamination of 29.6% with T. gondii in leafy green samples by real-time qPCR30. In another research conducted by Berrouch et al., the study examined vegetable samples, including carrot, coriander, lettuce, parsley, and radish, and identified a 16.6% contamination rate of vegetables with T. gondii using qPCR31. Similarly, Lass et al. from China utilized a real-time PCR assay and detected T. gondii DNA in 3.6% of the fresh vegetable samples32.
Vegetable contamination is a direct result of feline activity, which also causes contamination of the surrounding environment, most notably the soil and water. Since they travel freely and can release millions of oocysts of the parasite into the environment through their faeces, stray cats most likely play a significant role in the epidemiology of toxoplasmosis. These animals could wander into regions where fruit and vegetables are being grown and infect them with Toxoplasma oocysts in addition to contaminating the water and soil.
In the present study T. gondii DNA has been detected in different types of vegetable samples such as coriander Coriandrum sativum, fenugreek leaves Trigonella foenum-graecum, radish Raphanus sativus, cabbage Brassica oleracea, beetroot and leaves Beta vulgaris, spinach Spinacia oleracea, capsicum Capsicum annuum, sprinkle onion Allium cepa, mint Mentha, lettuce Lactuca sativa, parsley Petroselinum crispum, arugula Eruca sativa, rape Brassica napus and tomato & leaves Solanum lycopersicum. Similar to the types of vegetable found contaminated in present study, Lass et al. detected the presence of T. gondii DNA in lettuce, spinach, Chinese cabbage, red cabbage and broccoli and kale samples32. Berrouch et al. found maximum contamination of T. gondii in parsley, followed by coriander, carrot, lettuce and radish31. Furthermore, the surface characteristics may also influence parasitic attachment. Vegetables with rough surfaces are more prone to contamination compared to vegetables with smooth surfaces, such as radish and carrot33.
In current study, real-time PCR and real-time LAMP detected more positive samples for Toxoplasma gondii as compared to the conventional PCR. These results indicate high sensitivity of these techniques, that can detect even low concentration of parasite’s genomic DNA in the targeted samples. Similar results were seen by Koloren and Demirel, in which LAMP for B1 gene detected Toxoplasma gondii in 25% of natural water samples from different sources and nested PCR for 18srRNA detected T. gondii in only 11.7% of samples34. Additionally, no false positives were reported using molecular methods, as every sample which came positive by atleast two molecular techniques.
The prevalence of T. gondii in the Chandigarh region can be attributed to interconnected factors, including poor sanitation practices in slums and villages, which lead to environmental contamination by stray cat feces containing oocysts, and variations in agricultural methods, such as the use of untreated irrigation water and manure from potentially infected animals. Inefficiencies in water treatment and distribution infrastructure further contribute to oocyst persistence in treated water, especially in rural and underserved areas. Additionally, the region’s warm and humid climate creates favorable conditions for oocyst survival, increasing the risk of contamination through water, soil, and vegetables.
The isolates from our study showed > 98% sequencing identity with the other T. gondii B1 gene sequences of cat, sheep, pigs isolates from Iran, India, Mexico and with patient isolate from North India available on the database with accession number LC057646.1, LC057649.1, MK521885.1, MK521884.1, MK521883.1, MK521882.1, MK521881.1, KX270388.1, KX270387.1 etc. Thakur et al. 2019 performed phylogenetic analysis and showed that the positive pig samples for T. gondii from Chandigarh region showed clustering with the VEG type III strain which is similar to findings from current study35. Low levels of B1 gene polymorphism did not, however, allow for the differentiation of the clonal lineages of the several isolated strains. To identify the genotypes of T. gondii circulating in animal and environmental matrices in India, further research is needed that targets one or more loci of the parasite. Phylogenetic analysis will be crucial for this effort, as it reveals the genetic diversity within the parasite population and helps in understanding its evolution, transmission patterns, and the potential emergence of new strains or lineages36.
Conclusion
This study provides the first comprehensive assessment of Toxoplasma gondii contamination in environmental water, soil, and vegetables in Chandigarh, India, using advanced molecular techniques. The findings highlight significant public health concerns, emphasizing the need for improved sanitation, water treatment, and food safety measures to mitigate the risk of toxoplasmosis. While the study confirms the presence of T. gondii in various environmental sources, further research is required to investigate seasonal variations and the correlation between environmental contamination and human infection rates. Future studies should also explore genetic diversity and emerging strains to enhance surveillance and control strategies. Strengthening public awareness and preventive measures will be crucial in reducing the transmission of T. gondii and its associated health risks.
