Abstract
Early and accurate prediction of neurological outcomes in comatose patients following cardiac arrest is critical for informed clinical decision-making. Existing studies have predominantly focused on EEG for assessing brain injury, with some exploring ECG data. However, the integration of EEG, ECG, and clinical features remains insufficiently investigated, and its potential to enhance predictive accuracy has not been fully established. Moreover, the limited interpretability of current models poses significant barriers to clinical application. Using the I-CARE database, we analyzed EEG, ECG, and clinical data from comatose cardiac arrest patients. After rigorous preprocessing and feature engineering, machine learning models (Logistic Regression, SVM, Random Forest, and Gradient Boosting) were developed. Performance was evaluated through AUC-ROC, accuracy, sensitivity, and specificity, with SHAP applied to interpret feature contributions. Our multi-modal model outperformed single-modality models, achieving AUC values from 0.75 to 1.0. Notably, the model’s accuracy peaked at a critical point within the 12–24 h window (e.g., 18 h, AUC = 1.0), surpassing EEG-only (AUC 0.7–0.8) and ECG-only (AUC < 0.6) models. SHAP identified Shockable Rhythm as the most influential feature (mean SHAP value 0.17), emphasizing its role in predictive accuracy. This study presents a novel multi-modal approach that significantly enhances early neurological outcome prediction in critical care. SHAP-based interpretability further supports clinical applicability, paving the way for more personalized patient management post-cardiac arrest.
Similar content being viewed by others
Introduction
Cardiac arrest (CA) is a critical global health concern, affecting over 1 million individuals annually with persistently high mortality and morbidity rates1. Despite advancements in cardiopulmonary resuscitation (CPR) and advanced life support, the overall hospital discharge rate remains under 10%2. Even among those achieving return of spontaneous circulation (ROSC), nearly two-thirds experience severe neurological deficits, and about one-third remain comatose after 72 h3. Thus, preserving neurological function has become a focal point in post-resuscitation care, making early, accurate prognostication essential for guiding targeted interventions or decisions on withdrawing life support. The European Resuscitation Council (ERC) and the European Society of Intensive Care Medicine (ESICM) advocate for a multi-modal approach, combining clinical assessments, neurophysiology, biomarkers, and neuroimaging4. However, while effective in specificity, these methods often lack sensitivity, resulting in prognostic uncertainty in up to 68% of cases5. This emphasizes the need for more sensitive tools to better predict neurological outcomes and improve patient care.
Electroencephalography (EEG) is widely utilized in post-cardiac arrest monitoring due to its real-time, portable, and quantitative capabilities6. Recent studies demonstrate that quantitative EEG (QEEG) features are highly effective in predicting neurological outcomes, with significant accuracy; for example, Lee et al. reported an AUC of 0.887. Research by Ghassemi et al. highlights that temporal EEG features, which incorporate dynamic patterns, enhance prognostic models, outperforming static features (AUC = 0.83 vs. 0.79)8. Additionally, biomarkers like alpha-power spectral density have demonstrated strong predictive value for positive outcomes, achieving an AUC of 0.903 in studies by Kim et al.9. However, using EEG as a standalone prognostic tool in clinical settings is limited due to variability in feature selection, patient-specific factors (e.g., comorbidities, medication), and technical challenges like artifact contamination. These issues impact its consistency and generalizability in clinical workflows, suggesting that an integrative approach incorporating additional physiological indicators may be necessary for a more comprehensive assessment of neurological outcomes.
Recent research has increasingly focused on integrating electrocardiography (ECG), especially heart rate variability (HRV) analysis, with EEG to enhance the prediction of neurological outcomes after cardiac arrest. HRV metrics, reflecting autonomic nervous system (ANS) activity, have been linked to brain function, with specific markers such as very-low-frequency (VLF), low-frequency (LF) power, and the LF/HF ratio correlating with improved neurological outcomes10,11. Studies have also identified ECG abnormalities like QTc prolongation and arrhythmias as predictors of severe brain injury and higher mortality rates, particularly in traumatic brain injury (TBI) patients12,13. However, the limitation of ECG-based methods is that they primarily offer indirect measures of brain function by reflecting ANS activity, rather than capturing direct neurophysiological changes, especially during critical phases like ischemia and reperfusion14. To bridge this gap, recent studies have explored brain-heart coupling, examining the interplay between EEG frequency bands and HRV. For instance, Hermann et al. found that disrupted EEG-HRV interactions were significantly associated with poor neurological outcomes (p < 0.02), highlighting the potential of coupling these modalities15. Although these findings suggest the potential of coupling studies, such research typically focuses on specific interaction patterns without leveraging the full range of data available from a comprehensive multi-modal approach. To date, no studies have used both EEG and ECG features at a granular level to predict neurological outcomes post-cardiac arrest. Such a multi-modal integration could provide more nuanced patient insights, leading to more accurate and holistic assessments.
Despite the significant potential of integrating EEG and ECG data to enhance prognostic accuracy, a critical challenge lies in the interpretability of the sophisticated machine learning models employed for such analyses. While these models often achieve impressive predictive performance, their inherent “black box” nature limits their clinical applicability, particularly in intensive care units (ICUs) where crucial decisions—such as whether to initiate or withdraw life-sustaining treatments—require transparency and clear justifications. This opacity may hinder clinicians’ trust and acceptance, thereby impeding the integration of these models into routine practice.
In this study, we developed a multi-modal prognostic model that synthesizes EEG, ECG, and clinical data to improve outcome predictions following cardiac arrest. Our approach goes beyond mere predictive accuracy by incorporating SHapley Additive Explanations (SHAP) to elucidate the contribution of each feature, thereby enhancing model interpretability. This integration of interpretability ensures that the model is not only accurate but also transparent, fostering clinician trust. By leveraging the combined strengths of multi-modal data, our framework provides a robust, interpretable solution that aligns with clinical needs, ultimately supporting evidence-based decision-making in critical care settings. This approach holds significant promise for enhancing prognostic accuracy while maintaining the transparency necessary for clinical adoption.
Materials and methods
Datasets
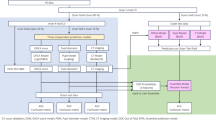
This study utilized data from the International Cardiac Arrest Research Consortium (I-CARE) database16, encompassing clinical, EEG, and ECG recordings from comatose patients post-cardiac arrest, collected across seven academic institutions in the United States and Europe. The cohort included adult patients who experienced either in-hospital or out-of-hospital cardiac arrest, achieved return of spontaneous circulation (ROSC), but remained comatose (Glasgow Coma Score ≤ 8). EEG monitoring commenced within hours of cardiac arrest and was sustained based on clinical requirements, with ECG recordings incorporated when available. Clinical data encompassed demographics, cardiac arrest specifics, ROSC duration, and targeted temperature management parameters, while neurological outcomes were evaluated using the Cerebral Performance Category (CPC) scale. Outcome data were acquired through phone interviews conducted at six months or chart review within three to six months post-ROSC. For a detailed workflow, refer to Fig. 1.
Ethical statement
The study received approval from the Institutional Review Boards (IRBs) of all participating institutions, including the Partners Healthcare IRB (#2013P001024) for the institutions in the United States. Due to the retrospective nature of the study and the use of anonymized data, informed consent was waived.
Pre-processing
EEG data were imported into the EEGLAB environment and re-referenced to a common average17. ECG data were synchronized and merged with EEG data for multi-modal analysis. A band-pass filter (0.5 to 30 Hz) was applied to remove artifacts, and the data were down-sampled to 100 Hz18. The combined dataset underwent manual artifact removal by trained reviewers. ECG-specific processing involved an additional band-pass filter (0.5 to 20 Hz), with QRS complexes detected using the Pan-Tompkins algorithm. Manual corrections of R-peaks and linear interpolation of ectopic beats were performed to refine R-R intervals19
Feature engineering
The feature engineering process in this study comprises three key steps: unimodal feature extraction, feature selection, and feature fusion20. These steps are designed to optimize the multi-modal model for predicting neurological outcomes in post-cardiac arrest patients
Feature extraction
Clean 20-minute segments of EEG and ECG data were used for analysis. EEG features included time-domain (mean, variance), frequency-domain (power spectral density, band power, coherence), and nonlinear metrics (fractal dimension: Higuchi fractal dimension, Shannon entropy, Lempel-Ziv complexity). ECG features focused on HRV metrics such as NN50, pNN50, HF, LF, SD1, SD2, and indices like cardiac sympathetic index (CSI) and cardiac vagal index (CVI). Clinical features (age, sex, ROSC duration, hypothermia status, shockable rhythm) were also integrated to enhance the model’s predictive capabilities21,22,23.
Feature selection
To optimize classification performance, this study employed a multi-step feature engineering strategy encompassing polynomial feature expansion and L1 regularization (Lasso) logistic regression24. Initially, polynomial expansion was applied to generate interaction and quadratic terms, enabling the model to capture non-linear interactions between physiological signals (EEG, ECG) and clinical features. Subsequently, L1 regularization was utilized for feature selection across both unimodal (EEG or ECG) and multi-modal (EEG + ECG) datasets, effectively shrinking irrelevant feature coefficients to zero and retaining only the most informative predictors. For unimodal datasets, this approach filtered redundant features, while for multi-modal datasets, principal component analysis (PCA) was employed post-L1 selection to reduce dimensionality while preserving 95% of the variance, thereby mitigating multicollinearity and enhancing computational efficiency25,26
Feature fusion
The study implemented feature-level fusion (FLF) to integrate EEG, ECG, and clinical data into a unified feature vector27,28. By amalgamating features from distinct modalities, FLF effectively captures complementary information, thereby enhancing the model’s ability to discern complex interdependencies between neurological, cardiac, and clinical parameters. This integrative approach significantly improves the predictive accuracy of neurological outcomes post-cardiac arrest, providing a comprehensive and robust assessment framework
Classification
Data balancing
To address the class imbalance inherent in the dataset, where patients with a CPC score of 1–2 (good neurological outcomes) are significantly fewer than those with a CPC score of 3–5 (poor neurological outcomes), the Synthetic Minority Over-sampling Technique (SMOTE) was employed during the training process. SMOTE, implemented using Python’s sklearn toolkit29, was used to generate synthetic samples for the minority class (CPC 1–2) by setting the parameter K = 5 (nearest neighbors). This technique ensures a balanced distribution between the positive and negative classes, improving the model’s ability to learn from both classes equally. To avoid overfitting, SMOTE was applied independently to each training fold and excluded before cross-validation, ensuring that synthetic samples did not influence the validation process.
Model selection and hyperparameter tuning
In this study, we systematically evaluated a range of machine learning algorithms, including Logistic Regression (LR), Support Vector Machine (SVM), Random Forest (RF), and Gradient Boosting (GBOOST). These algorithms are well-established for their ability to handle multi-modal data and tackle complex classification tasks30.
LR was selected as a baseline model due to its simplicity, interpretability, and effectiveness in linear classification problems. It serves as a reference for comparing more complex models, particularly when feature interactions are limited. SVM, on the other hand, was chosen for its capacity to define decision boundaries in high-dimensional spaces and its robustness in managing non-linear relationships. This is crucial for complex data types like EEG and ECG, as demonstrated in emotion recognition tasks involving multi-modal physiological signals31. RF, an ensemble learning technique, was employed for its ability to mitigate overfitting and perform automatic feature selection, making it ideal for handling multi-dimensional data. Its success in seizure classification and epilepsy localization further highlights its suitability for this study32. Lastly, GBOOST was incorporated for its capacity to build weak learners sequentially, progressively correcting errors from previous models. It is particularly effective in tasks involving intricate patterns and interactions, such as those found in multi-modal datasets used in seizure prediction and emotion recognition33,34.
The dataset was split into a training set and a test set using stratified sampling in an 80:20 ratio to preserve the original class distribution. This ensures proportional representation of each class, minimizing potential biases during model training and evaluation. The models were trained on the 80% training set and evaluated on the 20% test set to rigorously assess their generalization capabilities. Given the class imbalance in the dataset, all models incorporated a class_weight=’balanced’ strategy to address this issue. Additionally, the performance of each classifier was optimized using the Optuna framework for hyperparameter tuning. The Tree-structured Parzen Estimator (TPE) algorithm, employed by Optuna, efficiently explores the hyperparameter space and identifies optimal configurations35.
Performance evaluation metrics
Model robustness was assessed using 10-fold cross-validation with each fold serving as an independent test set once. This iterative process minimized data leakage by ensuring patient data was restricted to either the training or test fold. Key performance metrics, including AUC-ROC, accuracy, sensitivity, specificity, and F1 score, were reported with 95% confidence intervals to validate the model’s generalizability.
Interpretation
In this study, SHAP analysis were used in this study to interpret model predictions, clarifying the influence of specific features on patient outcomes and enhancing clinical applicability. SHAP, grounded in cooperative game theory, assigns each feature a contribution score indicating its effect on the prediction. Positive SHAP values increase the likelihood of poor outcomes, while negative values suggest favorable prognoses. By providing both global and individual prediction insights, SHAP supports clinicians in comprehending model behavior and its implications for patient care36.
Study endpoints
The primary endpoint of this study is the classification of neurological outcomes based on the CPC scale, which ranges from 1 (full recovery) to 5 (death). A good neurological outcome is defined as a CPC score of 1 or 2, while a poor neurological outcome corresponds to a CPC score of 3, 4, or 5. The classification task is framed as a binary prediction problem, where the model distinguishes between good (CPC 1–2) and poor (CPC 3–5) outcomes using features derived from EEG, ECG, and clinical data.
Statistical analysis
To assess differences among groups based on the 16 SHAP-identified features, robust statistical methods were applied. Continuous variables were analyzed using the Mann-Whitney U test, and associations with categorical variables, such as “Shockable Rhythm,” were evaluated using the Chi-Square test. To mitigate type I errors from multiple comparisons, the Benjamini-Hochberg correction was used with a significance threshold of p < 0.05. Conducted in SPSS, these analyses validated the discriminative power of key features, reinforcing the predictive model’s reliability and interpretability.
Results
Population description
In this study, data were selectively included based on the study objectives, retaining only patients who had both EEG and ECG data available. As a result, a total of 277 patients were included in the analysis (See supporting, Table 1 for details). The mean age was 61.04 ± 16.52 years, with 69.2% female. Of these, 88 patients had good neurological outcomes, while 178 had poor outcomes. The good outcome group was younger (mean age 56.49 ± 14.04) compared to the poor outcome group (63.15 ± 17.18). Shockable rhythms were more common in the good outcome group (69.3% vs. 31.6%), with similar rates of targeted temperature management (27.3% vs. 27%).
Classification performance
After pre-processing, three types of features were extracted: EEG, ECG, and clinical data. These features were then combined into three distinct scenarios for comparative analysis: (1) all features combined (EEG + ECG + clinical), (2) EEG features alone, and (3) ECG features alone. We employed four machine learning models: Logistic Regression (LR), Support Vector Machine (SVM), Random Forest (RF), and Gradient Boosting (GBOOST) to evaluate the classification performance across these scenarios.
Across all models, the multi-modal integration of EEG + ECG + clinical features (FLF) consistently provides the best performance, with AUC scores ranging from 0.75 to 1.0, and in some time periods, reaching a perfect score of 1.0.
Specifically, Fig. 2A (LR model) shows that the multi-modal approach yields high AUC values, especially in the 12–24 h and 49–72 h time windows. In contrast, EEG features alone exhibit more variability in performance, with AUC values ranging from 0.5 to 0.9, and ECG features alone generally demonstrate limited predictive power, with AUCs consistently below 0.6. Similarly, Fig. 2B (SVM model) demonstrates that the multi-modal approach (FLF) consistently outperforms both EEG only and ECG only features, with AUC scores between 0.8 and 1.0 across most time windows. EEG only features show moderate AUC values around 0.7 to 0.8 during the 25–48 h and 49–72 h windows, while ECG only consistently performs poorly, with AUC values typically below 0.6. In Fig. 2C (RF model), the multi-modal integration of EEG + ECG + clinical features (FLF) shows substantial variability, occasionally achieving AUC values close to 1.0, particularly in the 25–48 h time window. EEG features alone perform consistently with AUCs above 0.7 during the 49–72 h window, although their performance fluctuates in other periods. ECG features alone show AUC scores generally below 0.6, reflecting their limited utility as standalone predictors. Finally, Fig. 2D (GBOOST model) illustrates that the FLF approach again achieves superior AUC scores, particularly within the 12–48 h windows, where AUCs range from 0.75 to 1.0. EEG features alone show moderate performance, with AUC values around 0.7, while ECG features alone consistently exhibit weak predictive power, with AUCs remaining below 0.6 across all time windows.
Although performance across all time points is relevant, the 24-hour and 36-hour time windows are highlighted due to their consistently superior performance across all models using the multi-modal integration of EEG, ECG, and clinical features (FLF). These time points achieved the highest accuracy scores, consistently approaching 1.0, which demonstrates the clear advantage of the FLF approach in predictive modeling. As shown in Table 2, we emphasize these two time points by presenting additional metrics, further supporting the robustness of FLF.
At the 24-hour time window, FLF outperforms both EEG-only and ECG-only features across all models. The SVM model achieves an accuracy of 0.853, surpassing both EEG-only (accuracy: 0.756) and ECG-only (accuracy: 0.536) models. Similarly, the LR model shows a significant performance boost with FLF, attaining an accuracy of 0.829, again outperforming EEG-only (accuracy: 0.707) and ECG-only (accuracy: 0.585). The RF model also demonstrates the strength of FLF, with an accuracy of 0.707, compared to lower performance from EEG-only (accuracy: 0.609) and ECG-only (accuracy: 0.585). The GBOOST model follows a similar trend, with FLF yielding an accuracy of 0.731, surpassing both EEG-only (accuracy: 0.658) and ECG-only (accuracy: 0.512). At the 36-hour time point, FLF consistently achieves perfect performance across all models, with accuracy scores reaching 1.0. These results further highlight the superiority of the multi-modal approach in predicting outcomes. This reinforces the crucial role of multi-modal data fusion in improving predictive power, particularly at the 24-hour and 36-hour time points. For a comprehensive analysis across all time points, including those not highlighted in this summary, please refer to the complete dataset in the submitted Excel file.
SHAP analysis
In this study, SHAP analysis was employed to assess the contribution of individual features from a multi-modal dataset (including EEG, ECG, and clinical data) to the predictions made by the machine learning model. To ensure scientific rigor and reliability of the results, the Random Forest (RF) model was chosen as the primary analysis tool, owing to its superior performance across various evaluation metrics, such as accuracy and AUC. By utilizing the RF model, SHAP analysis provided valuable insights into how different physiological and clinical variables drive model predictions. Figure 3 highlights that “Shockable Rhythm” had the highest mean SHAP value (~ 0.17), indicating its strong influence on outcomes, while features like HiguchiFD-2, HiguchiFD-7, and SD2 had moderate effects (mean SHAP values of 0.02 to 0.03). The summary plot further illustrates the consistent positive impact of “Shockable Rhythm,” while other features, such as HiguchiFD metrics, displayed context-dependent effects ranging between − 0.1 and 0.1. Minimal influence was observed for features like LF and CVI, with SHAP values below 0.01.
Overall, the SHAP analysis confirms the dominant role of clinical features, particularly “Shockable Rhythm,” in model predictions, while EEG and ECG data offer complementary context-specific contributions. These results demonstrate that integrating multi-modal data significantly enhances model robustness and predictive accuracy in clinical decision-making.
SHAP based Feature ranking of top 16 features (a) SHAP feature ranking (b) SHAP summary plot. Note: The x-axis represents the shapley values while the y axis represents the included features ranking. Each blue dot corresponds to a lower magnitude of the feature for different samples, while the red dots indicate higher magnitudes of the features.
Statistical comparison of differences
Statistical tests validated the significance of key features identified via SHAP analysis. According to Supporting Table 2, Mann-Whitney U and Chi-Square tests showed that while some ECG features (e.g., SD2, Mean_RR) lacked statistical significance (p > 0.05), several EEG features (e.g., HiguchiFD_2, HiguchiFD_7) were highly significant (adjusted p < 0.001). Additionally, the clinical feature “Shockable Rhythm” was associated with better outcomes (p = 0.004). These findings emphasize that integrating EEG and clinical data significantly enhances predictive accuracy, even when ECG features alone are less informative, thereby improving model robustness and clinical applicability.
Discussion
In this study, we developed a machine learning model integrating multi-modal data (EEG, ECG, and clinical information) to predict neurological outcomes in post-cardiac arrest patients. The results demonstrate that combining all multi-modal features significantly improves predictive accuracy compared to single-modality approaches. Notably, during early critical time windows (12–24 and 25–48 h), the model achieved AUC values up to 1.0, outperforming models based solely on EEG (AUC 0.7–0.8) or ECG (AUC < 0.6). This highlights the importance of early, comprehensive assessments for guiding clinical interventions.
Model performance evaluation
This study builds upon previous research that has employed machine learning models to predict neurological outcomes in patients following cardiac arrest. Such as those by Maschke et al. and Amorim et al., have primarily focused on developing predictive models using EEG data alone. These single-modality approaches demonstrated moderate predictive performance, with AUC values typically ranging from 0.7 to 0.837,38. Similarly, studies by Sung et al. and Kim et al. investigated ECG-based features, like HRV, for outcome prediction; however, their models generally yielded lower AUC scores, often below 0.79,39. Our study extends these findings by integrating multi-modal data—encompassing EEG, ECG, and clinical features—into a unified machine learning framework. This multi-modal approach allows for a more comprehensive representation of the patient’s condition by leveraging the complementary strengths of each data type, thereby enhancing overall prediction accuracy. The results show that our model, incorporating all multi-modal features, achieved higher AUC values, ranging from 0.75 to 1.0, compared to single-modality models. This underscores the advantage of multi-modal integration in capturing a broader spectrum of factors that influence neurological outcomes.
A particularly notable finding in our study is the model’s performance across different time windows. The classification analyses revealed that integrating multi-modal data significantly enhances predictive accuracy, especially in the early time windows (12–24 h and 25–48 h), where AUC values reached 1.0 and 0.85, respectively. This is a substantial improvement over single-modality models based on EEG (AUC 0.7–0.8) or ECG (AUC below 0.6). Our results align with and extend the findings of Uslenghi et al., who emphasized the importance of early time-window analysis for reliable predictions in post-cardiac arrest patients40. The ability of our model to achieve higher predictive accuracy in early time windows underscores the critical value of early multi-modal assessments in clinical decision-making. Moreover, our approach’s ability to capture the temporal dynamics of various physiological signals is critical, as highlighted by Ghassemi et al.and Dai et al., who noted that the predictive value of certain physiological markers, such as EEG complexity, may change over time due to evolving pathophysiological states8,41.
By employing advanced machine learning models such as LR, SVM, RF, and GBOOST, our approach handles complex, high-dimensional data better than traditional methods. These models excel at capturing non-linear relationships, aligning with advancements in AI-driven critical care as noted by Callaway et al. and Nolan et al.42,43. Overall, integrating multi-modal data and focusing on early assessments significantly enhances prediction accuracy, offering a robust foundation for future research and clinical applications.
Explainable results
To elucidate the contribution of each feature to predicting neurological outcomes after cardiac arrest, we employed SHAP to interpret their impact on the model’s predictions. “Shockable Rhythm,” which refers to the initial presentation of shockable rhythms like ventricular fibrillation (VF) or pulseless ventricular tachycardia (VT), emerged as the most influential feature with the highest mean SHAP value. This finding is consistent with established clinical guidelines by Callaway et al. and Nolan et al., which emphasize that patients with initial shockable rhythms have a significantly better prognosis than those with non-shockable rhythms like asystole or pulseless electrical activity (PEA)42,43. The prominence of Shockable Rhythm in our model underscores the crucial role of timely defibrillation and early resuscitation interventions in improving neurological outcomes, reinforcing its value in clinical decision-making44.
In addition, our SHAP analysis also highlighted the importance of EEG-based Higuchi Fractal Dimension (HiguchiFD) metrics and HRV measures as significant predictors. HiguchiFD, which quantifies EEG signal complexity, has been validated as a marker for brain activity and injury severity after resuscitation21. Notable HiguchiFD features in our model, such as HiguchiFD-2 (channel FP2) and HiguchiFD-7 (channel P3), point to the relevance of specific brain regions. The FP2 electrode, associated with the frontal lobe, is linked to cognitive functions and decision-making processes crucial for recovery post-brain injury45. Similarly, P3 in the parietal lobe is related to sensory integration and working memory, which are vital for functional recovery and neural reorganization after cardiac arrest46. The significant SHAP values for these features suggest that neural complexity in these regions reflects key processes of neuroplasticity and network adaptation critical to recovery.
HRV features, including SD2, mean_RR, and SDNN, also emerged as important indicators, reflecting autonomic regulation during post-arrest recovery. Reduced HRV is a known predictor of poor outcomes, indicating autonomic dysfunction and impaired recovery potential47,48. Our findings, where mean_RR and SDNN show substantial SHAP values, reinforce the value of integrating HRV metrics for early risk stratification and improving the model’s predictive capability. By combining clinical indicators like the initial shockable rhythm with EEG complexity and HRV measures, our model offers a robust, multi-modal approach to neurological prognostication, supporting more personalized and effective interventions for cardiac arrest patients.
Clinical relevance
This study highlights the efficacy of integrating multi-modal data (EEG, ECG, and clinical variables) to enhance early-stage neurological outcome predictions in post-cardiac arrest patients, aligning with the European Resuscitation Council’s guidelines for a multi-modal prognostic approach43. Utilizing SHAP analysis significantly improves model interpretability, allowing clinicians to better understand each feature’s contribution to predictions49. This transparency is crucial for fostering trust in AI-assisted decision-making, particularly in critical interventions where the decision to continue or withdraw life-sustaining therapies is based on individualized risk assessments45. The methodology extends beyond cardiac arrest care, with potential applications in stroke management and anesthesia, by leveraging comprehensive physiological data to inform more tailored treatment strategies50.
Limitations and future directions
While our study yielded promising outcomes, certain limitations must be addressed for broader validation. The relatively small sample size restricts the generalizability and statistical power of our findings, highlighting the need for larger, diverse cohorts in future research. Although multi-modal data integration has improved predictive accuracy, challenges remain in synchronizing diverse data streams. Future studies should explore advanced data fusion methods to better capture temporal dynamics. The current use of static time windows may not fully capture dynamic physiological changes; thus, adaptive time-windowing techniques that adjust model parameters in real-time could enhance personalized interventions. Furthermore, incorporating additional signals like blood pressure variability and respiratory patterns could further improve neurological outcome assessments. Addressing these gaps will enhance the framework’s precision and clinical relevance in critical care settings.
Conclusion
Leveraging advanced machine learning models allowed us to effectively capture the intricate, nonlinear interactions between various features, establishing a solid foundation for precise and timely predictions in critical care environments.Multi-modal integration plays a crucial role in optimizing patient prognosis and tailoring interventions in post-cardiac arrest management, significantly improving patient outcomes and promoting more personalized care in post-arrest management.
Data availability
The data used in this study is derived from the International Cardiac Arrest Research Consortium (I-CARE) database, which includes clinical, EEG, and ECG data collected from seven academic institutions in the United States and Europe. Access to this dataset is subject to approval from the I-CARE database administrators. For further inquiries regarding data access, please contact Haojun Fan at fanhaojun999@126.com. All data are available upon reasonable request and in compliance with the relevant data-sharing policies of the I-CARE consortium.
References
Perkins, G. D. et al. Brain injury after cardiac arrest. Lancet 398 (10307), 1269–1278 (2021).
Perkins, G. D. et al. European resuscitation Council guidelines 2021: executive summary [published correction appears in resuscitation. ;163:97–98. (2021).
Callaway, C. W. et al. Part 8: Post-cardiac arrest care: 2015 American heart association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care [published correction appears in circulation. ;136(10):e197. (2017).
Nolan, J. P. et al. European resuscitation Council and European society of intensive care medicine 2015 guidelines for post-resuscitation care [published correction appears in intensive. Care Med. 42 (3), 488–489 (2016).
Sandroni, C., D’Arrigo, S. & Nolan, J. P. Prognostication after cardiac arrest. Crit Care. ;22(1):150. Published 2018 Jun 5. (2018).
Sandroni, C., Cronberg, T. & Hofmeijer, J. EEG monitoring after cardiac arrest. Intensive Care Med. 48 (10), 1439–1442 (2022).
Lee, S. et al. Quantitative EEG predicts outcomes in children after cardiac arrest. Neurology 92 (20), e2329–e2338 (2019).
Ghassemi, M. M. et al. Quantitative electroencephalogram trends predict recovery in hypoxic-ischemic encephalopathy. Crit. Care Med. 47 (10), 1416–1423 (2019).
Kim, M. J., Kim, Y. J., Yum, M. S. & Kim, W. Y. Alpha-power in electroencephalography as good outcome predictor for out-of-hospital cardiac arrest survivors. Sci. Rep. 12 (1), 10907 (2022). Published 2022 Jun 28.
Dietrichs, E. S., Tveita, T., Myles, R. & Smith, G. A novel ECG-biomarker for cardiac arrest during hypothermia. Scand. J. Trauma. Resusc. Emerg. Med. 28 (1), 27 (2020). Published 2020 Apr 10.
Benghanem, S. et al. Heart rate variability for neuro-prognostication after CA: insight from the Parisian registry. Resusc. Published Online June 24, (2024).
Lenstra, J. J. et al. la The association of early electrocardiographic abnormalities with brain injury severity and outcome in severe traumatic brain injury. Front Neurol. ;11:597737. Published 2021 Jan 8. (2021).
Martin, S. D. et al. Heart rate variability in children with moderate and severe traumatic brain injury: a prospective observational study.J. Pediatric Intensive Care.
Rohaut, B. et al. multi-modal assessment improves neuroprognosis performance in clinically unresponsive critical-care patients with brain injury. Nat. Med. 30 (8), 2349–2355 (2024).
Hermann, B. et al. Aberrant brain-heart coupling is associated with the severity of post cardiac arrest brain injury. Ann. Clin. Transl Neurol. 11 (4), 866–882 (2024).
Amorim, E. et al. I-CARE: international cardiac arrest research consortium database (version 2.0). PhysioNet. (2023).
Delorme, A. & Makeig, S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods. 134 (1), 9–21 (2004).
Zheng, W. L. et al. Predicting neurological outcome from electroencephalogram dynamics in comatose patients after cardiac arrest with deep learning. IEEE Trans. Biomed. Eng. 69 (5), 1813–1825 (2022).
Rangsungnoen, S., Chanbenjapipu, P., Mathuradavong, N. & Suwanprasert, K. A hybrid signal processing of RR intervals from QTc variation searching arrhythmia and improving heart rate variability assessment in acute large artery ischemic stroke. J. Med. Eng. 2016, 9823026 (2016).
Ramadan, M. A. et al. multi-modal machine learning approach for emotion recognition using physiological signals. Biomed. Signal Process. Control, 96. (2024).
Dai, C. et al. Combining early post-resuscitation EEG and HRV features improves the prognostic performance in cardiac arrest model of rats. Am. J. Emerg. Med. 36 (12), 2242–2248 (2018).
Tian, F. et al. Surge of corticocardiac coupling in SHRSP rats exposed to forebrain cerebral ischemia. J. Neurophysiol. 121 (3), 842–852 (2019).
Sung, C. W., Shieh, J. S. & Jaw, F. S. Machine learning analysis of heart rate variability for the detection of seizures in comatose cardiac arrest survivors. IEEE Access. 8, 160515–160525 (2020).
Cui, X. et al. Adaptive LASSO logistic regression based on particle swarm optimization for Alzheimer’s disease early diagnosis. Comput. Methods Programs Biomed. 207, 106177 (2021).
Jomthanachai, S., Wong, W. P. & Khaw, K. W. An Application of Machine Learning Regression To Feature Selection: a Study of Logistics Performance and Economic Attribute (Neural Computing and Applications, 2022).
El Guide, M., Jbilou, K. & Koukouvinos, C. Comparative Study of L1 Regularized Logistic Regression Methods for Variable Selection (Communications in Statistics - Simulation and Computation, 2022).
Huddar, M. G., Sannakki, S. S. & Manjunath, S. Multi-level feature optimization and multi-modal contextual fusion for sentiment analysis and emotion classification. Comput. Intell. 36 (2), 440–452 (2020).
Wang, X., Xu, M., Xiong, X. & Ning, C. Remote sensing scene classification using heterogeneous feature extraction and multi-level fusion. IEEE Access. 8, 211814–211825 (2020).
Elreedy, D., Atiya, A. F. & Kamalov, F. A theoretical distribution analysis of synthetic minority oversampling technique (SMOTE) for imbalanced learning. Mach. Learn., (2024).
Brugnara, G., Neuberger, U., Mahmutoglu, M. A., Foltyn, M., Herweh, C., Nagel, S., Heiland, S. Multi-modal predictive modeling of endovascular treatment outcome for acute ischemic stroke using machine-learning. Stroke, 2020.
Sharma, R., Pachori, R. B. & Sircar, P. Seizures classification based on higher order statistics and deep neural network. Biomed. Signal Process. Control. 59, 101921 (2020).
Sharma, R. Automated human emotion recognition using hybrid approach based on sensitivity analysis on residual time-frequency plane with online learning algorithm. Biomed. Signal Process. Control. 84, 1–10 (2023).
Sharma, R., Pachori, R. B. & