Abstract
Fuchs endothelial corneal dystrophy (FECD) is a bilateral, progressive corneal endothelial disease characterized by the formation of extracellular matrix (ECM) excrescences called guttae. This study integrated proteomic and transcriptomic analyses to elucidate the molecular composition and spatial organization of ECM proteins in the Descemet membrane (DM) of FECD patients. Through shotgun proteomics of FECD-derived DM specimens and RNA sequencing data from FECD (n = 10) and control (n = 7) corneal endothelial cells, we identified 19 significantly upregulated molecules in FECD, including 13 ECM-related proteins. Gene Ontology and Reactome analyses revealed ECM-related pathways as central to FECD pathology. Immunofluorescence analyses of flat-mounted and cross-sectional specimens from FECD patients undergoing Descemet membrane endothelial keratoplasty (DMEK) and controls demonstrated distinct spatial patterns for six ECM proteins. Fibronectin and collagen VI α1 were detected on the outer surfaces of guttae, matrilin-3 and biglycan localized around guttae, while LTBP2 and tenascin were strongly associated with the posterior fibrillar layer (PFL). The peripheral corneal regions predominantly exhibited scattered guttae, whereas the central region displayed buried guttae encapsulated by ECM deposition. This study comprehensively examined ECM protein expression patterns, revealing distinct spatial distributions across guttae, PFL, and surrounding DM regions. These findings suggest that clinical assessments should incorporate both guttae confluence and the presence of ECM-rich PFL to achieve a more comprehensive understanding of FECD progression, thereby informing more accurate staging and optimal surgical planning.
Similar content being viewed by others
Introduction
Fuchs endothelial corneal dystrophy (FECD) is a bilateral, progressive corneal endothelial disease characterized by the formation of excrescences, called guttae, in the central cornea1,2,3. These guttae are initially detected and diagnosed through slit-lamp microscopy1,2,3. As the disease progresses, the number of guttae increases, particularly in the central cornea, leading to a condition known as “confluent guttae”4. When the disease advances to Grade 4 (2–5 mm confluent guttae) or Grade 5 (> 5 mm confluent guttae)5,6, visual function deteriorates due to light scattering caused by the guttae7,8,9. In advanced stages, progressive corneal endothelial dysfunction leads to corneal edema, resulting in further vision loss4.
While clinical grading systems focus on the extent of guttae confluence5,6, histological studies have revealed that disease progression involves both the confluence of guttae and their coverage by extracellular matrix (ECM) components, specifically the posterior fibrillar layer (PFL)10,11,12,13,14. The Descemet membrane (DM) in normal corneas, which is continuously secreted by corneal endothelial cells throughout life, consists of two distinct layers: the embryonically-derived anterior banded layer (ABL) and the post-natal posterior non-banded layer (PNBL)15,16. In FECD, while the ABL structure remains preserved, the PNBL becomes attenuated or absent. Additionally, a posterior banded layer develops, characterized by the formation of guttae excrescences15,16. As the disease advances, a distinct PFL forms between the corneal endothelium and DM, particularly in regions where endothelial cells become depleted. Under slit-lamp biomicroscopy examination, the PFL presents as indistinct guttae-like excrescences with blurred boundaries and a whitish membranous appearance on the DM. Despite its clinical significance, the molecular composition and pathophysiological role of the PFL in disease progression remain incompletely characterized15,16. Understanding these histological changes is essential for accurate disease staging, as assessment based solely on guttae confluence, without considering the presence and characteristics of the PFL, may lead to incomplete evaluation of the underlying pathological alterations.
Recent surgical advances, including Descemet membrane endothelial keratoplasty (DMEK)17,18,19 and Descemetorhexis stripping only (DSO)20,21,22,23, now allow for intervention in FECD even before the onset of clinical corneal edema, primarily by targeting guttae and PFL removal. Given these developments, a comprehensive understanding of the histological characteristics and detailed composition of guttae and the PFL in FECD has become increasingly important. This knowledge is essential for clinicians to accurately assess patient condition and determine surgical timing11,12,13, while also providing valuable insights for understanding the disease pathogenesis and for developing potential pharmacological treatments. We hypothesized that ECM-related molecules are differentially expressed in the DM of FECD patients compared to controls, and that these molecules exhibit distinct spatial distribution patterns that correlate with disease progression. Understanding these molecular alterations and their spatial organization could provide insights into FECD pathogenesis and help establish more accurate clinical assessments.
In the current study, we employed proteomics and transcriptome analyses to perform a comprehensive characterization of the ECM components deposited in the DM of FECD patients. We also used immunohistochemistry to investigate the spatial distribution of these components to obtain a better understanding of their expression patterns in relation to FECD disease progression.
Results
Identification of overexpressed molecules in FECD DM
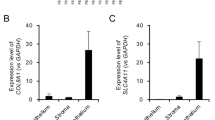
Our previous shotgun proteomics analysis identified 200 proteins in the DM of FECD patients14. Although shotgun proteomics provides comprehensive protein expression profiles, a direct comparison of protein expression levels between FECD and control DM specimens is not feasible using this approach. Therefore, we analyzed RNA sequencing data to identify the highly expressed molecules in DM associated with pathological CECs in FECD (Fig. 1). Among the 200 proteins identified, 19 were differentially expressed in FECD CECs compared with the controls (Table 1). Notably, 13 of these 19 molecules were ECM-related proteins (Supplementary Table 1, Supplementary Fig. 1). Analysis of RNA sequencing data revealed that all 13 ECM-related molecules demonstrated statistically significant upregulation in FECD specimens compared to control tissues. None of the investigated molecules exhibited decreased expression in FECD samples relative to controls.
Workflow for identifying upregulated extracellular matrix (ECM) proteins in Descemet membrane (DM) from patients with Fuchs endothelial corneal dystrophy (FECD). Shotgun proteomics identified 200 proteins expressed in DM from FECD patients. Shotgun proteomics does not allow quantitative evaluation of expression levels; therefore, RNA-Seq analysis was utilized to identify molecules significantly upregulated in CECs from FECD patients compared to non-FECD subjects, yielding 19 molecules. Gene ontology and pathway enrichment analyses were conducted on these molecules. Among the 19 molecules, 13 were ECM-related proteins. To validate their expression and examine their distribution, immunostaining was performed on DM containing CECs from FECD patients.
Gene ontology enrichment analysis and reactome analysis for overexpressed molecules identified in DMs of FECD
GO analysis and Reactome analysis were conducted on the 19 identified overexpressed molecules. GO Biological Process (BP) terms showed significant enrichment in synapse organization, positive regulation of nervous system development, gastrulation, and formation of primary germ layers, along with multiple ECM-related pathways. Analysis of Cellular Component (CC) and Molecular Function (MF) terms revealed significant alterations in multiple ECM-related molecules, particularly in collagen-containing extracellular matrix and extracellular matrix structural constituents (Fig. 2A). Consistent with these findings, the Reactome analysis revealed a notable association with ECM-related signals, including ECM organization, ECM proteoglycans, degradation of ECM, and non-integrin membrane–ECM interactions (Fig. 2B).
Functional enrichment analysis of differentially expressed proteins in the Descemet membrane of Fuchs endothelial corneal dystrophy (FECD) patients. (A) Gene Ontology (GO) enrichment analysis was conducted on 19 proteins that demonstrated both elevated protein abundance in shotgun proteomics and increased transcript levels in RNA sequencing analyses of FECD specimens compared to controls. The analysis encompassed three GO domains: Biological Process, Cellular Component, and Molecular Function. The enriched GO terms are presented on the y-axis, with corresponding gene ratios displayed on the x-axis. GO analysis performed using genes with adjusted p-values < 0.05 as input data, displaying the top 15 GO terms ranked by gene ratio among terms with p-values < 0.05. Statistical significance is represented by a color gradient, where increasing intensity of red indicates higher statistical significance (−log10[P-value]). (B) Reactome pathway analysis revealed significantly enriched biological pathways. Enriched pathways are displayed on the y-axis, with corresponding gene ratios on the x-axis. The statistical significance is indicated by a blue color gradient, where darker blue represents higher significance (-log10[P-value]).
Immunofluorescence analysis of ECM proteins in flat-mounted DM
In control specimens, perinuclear fibronectin (FINC) staining was occasionally observed in a few cells (white arrows, Fig. 3A). In FECD specimens, FINC was localized to the outer surface of the guttae (asterisks). Matrilin-3 (MATN3) immunoreactivity was absent in control specimens but showed punctate distribution around the guttae in FECD specimens (Fig. 3B). Biglycan (PGS1) was undetectable in control specimens but exhibited intense staining in fibrillar structures surrounding the guttae in FECD specimens (Fig. 3C). Staining for latent transforming growth factor beta binding protein-2 (LTBP2) was absent in control specimens but indicated strong immunoreactivity in fine fibrillar structures around the guttae in FECD specimens (Fig. 3D). Collagen alpha-1(VI) chain (COL6A1) was not detected in control specimens but was present on the surfaces of the guttae and adjacent matrix in FECD specimens (Fig. 3E). Tenascin (TENA) immunoreactivity was absent in control specimens but was present in the matrix surrounding the guttae in FECD specimens (Fig. 3F). All staining experiments were performed in triplicate using three control and four FECD specimens. All observations were conducted within the central 3 mm diameter zone of the cornea. While all four FECD specimens exhibited confluent guttae and were characterized by ECM-covered guttae in the buried guttae zone, some areas contained typical scattered guttae that were not covered by ECM. Technical controls using nonspecific rabbit and mouse IgG were consistently negative (data not shown).
Identification by immunostaining of ECM-specific markers in Descemet membranes from FECD patients. Immunofluorescence analysis of six ECM proteins, identified through shotgun proteomics and shown to be upregulated at the mRNA level in FECD samples, was performed on flat-mounted Descemet membranes. The analyzed proteins were (A) Fibronectin (FINC), (B) Matrilin-3 (MATN3), (C) Biglycan (PGS1), (D) Latent TGF-β Binding Protein-2 (LTBP2), (E) Collagen 6α1 (COL6A1), and (F) Tenascin (TENA). White arrows indicate cytoplasmic Fibronectin expression in control specimens (A), while asterisks denote guttae in FECD samples. Nuclei were counterstained with 4′,6-diamidino-2-phénylindole (DAPI) (blue). Control specimens were obtained from healthy Descemet membranes of non-FECD subjects. Three healthy and four FECD samples were examined per protein. Images were acquired from the central region of both control and FECD Descemet membranes using epifluorescence microscopy. Scale bar = 20 μm.
Immunofluorescence analysis of ECM proteins in cross-sectional DM
In control specimens, FINC immunoreactivity was localized to the anterior banded layer (ABL), whereas FECD specimens showed additional intense staining in the PFL (Fig. 4A). MATN3 showed weak punctate staining in the ABL of control specimens but was distributed throughout the DM, with additional punctate patterns, in FECD specimens (Fig. 4B). PGS1 was undetectable in control specimens but present in the PFL of FECD specimens, particularly in the ECM covering buried guttae (Fig. 4C). LTBP2 was absent in control specimens but showed strong immunoreactivity in the PFL of FECD specimens (Fig. 4D). COL6A1 was present in the ABL of control specimens and showed additional localization to guttae surfaces in FECD specimens (Fig. 4E). TENA was undetectable in control specimens but present in the PFL of FECD specimens, predominantly in the matrix surrounding the buried guttae (Fig. 4F). All analyses were conducted within the central 3 mm diameter zone of the cornea, and representative images from these regions are shown. Technical controls using nonspecific rabbit and mouse IgG were consistently negative (data not shown).
Immunofluorescence analysis of extracellular matrix proteins on cross-sectional Descemet membrane (DM). In parallel with the flat-mounted DMs, immunofluorescence staining was also performed on cross-sectional DMs for the same six proteins examined in Fig. 3: (A) Fibronectin (FINC), (B) Matrilin-3 (MATN3), (C) Biglycan (PGS1), (D) Latent Transforming Growth Factor Beta Binding Protein-2 (LTBP2), (E) Collagen 6 alpha chain 1 (COL6A1), and (F) Tenascin (TENA). Nuclei were counterstained with DAPI (blue). Control samples were obtained from healthy DMs of non-FECD subjects, with three healthy and three FECD samples examined per protein. All images from control samples were captured from the 8 mm center of the DM. Asterisks indicate guttae. Images were taken at either 40× or 60× magnification, with scale bars representing 25 μm.
Spatial distribution of ECM proteins in FECD DM
Semi-thin cross-sections from healthy and FECD specimens are shown in Fig. 5A. Control DM appeared thin and uniform, with a distinct ABL and a posterior non-banded layer (PNBL) (Fig. 5A, right). FECD specimens exhibited two distinct regions: a typical guttae zone (Fig. 5A, middle) and a buried guttae zone characterized by ECM-covered guttae (Fig. 5A, left). Flat-mount analysis of FECD specimens revealed typical guttae predominantly in the peripheral region, whereas buried guttae were concentrated centrally (Fig. 5B). A schematic cross-sectional representation of DM (Fig. 5C) illustrates the structural differences between the FECD and control specimens, highlighting the buried guttae and scattered zones. The localization of six ECM proteins analyzed by immunofluorescence was mapped to their respective histological layers.
Spatial distribution of abnormal extracellular matrix on the Descemet membrane (DM) in FECD. (A) Histology of the DM in healthy and FECD subjects. Histological analysis was performed on semi-thin cross sections stained with toluidine blue. Red arrows indicate posterior fibrillar layers (PFLs) covering the guttae, while black arrows point to cell nuclei. Scale bar: 10 μm. (B) Distribution of scattered and buried guttae on an 8 mm flat-mounted DM of FECD. Scattered guttae, which are considered “typical,” have a high-contrast, black outline, whereas buried or semi-buried guttae have a less defined outline. Scale bar: 50 μm. (C) Schematic representation of the distribution of overexpressed and abnormal ECM on FECD DM. Normal DM of a non-FECD subject is also shown. Key features include guttae (G), PFL, posterior banded layer (PBL), posterior non-banded layer (PNBL), and anterior banded layer (ABL). ECM proteins identified by immunostaining in the present study are shown in blue text, while previously reported protein expressions are indicated in black text.
Discussion
In the current study, we employed shotgun proteomics and RNA-Seq analyses to identify molecules that are differentially expressed in the DM of FECD patients compared to non-FECD controls. Our comprehensive investigation focused on ECM molecules involved in guttae and PFL formation, with subsequent immunohistochemical validation to elucidate the structural abnormalities in the DM of FECD patients. Given that protein expression is regulated through multiple cellular mechanisms, including transcriptional control, post-transcriptional modification, translational regulation, and post-translational modifications, we initially utilized shotgun proteomics to identify proteins actually deposited in the DM. We then conducted RNA-Seq analysis to narrow down molecules with elevated gene expression in the CECs of FECD patients.
Our systematic proteomic analysis identified 19 distinct proteins, including 13 ECM-related molecules. Previous studies have established associations between FECD and several proteins, including fibronectin, collagens type I, III, and IV, clusterin, and transforming growth factor beta-induced protein11,12,24,25,26. The present study expands this list by identifying additional ECM proteins. Notably, our previous shotgun proteomics analysis of isolated DM identified hemoglobin α, SRPX2, tenascin-C, and hemoglobin γ/δ/β as FECD-specific proteins14. All of these proteins were among the 13 ECM molecules identified in the current study, which also integrated transcriptome analysis of CECs. This concordance between our previous DM proteomics and current cellular transcriptome findings not only validates our comprehensive analytical approach, but also supports the hypothesis that dysfunctional CECs are the primary source of aberrant ECM deposition in FECD.
Multiple studies have demonstrated that visual deterioration in FECD can be attributed to corneal guttae, even in the absence of stromal edema. For example, Wacker et al. reported elevated posterior corneal high-order aberrations in mild FECD, with increased anterior and posterior corneal backscatter across all disease severities9. Watanabe et al. established significant correlations between the area ratio of corneal guttae and visual quality parameters8, while Shah et al. demonstrated that central corneal densitometry and severity of guttae independently predicted decreased visual acuity, even after adjusting for corneal edema27. Current clinical assessments primarily rely on slit-lamp microscopy and severity grading systems that evaluate the extent of “confluent guttae” and presence of corneal edema. While historical documentation indicates that guttae begin as focal excrescences and progress to being covered by PFL15, existing clinical grading systems only describe “confluent guttae,” without distinguishing between increased guttae density and PFL formation. Although this distinction may be challenging to make using slit-lamp examination alone, clinicians need to understand the pathological progression of guttae morphology for a more accurate assessment of FECD. Scheimpflug imaging has been proposed to provide a more precise evaluation of guttae density and PFL distribution12. Our comprehensive molecular expression profiling and spatial distribution analysis may help to improve these objective measurements, thereby potentially facilitating more informed surgical timing decisions for DMEK or DSO procedures and enhancing prognostic predictions.
Recent studies have demonstrated that guttae contribute to disease progression through multiple mechanisms beyond their direct effect on light scattering. For instance, guttae can actively influence CEC behavior in a size-dependent manner. Specifically, while small guttae have minimal impact on cellular phenotype, medium-sized guttae induce the formation of rosette-like structures with upregulation of endothelial-mesenchymal transition (EMT) and senescence markers28. Large guttae further escalate these pathological changes by inducing rosette formation and promoting EMT, senescence, and cellular apoptosis28. Our previous research has demonstrated that ECM proteins, which are primary components of guttae, can trigger the unfolded protein response when overexpressed in CECs29,30. Using disease-specific cellular models derived from FECD patients, we showed that this unfolded protein response induces cell death through endoplasmic reticulum stress pathways29,30. These findings suggest that ECM proteins play a central role in FECD pathogenesis, not only through the formation of guttae and PFL, but also by directly influencing CEC phenotype and survival. This creates a vicious cycle3 wherein abnormal ECM accumulation leads to cellular dysfunction, which in turn may promote further ECM deposition and disease progression.
The limitations of the present study include the relatively small sample size for shotgun proteomics analysis and the lack of information regarding the status of TCF4 trinucleotide repeat expansion—the most common genetic mutation in FECD patients. Future studies with larger sample sizes, stratified by TCF4repeat expansion status, will be necessary to elucidate the relationship between genetic mutations and ECM expression patterns. Additionally, our validation studies were limited to four FECD samples, all of which were classified as grade 6 severity with corneal edema6, precluding analysis stratified by disease grade—an important area for future investigation. While we identified 13 overexpressed ECM proteins in the present study, further investigation is needed to determine their specific roles in modulating the CEC phenotype and cell death, as discussed above. These investigations could provide valuable insights into disease mechanisms and potential therapeutic targets.
In conclusion, our study has characterized the ECM proteins present in guttae and PFL and demonstrated their distinct spatial distribution patterns through immunohistochemical analysis. These findings have important clinical implications, as they suggest that ophthalmologists should consider both the presence of guttae and the formation and extent of PFL when assessing FECD severity. This enhanced understanding of ECM distribution patterns could lead to improved clinical decision making and more precise timing of surgical interventions.
Methods
Ethics statement
All corneal tissues in this study were handled in accordance with the Declaration of Helsinki ethical guidelines. The RNA sequencing analyses were approved by the Ethical Review Committee at Friedrich-Alexander Universität Erlangen-Nürnberg (FAU; approval number: 140_20 B) and the Committee on Human Research Ethics at Doshisha University (approval numbers: 20009 and 20032). DM specimens for immunofluorescence and histological analyses were obtained in Saint-Étienne, France, in compliance with French ethical regulations. Control corneas from healthy donors (non-FECD) were provided by the Laboratory of Anatomy, Faculty of Medicine, Saint-Étienne, for research purposes. The scientific utilization of these corneal specimens was approved by the Committee on Human Research Ethics at Jean Monnet University (approval number: 42-23-09). FECD specimens were collected as surgical waste from the operating theater. Written informed consent was obtained from all FECD patients at the University Hospital of Saint-Étienne. The BiiO laboratory, authorized by the French Ministry of Higher Education, Research, and Innovation (Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation, MESRI; dossier number: DC-2023–5458), conducted the experimental procedures on these human corneal and DM specimens without requiring additional ethical review. All procedures adhered to the principles outlined in the Declaration of Helsinki for biomedical research involving human tissues.
Identification and confirmation of differentially expressed genes
RNA sequencing data from control corneal endothelial cells (CECs) were obtained from seven healthy individuals (three males and four females of Caucasian descent; age range, 48–69 years), as previously reported31. FECD samples were collected from ten patients (five males and five females of Caucasian descent; age range, 53–79 years)32. Study participants were selected from FECD patients with grades 4–6 severity according to the Modified Krachmer grading system6 who met the criteria for DMEK surgery. Patients with clinically significant corneal edema detected by slit-lamp biomicroscopy were excluded to ensure optimal corneal endothelial cell harvesting. Differential gene expression between FECD and control samples was analyzed using DESeq233 (Bioconductor version 3.14; https://www.bioconductor.org/) with RSEM34. During quality control, genes were excluded if they showed ‘NA’ in adjusted P-values from the Wald test, indicating low expression levels, or if the Benjamini-Hochberg adjusted P-value computation failed. We then examined the expression variance of genes and proteins that were common to both the RNA sequencing and shotgun proteomics datasets. Differentially expressed genes within this overlapping set were defined as those with |log2 fold change| ≥ 1 and adjusted P-value < 0.05.
Gene ontology (GO) enrichment and reactome analysis
Gene identifiers were converted using the biomaRt package (version 2.58.2). Enrichment analyses were performed using the clusterProfiler package (version 4.10.0) with the org.Hs.eg.db annotation database (version 3.18.0). GO terms were considered significantly enriched at P< 0.05. The major GO terms in the Biological Process, Cellular Component, and Molecular Function categories were visualized using the ggplot2 package (version 3.4.4) in R. Reactome analysis35,36 was conducted using the ReactomePA package (version 1.46.0) in R, and the results were visualized using ggplot2. Pathways with P-value < 0.05 were considered significant.
Immunofluorescence analysis
Immunofluorescence analysis was performed on DM specimens obtained from three healthy donors (one 77-year-old male and two females aged 76 and 104 years) and four FECD patients (three females aged 58, 64, and 77 years, and one 74-year-old male). All FECD specimens were obtained from patients classified as grade 6 according to the Modified Krachmer grading system6, characterized by confluent central guttae with mild corneal edema observable by slit-lamp biomicroscopy. The degree of edema allowed relatively clear visualization of the pupillary area and iris detail. Each DM specimen was divided into two portions: three-quarters for flat-mounted tissue analysis and one-quarter for cross-sectional analysis. For cross-sections, specimens were embedded in Optimal Cutting Temperature (OCT) compound (Sakura Finetek Japan, Tokyo, Japan) and sectioned at 6 μm thickness. All specimens were rehydrated in phosphate-buffered saline (PBS), permeabilized with 0.5% Triton X-100 (EuroMedex, Souffelweyersheim, France), and blocked with 2% heat-inactivated goat serum and 2% bovine serum albumin in PBS.
The following primary antibodies (diluted 1:500) were used for immunofluorescence studies: anti-fibronectin (F3648, Sigma-Aldrich, Burlington, MA), anti-matrilin-3 (ab106388, Abcam, Cambridge, UK), anti-biglycan (sc-100857, Santa Cruz Biotechnology, Dallas, TX), anti-latent transforming growth factor beta binding protein 2 (LTBP2; PAB-630H001, Cloud-Clone Corp, Katy, TX), anti-collagen type VI α1 chain (sc-47712, Santa Cruz Biotechnology), and anti-tenascin-C (ab3970, Abcam). Antibody specificity was verified using nonspecific rabbit and/or mouse immunoglobulin G (IgG; Zymed, Carlsbad, CA, USA) as the primary antibody controls. Secondary antibodies (Alexa Fluor 488 goat anti-mouse [A32723] or Alexa Fluor 555 goat anti-rabbit [A32732], both from Thermo Fisher Scientific, Waltham, MA) were applied at 1:1000 dilution. Following a 60 min incubation at 37 °C, the specimens were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) and washed three times with PBS between steps. Samples were mounted using fluorescence mounting medium (NB-23–00158-2, Neo Biotech, Nanterre, France) and visualized with an Olympus IX81 epifluorescence inverted microscope (Olympus, Tokyo, Japan) equipped with cellSens imaging software (cellSens Dimension, Olympus, Germany).
Histological analysis of semi-thin sections
The endothelium-DM complexes from healthy subjects and FECD patients were fixed in a solution containing 1% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) and 0.5% paraformaldehyde (Thermo Fisher Scientific) in 0.02 M Na2HPO4 (Prolabo, Paris, France)/0.08 M KH2PO4 (Sigma-Aldrich) buffer (pH 7.4). Specimens were postfixed with 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) in 0.1 M cacodylate buffer for 1 h, dehydrated through a graded ethanol series, and embedded in Epon resin (Oxford Instruments, Oxford, UK). Semi-thin sections (2 μm) were cut using an ultramicrotome (RMC MTXL, Leica Microsystems, Wetzlar, Germany), stained with 1% toluidine blue, and observed using an optical microscope equipped with a DP 26 camera (Olympus, Tokyo, Japan).
Data availability
Corneal endothelial transcriptome data from patients with FECD and healthy controls were obtained from the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (SRA) under accession numbers DRA01023731 and DRA01507832. Shotgun proteomics data for corneal endothelial proteins from FECD patients and healthy controls were retrieved from the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository under accession ID MSV00009107814.
References
Eghrari, A. O., Riazuddin, S. A. & Gottsch, J. D. Fuchs corneal dystrophy. Prog. Mol. Biol. Transl. Sci. 134, 79–97. https://doi.org/10.1016/bs.pmbts.2015.04.005 (2015).
Fautsch, M. P. et al. TCF4-mediated Fuchs endothelial corneal dystrophy: Insights into a common trinucleotide repeat-associated disease. Prog. Retin. Eye Res. 81. https://doi.org/10.1016/j.preteyeres.2020.100883 (2021).
Ong Tone, S. et al. Fuchs endothelial corneal dystrophy: The vicious cycle of Fuchs pathogenesis. Prog. Retin. Eye Res. 80. https://doi.org/10.1016/j.preteyeres.2020.100863 (2021).
Weisenthal, R. & Streeten, B. Descemet’s membrane and endothelial dystrophies. Cornea Third Ed. 1, 845–864 (2011).
Krachmer, J. H., Purcell, J. J. Jr., Young, C. W. & Bucher, K. D. Corneal endothelial dystrophy. A study of 64 families. Arch. Ophthalmol. 96, 2036–2039 (1978).
Louttit, M. D. et al. A multicenter study to map genes for Fuchs endothelial corneal dystrophy: Baseline characteristics and heritability. Cornea 31, 26–35. https://doi.org/10.1097/ICO.0b013e31821c9b8f (2012).
Patel, S. V., Baratz, K. H., Hodge, D. O., Maguire, L. J. & McLaren, J. W. The effect of corneal light scatter on vision after Descemet stripping with endothelial keratoplasty. Arch. Ophthalmol. (Chicago Ill.: 1960) 127, 153–160 (2009).
Watanabe, S. et al. Relationship between corneal guttae and quality of vision in patients with mild Fuchs’ endothelial corneal dystrophy. Ophthalmology 122, 2103–2109. https://doi.org/10.1016/j.ophtha.2015.06.019 (2015).
Wacker, K., McLaren, J. W., Amin, S. R., Baratz, K. H. & Patel, S. V. Corneal high-order aberrations and backscatter in Fuchs’ endothelial corneal dystrophy. Ophthalmology 122, 1645–1652. https://doi.org/10.1016/j.ophtha.2015.05.005 (2015).
Weller, J. M. et al. Extracellular matrix alterations in late-onset Fuchs’ corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 55, 3700–3708. https://doi.org/10.1167/iovs.14-14154 (2014).
Hribek, A. et al. Fibrillar layer as a marker for areas of pronounced corneal endothelial cell loss in advanced Fuchs endothelial corneal dystrophy. Am. J. Ophthalmol. 222, 292–301 (2021).
Hribek, A. et al. Scheimpflug backscatter imaging of the fibrillar layer in Fuchs endothelial corneal dystrophy. Am. J. Ophthalmol. 235, 63–70 (2022).
Ozer, O. et al. Correlation of clinical fibrillar layer detection and corneal thickness in advanced Fuchs endothelial corneal dystrophy. J. Clin. Med. 11. https://doi.org/10.3390/jcm11102815 (2022).
Nakagawa, T. et al. Shotgun proteomics identification of proteins expressed in the Descemet’s membrane of patients with Fuchs endothelial corneal dystrophy. Sci. Rep. 13, 10401. https://doi.org/10.1038/s41598-023-37104-1 (2023).
Bourne, W. M., Johnson, D. H. & Campbell, R. J. The ultrastructure of Descemet’s membrane. III. Fuchs’ dystrophy. Arch. Ophthalmol. 100, 1952–1955. https://doi.org/10.1001/archopht.1982.01030040932013 (1982).
Waring, G. O. 3rd, Bourne, W. M., Edelhauser, H. F. & Kenyon, K. R. The corneal endothelium. Normal and pathologic structure and function. Ophthalmology 89, 531–590 (1982).
Melles, G. R., Ong, T. S., Ververs, B. & van der Wees, J. Descemet membrane endothelial keratoplasty (DMEK). Cornea 25, 987–990. https://doi.org/10.1097/01.ico.0000248385.16896.34 (2006).
Kruse, F. E., Schrehardt, U. S. & Tourtas, T. Optimizing outcomes with Descemet’s membrane endothelial keratoplasty. Curr. Opin. Ophthalmol. 25, 325–334. https://doi.org/10.1097/ICU.0000000000000072 (2014).
Price, M. O., Gupta, P., Lass, J. & Price, F. W. Jr. EK (DLEK, DSEK, DMEK): New frontier in cornea surgery. Annu. Rev. Vis. Sci. 3, 69–90. https://doi.org/10.1146/annurev-vision-102016-061400 (2017).
Garcerant, D. et al. Descemet’s stripping without endothelial keratoplasty. Curr. Opin. Ophthalmol. 30, 275–285. https://doi.org/10.1097/ICU.0000000000000579 (2019).
Blitzer, A. L. & Colby, K. A. Update on the surgical management of Fuchs endothelial corneal dystrophy. Ophthalmol. Ther. 9, 757–765. https://doi.org/10.1007/s40123-020-00293-3 (2020).
Din, N. et al. Surgical management of Fuchs endothelial corneal dystrophy: A treatment algorithm and individual patient meta-analysis of Descemet stripping only. Cornea 41, 1188–1195. https://doi.org/10.1097/ICO.0000000000002975 (2022).
Rizk, M. et al. Long-term follow-up of Descemet stripping only: Data up to 7 years postoperatively. Cornea 43, 1245–1248. https://doi.org/10.1097/ICO.0000000000003449 (2024).
Jurkunas, U. V. et al. Increased clusterin expression in Fuchs’ endothelial dystrophy. Invest. Ophthalmol. Vis. Sci. 49, 2946–2955. https://doi.org/10.1167/iovs.07-1405.iovs.07-1405 (2008).
Jurkunas, U. V., Bitar, M. & Rawe, I. Colocalization of increased transforming growth factor-beta-induced protein (TGFBIp) and clusterin in Fuchs endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 50, 1129–1136. https://doi.org/10.1167/iovs.08-2525.iovs.08-2525 (2009).
Kuot, A. et al. Association of TCF4 and CLU polymorphisms with Fuchs’ endothelial dystrophy and implication of CLU and TGFBI proteins in the disease process. Eur. J. Hum. Genet. 20, 632–638. https://doi.org/10.1038/ejhg.2011.248 (2012).
Shah, K., Eghrari, A. O., Vanner, E. A., O’Brien, T. P. & Koo, E. H. Scheimpflug corneal densitometry values and severity of guttae in relation to visual acuity in Fuchs endothelial corneal dystrophy. Cornea 41, 692–698. https://doi.org/10.1097/ICO.0000000000002762 (2022).
Kocaba, V. et al. Association of the Gutta-Induced microenvironment with corneal endothelial cell behavior and demise in Fuchs endothelial corneal dystrophy. JAMA Ophthalmol. 136, 886–892. https://doi.org/10.1001/jamaophthalmol.2018.2031 (2018).
Okumura, N. et al. Sustained activation of the unfolded protein response induces cell death in Fuchs’ endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 58, 3697–3707. https://doi.org/10.1167/iovs.16-21023 (2017).
Okumura, N. et al. Activation of TGF-β signaling induces cell death via the unfolded protein response in Fuchs endothelial corneal dystrophy. Sci. Rep. 7. https://doi.org/10.1038/s41598-017-06924-3 (2017).
Tokuda, Y. et al. Transcriptome dataset of human corneal endothelium based on ribosomal RNA-depleted RNA-Seq data. Sci. Data 7. https://doi.org/10.1038/s41597-020-00754-1 (2020).
Nakagawa, T. et al. RNA-Seq-based transcriptome analysis of corneal endothelial cells derived from patients with Fuchs endothelial corneal dystrophy. Sci. Rep. 13. https://doi.org/10.1038/s41598-023-35468-y (2023).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Li, B. & Dewey, C. N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12. https://doi.org/10.1186/1471-2105-12-323 (2011).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462. https://doi.org/10.1093/nar/gkv1070 (2016).
Acknowledgements
This work was supported by JSPS KAKENHI (Grant Numbers: 18K09464 to N.O. and 21K06840 to Y.T.) and AMED (Grant Number: JP22ek0109590). The authors thank Mr. Florian Bergandi (Anatomy Laboratory, Faculty of Medicine, Saint-Étienne) and Mrs. Chantal Perrache (BiiO Laboratory) for their technical assistance.
Author information
Authors and Affiliations
Contributions
N.K., G.T., and N.O. conceived and designed the study. S.I, H.V., T.Y., T.N., Y.T, T.N. Y.T. M.N., T.T., U.S., F.F., I.A., and Z.H.performed the experiments. S.I. and N.O. drafted the article, and all authors revised the paper and approved the final version to be published.
Corresponding authors
Ethics declarations
Competing interests
Naoki Okumura and Noriko Koizumi are co-founders of ActualEyes Inc., which is currently developing a pharmaceutical therapy for treating Fuchs endothelial corneal dystrophy. However, this affiliation has no influence on the data analysis, results, or conclusions presented in this study. Other authors do not have a competing financial interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Inagaki, S., Vaitinadapoule, H., Yuasa, T. et al. Comprehensive identification of dysregulated extracellular matrix molecules in the corneal endothelium of patients with Fuchs endothelial corneal dystrophy. Sci Rep 15, 14654 (2025). https://doi.org/10.1038/s41598-025-91850-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-91850-y