Methods
This prospective cross-sectional study was conducted in Chandigarh, a city in North India, having a predominantly urban population. The present study was ethically approved by the Institutional ethics committee of the Postgraduate Institute of Medical Education and Research, Chandigarh (Approval No: INT/IEC/2021/SPL-960). All the sampling was done strictly as per the guidelines.
Water sampling
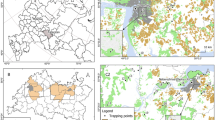
Sampling Area and collection: A total of 100 water samples were collected randomly from different drinking and public water sources of Chandigarh region. The study was done from December 2020 December 2022. Sampling was conducted from each phase—Phase I, Phase II, and Phase III—in the Chandigarh region, as well as from villages within and around the area. This approach ensured that water samples were collected from every phase and villages of the city. Water samples were collected from different resources such as from ponds, lakes, tap water from household, public taps located in different schools, government offices and public parks, tube wells etc. (Table 4).
Sample processing
A total of 10 L of water samples was collected from each site in plastic bottles/ containers37. All the samples were transported to the department of Medical Parasitology for further processing. The water samples which were having less sediment and collected from the tap water located in schools, public parks, govt. offices, houses etc. were processed via filtration method38,39. Millipore water assembly was used with 0.22 μm of filter paper (Millipore). Ten litres of water samples was collected from each site and passed through the filtration assembly (5–6 h). Afterwards, filter paper was dipped in PBS solution (25 µL Phosphate buffer saline + 0.5 µL Tween 20) in 15 mL falcon for overnight. The pellet obtained were transferred into the new 1.5 mL micro centrifuge tube. Processing of dirty and pond water was done using the aluminium sulphate flocculation method34,40. Ten liters of homogenized water samples collected from ponds and lakes were treated with 50–100 µL of Al₂(SO₄)₃ solution for flocculation. The resulting pellets were washed twice with distilled water before being analysed for the presence of oocysts. These pellets, obtained through filtration and flocculation, underwent DNA extraction using the Qiagen Blood and Tissue Kit Qiagen, Germany (Cat. No. 69504), with modifications to the manufacturer’s protocol as described41. The modifications included: subjecting samples to 5 freeze (in liquid nitrogen) and thaw cycles (in a water bath at 70 °C), extending the incubation time to 2–3 h for sample lysis using tissue lysis buffer and AL, eluting the DNA in 35–40 µL of elution buffer (AE) to increase the concentration of DNA in the samples. DNA obtained from the samples were stored at -20oC until used.
Soil sampling
Sampling Area and collection: Sampling sites consist of public parks, urban slum areas, cattle sheds, hospitals, public schools, villages located in the outer boundary of the Chandigarh city, India. A total of 100 samples were collected over the period of two years from December 2020-December 2022 (Fig. 3). Sampling was done from every phase and nearby areas of the city to ensure the random sampling from the area i.e., Phase I, Phase II, Phase III, villages located in and around the Chandigarh region. A total of 20 g of soil was collected in sterile polythene bags from almost 5 cm below the surface layer with the help of a stainless-steel scoop. After collection, it was air dried for 24 h by spreading it on brown paper at room temperature. Air-dried samples were passed through a 20 mm mesh sieve to remove the large particles present in the soil that can hinder the DNA extraction process. After sieving, soil samples were stored in sterile uricols at 4 °C for further use42,43.
DNA extraction
DNA extraction was done using the soil DNA extraction of NucleoSpin Soil Kit (MACHEREY-NAGEL GmbH & Co. KG, Valencienner Str. 11, 52355 Dueren, Germany) and GCC biotech kit (India Pvt. Ltd. Kolkata). Soil DNA extraction was done using these kit with slight modifications in the manufacturer’s protocol. The modifications were as follows: (a) 300 mg of soil sample was used instead of the 250 mg specified in the protocol. (b) Among the two lysis buffers provided in the NucleoSpin Soil Kit (SL1 and SL2), SL2 was chosen for DNA extraction due to its superior performance when combined with Enhancer SX. (c) The centrifugation speed was increased to 12,000 rpm throughout all steps to achieve optimal results and high-quality output.
Vegetable sampling
Sample collection
Vegetable samples were collected from different areas of Chandigarh region such as farms (organic and other), street vendors, shops in the market, vegetable market, kitchen gardens etc. The sampling was done for a period of two years i.e., from December 2020 to December 2022. Different types of vegetables were collected such as Coriandrum sativum (coriander), Beta vulgaris (beetroot greens), Trigonella foenum-graecum (methi), Brassica oleracea (cabbage), Raphanus sativus (radish), Mentha (mint), Spinacia oleracea (spinach), Lactuca sativa (lettuce), Capsicum annuum (green chili), Beta vulgaris var. cicla (Swiss chard), Petroselinum crispum (parsley), Allium cepa (scallion), Eruca sativa (arugula), Brassica napus (rape), Solanum lycopersicum (Tomatoes and leaves) etc. Randomized sampling was done from Kaimwala, Dhanas, vegetable markets/shops in sector 26 & 27 and villages of Khuda ali sehar, Vikas nagar, from street vendors in sectors (38, 28,30,31, 32, 33, 34 etc.), Nayagaon, Khuda lohra, Maloya, Sarangpur, Manimajra, Khuda jassu (all villages), areas nearby hospitals, etc. A total of 500 vegetables samples were collected from various areas of the city as detailed above (Supplementary Table 1).
Sample processing
In a zipper bags/ stomacher bag, 50 g of fresh leafy vegetables, and one of each root vegetable (radish, carrot etc.) were placed. Washing solution of about 200 mL which consists of 0.1% Tween 80 in 0.01 M phosphate-buffered saline (PBS; pH 7.4) was added to the bag. The bags were then packed firmly and placed on an orbital shaker (Hoefer, Holliston, MA, USA) for 15 min. The sample bags were then repeatedly manually shakenupside-down. After that wash solution from the bag was transferred to the 50 mL falcons by sieving it through 4-layer gauge filter. Ultracentrifuge was used to centrifuge the wash solution from the bag at 5,000 g for 20 min and extra fluid was dumped. Rewashing the food samples in the zipper bags with 0.01 M PBS solution for 15 min followed by the same centrifugation of the wash solution as previously. The pellets were spun at 1,000 g for 20 min after being transferred to 50 mL tubes in a centrifuge. Finally, micro centrifuge tubes were used to collect pellets44,45,46. DNA was extracted from the pellet obtained using Qiagen Blood and Tissue Kit (Qiagen, Germany) with some modifications in the manufacturer’s protocol as described in the processing of water samples.
Molecular analysis in environmental and fresh produce samples
Conventional PCR assay
All the water, soil and vegetables samples were subjected to conventional PCR assay for the housekeeping (18srRNA, small subunit rRNA) gene and Toxoplasma gondii specific glycerol-3-phosphate dehydrogenase, B1 gene. Reaction mixture was prepared by adding 12.5 µL of master mix (Promega Corporation, USA), 0.8 µL of forward primer, 0.8 µL of reverse primer and variable template DNA (Supplementary Table 2)47,48.
Real-time PCR assay
Real-Time PCR assay with SYBR Green based chemistry was performed on CFX96 real-time thermal cycler (Bio-Rad laboratories India Pvt.Ltd) machine in all samples for the detection of Toxoplasma gondii by targeting B1 gene. The master mix for the reaction contained, 5µL SYBR Green dye, 0.3µL forward primer & reverse primer respectively (Supplementary Table 2), nuclease free water and template DNA (50 ng/µL of all samples). The reaction was put in opaque real-time PCR strips (0.2 mL). Thermocycling conditions were initial heating at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 0.15 s. and annealing at 60 °C for 0.30 Sects23,49. Melt curve analysis (MCA) was also performed from 65 °C to 95 °C at default setting to keep a check for the formation of non-specific products such as primer-dimer etc. Results were analyzed using the Bio-Rad CFX manager software.
Real- time LAMP assay
Real-Time LAMP was performed in all the samples for detection of T. gondii. This technique was performed on the Genie III instrument by Optigene Pvt. Ltd. Hyderabad, India. Using real-time LAMP, two genes of T. gondii were targeted for the detection i.e., Glycerol-3-phosphate dehydrogenase, B1 and Toxoplasma gondii outer wall protein gene, TgOWP. The master mix contained 12.5 µL of Isothermal master mix (Optigene, Hyderabad, Cat. No. AL306), 1 µL of primer mix (40mM BIP, backward inner primer & FIP, forward inner primer, each and 10mM F3 & B3 each), template DNA (2–3 µL) and nuclease free water (Supplementary Table 2)50.
Sanger sequencing and phylogenetic analysis
The purified PCR products were sent for sanger sequencing at the Central Sophisticated Instrument Cell (CSIC), PGIMER, Chandigarh. For the sequencing, ABI 3730 DNA Analyzer were and sequencing was performed according to the standard protocol51. Further, Molecular Evolutionary Genetic Analysis version 11.0.10 (MEGA-X) programme was used to construct neighbour joining tree with the isolates (soil, water and vegetables) obtained from our study and previously published non-typing B1 gene sequences available on NCBI database52.
Statistical analysis
One way- Anova was performed for water samples to compares the positivity rate of samples with Toxoplasma gondii via different molecular techniques using GraphPad Prism 8.0.2 (263) software (GraphPad Software, Inc.). For soil samples, graph was prepared for positive samples distribution from different sources using Graph Pad Prism software.
Data availability
All the data is submitted to NCBI GenBank Database. Accession numbers are provided in the manuscript for the same.
References
Tenter, A. M., Heckeroth, A. R. & Weiss, L. M. Toxoplasma Gondii: from animals to humans. Int. J. Parasitol. 30, 1217–1258 (2000).
Singh, S., Munawwar, A., Rao, S., Mehta, S. & Hazarika, N. K. Serologic prevalence of Toxoplasma Gondii in Indian women of child bearing age and effects of social and environmental factors. PLoS Negl. Trop. Dis. 8, 2737. https://doi.org/10.1371/journal.pntd.0002737 (2014).
Dubey, J. P. & Beattie, C. P. Toxoplasmosis of animals and man. (1988).
Triviño-Valencia, J., Lora, F., Zuluaga, J. D. & Gomez-Marin, J. E. Detection by PCR of pathogenic protozoa in raw and drinkable water samples in Colombia. Parasitol. Res. 115, 1789–1797 (2016).
Krueger, W. S., Hilborn, E. D., Converse, R. R. & Wade, T. J. Drinking water source and human Toxoplasma gondii infection in the United States: a cross-sectional analysis of NHANES data. BMC Public. Health. 14, 1–10 (2014).
Awobode, H. O., Ohiolei, J. A., Adekeye, T. A., Adeyi, A. O. & Anumudu, C. I. Shedding proportion of Toxoplasma gondii-like oocysts in feral cats and soil contamination in Oyo State, Nigeria. Parasite Epidemiol. Control. 11, 00181. https://doi.org/10.1016/j.parepi.2020.e00181 (2020).
Shapiro, K. et al. Environmental transmission of Toxoplasma Gondii: oocysts in water, soil and food. Food Waterborne Parasit. 15, 00049. https://doi.org/10.1016/j.fawpar.2019.e00049 (2019).
Jones, J. L. et al. Toxoplasma Gondii infection in the United States: seroprevalence and risk factors. Am. J. Epidemiol. 154, 357–365 (2001). (2001).
Ekman, C. C. et al. Case-control study of an outbreak of acute toxoplasmosis in an industrial plant in the state of São Paulo, Brazil. Revista do Instituto De Med. Trop. De São Paulo. 54, 239–244 (2012).
Morais, R. D. A. P. B. et al. Outbreak of acute toxoplasmosis in the municipality of Ponta De Pedras, Marajó Archipelago, State of Pará, Brazil: clinical, laboratory, and epidemiological characteristics. Rev. Pan-Amazônica Saúde. 7, 10–10 (2016).
Tlamçani, Z., Lemkhenete, Z. & Lmimouni, B. E. Toxoplasmosis: The value of molecular methods in diagnosis compared to conventional methods. JMID 3, 93–99 (2013).
Stelzer, S. et al. Toxoplasma Gondii infection and toxoplasmosis in farm animals: risk factors and economic impact. FOOD WATERB PARASIT. 15, 00037. https://doi.org/10.1016/j.fawpar.2019.e00037 (2019).
Delgado, I. L. et al. The apicomplexan parasite Toxoplasma Gondii. Encyclopedia 2, 189–211 (2022).
Hall, S. M., Pandit, A., Golwilkar, A. & Williams, T. S. How do jains get Toxoplasma infection? Lancet 354, 486–487 (1999).
Sousa, S., Fernandes, M. & Correia Da Costa, J.M. Serotyping, a challenging approach for Toxoplasma gondii typing. Front. Med. 10, 1111509 (2023).
Fernández-Escobar, M. et al. Toxoplasma Gondii genotyping: a closer look into Europe. Front. Cell. Infect. Microbiol. 12, 842595 (2022).
Su, Y. J. et al. Geospatial epidemiology of Toxoplasma gondii infection in livestock, pets, and humans in China, 1984–2020. Parasitol. Res. 121, 743–750 (2022).
Chalana, M. Chandigarh: City and periphery. J. Plan. Hist. 14, 62–84 (2015).
Utaaker, K. S. et al. Goats in the city: prevalence of Giardia Duodenalis and Cryptosporidium spp. in extensively reared goats in northern India. Acta Vet. Scand. 59, 1–9 (2017).
Chhachhiya, V. Changing pattern of rape in Chandigarh: a geographical analysis. Int. J. Res. Soc. Sci. 7, 525–541 (2017).
Pinto-Duarte, V. A. et al. Detection of Giardia Duodenalis and Toxoplasma Gondii in soil and water samples in the Quindío River basin, Colombia. FOOD WATERB PARASIT. 28, 00175. https://doi.org/10.1016/j.fawpar.2022.e00175 (2022).
Hernandez-Cortazar, I. B. et al. Presence of Toxoplasma Gondii in drinking water from an endemic region in Southern Mexico. Fpd 14, 288–292 (2017).
Lass, A. et al. Investigation of Toxoplasma Gondii in wastewater and surface water in the Qinghai-Tibet Plateau, China using real-time PCR and multilocus genotyping. Sci. Rep. 12, 5428 (2022).
Ahmed, D. N., Muhsin, S. & Chyiad, A. Comparative study in detection of Toxoplasma Gondii on soil sample from Baghdad and Kut cities by using PCR. IJFMT 13, 4 (2019).
Haghparast-Kenari, B. et al. Isolation and genotypic characterization of Toxoplasma Gondii based on GRA6 gene from environmental soil samples in Mazandaran Province, North of Iran. IJP 15, 158 (2020).
Al-Malki, E. S. Toxoplasmosis: stages of the protozoan life cycle and risk assessment in humans and animals for an enhanced awareness and an improved socio-economic status. Saudi J. Biol. Sci. 28, 962–969 (2021).
Hartmann, K. et al. Toxoplasma Gondii infection in cats: ABCD guidelines on prevention and management. JFMS 15, 631–637 (2013).
Xia, N. et al. Seroprevalence and risk factors of Toxoplasma Gondii in urban cats from China. BMC Vet. Res. 18, 331 (2022).
Davies, R. H., Lawes, J. R. & Wales, A. D. Raw diets for dogs and cats: a review, with particular reference to microbiological hazards. JSAP 60, 329–339 (2019).
Berrouch, S. et al. Protozoan parasites and leafy greens in Marrakech: study of occurrence using a molecular method. Acta Parasitol. 67, 546–554 (2022).
Berrouch, S. et al. Cryptosporidium spp., Giardia duodenalis and Toxoplasma Gondii detection in fresh vegetables consumed in Marrakech, Morocco. Afr. Health Sci. 20, 1669–1678 (2020).
Lass, A. et al. First molecular detection of Toxoplasma Gondii in vegetable samples in China using qualitative, quantitative real-time PCR and multilocus genotyping. Sci. Rep. 9, 17581 (2019).
Kudah, C., Sovoe, S. & Baiden, F. Parasitic contamination of commonly consumed vegetables in two markets in Ghana. Ghana. Med. J. 52, 88–93 (2018).
Koloren, Z. & Demirel, E. Detection of Toxoplasma Gondii in Turkish river and drinking water samples by different PCR and LAMP methods. CLEAN 41, 963–968 (2013).
Thakur, R., Sharma, R., Aulakh, R. S., Gill, J. P. S. & Singh, B. B. Prevalence, molecular detection and risk factors investigation for the occurrence of Toxoplasma Gondii in slaughter pigs in North India. BMC Vet. Res. 15, 1–7 (2019).
Pinto-Ferreira, F. et al. Patterns of transmission and sources of infection in outbreaks of human toxoplasmosis. EID 25, 2177 (2019).
Wells, B. et al. Molecular detection of Toxoplasma Gondii in water samples from Scotland and a comparison between the 529 bp real-time PCR and ITS1 nested PCR. Water Res. 87, 175–181 (2015).
Galvani, A. T. et al. Real-time PCR detection of Toxoplasma Gondii in surface water samples in São Paulo, Brazil. Parasitol. Res. 118, 631–640 (2019).
Vieira, F. P. et al. Waterborne toxoplasmosis investigated and analysed under hydrogeological assessment: new data and perspectives for further research. MIOC 110, 929–935 (2015).
Gallas-Lindemann, C., Sotiriadou, I., Mahmoodi, M. R. & Karanis, P. Detection of Toxoplasma Gondii oocysts in different water resources by Loop mediated Isothermal amplification (LAMP). Acta Trop. 125, 231–236 (2013).
Adamska, M. Molecular detection of Toxoplasma Gondii in natural surface water bodies in Poland. J. Water Health. 16, 657–660 (2018).
Cong, W. et al. Prevalence, risk factors and genotype distribution of Toxoplasma gondii DNA in soil in China. Ecotoxicol. Environ. Saf. 189, 109999 (2020).
Du, F. et al. Survey on the contamination of Toxoplasma Gondii oocysts in the soil of public parks of Wuhan, China. Vet. Parasitol. 184, 141–146 (2012).
Ferreira, F. P. et al. The effect of water source and soil supplementation on parasite contamination in organic vegetable gardens. Rev. Bras. Parasitol. Vet. 27, 327–337 (2018).
Marchioro, A. A. et al. First detection of Toxoplasma gondii DNA in the fresh leafs of vegetables in South America. VBZD 16, 624–626 (2016).
Pineda, C. O., Temesgen, T. T. & Robertson, L. J. Multiplex quantitative PCR analysis of strawberries from Bogotá, Colombia, for contamination with three parasites. JFP 83, 1679–1684 (2020).
Jones, C. D., Okhravi, N., Adamson, P., Tasker, S. & Lightman, S. Comparison of PCR detection methods for B1, P30, and 18S rDNA genes of T. Gondii in aqueous humor. IOVS 41, 634–644 (2000).
Wang, Y., Wang, G., Zhang, D., Yin, H. & Wang, M. Detection of acute toxoplasmosis in pigs using loop-mediated isothermal amplification and quantitative PCR. Korean J. Parasitol. 51, 573 (2013).
Lin, M. H., Chen, T. C., Kuo, T. T., Tseng, C. C. & Tseng, C. P. Real-time PCR for quantitative detection of Toxoplasma Gondii. J. Clin. Microbiol. 38, 4121–4125 (2000).
Sotiriadou, I. & Karanis, P. Evaluation of loop-mediated isothermal amplification for detection of Toxoplasma Gondii in water samples and comparative findings by polymerase chain reaction and immunofluorescence test (IFT). Diagn. Microbiol. Infect. Dis. 62, 357–365 (2008).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. PNAS 74, 5463–5467 (1977).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. MBE 38, 3022–3027 (2021). Statements & Declarations Authors Information
Acknowledgements
We would like to acknowledge Postgraduate Institute of Medical Education and Research for providing the infrastructure and space to carry out this work.
Author information
Authors and Affiliations
Contributions
D. R.: Data curation, Writing- Original draft preparation, Investigation, Formal analysis, Software, Methodology, Investigation, Visualization P.D.: Validation, Resources, Writing - Review & Editing, Supervision D. S.: Writing - Review & Editing, Formal Analysis, Software C. K. B.: Sample collection R. S.: Funding acquisition, Writing - Review & Editing, Supervision, Project administration, Resources, Validation, Conceptualization.
Corresponding author
Ethics declarations
Ethics approval
The present study was ethically approved by the Institutional ethics committee of the Postgraduate Institute of Medical Education and Research, Chandigarh (Approval No: INT/IEC/2021/SPL-960).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rattan, D., Datta, P., Sharma, D. et al. First report of molecular detection and phylogenetic analysis of Toxoplasma Gondii in soil, water and vegetables from Chandigarh city, India. Sci Rep 15, 10537 (2025). https://doi.org/10.1038/s41598-025-90469-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-90469-3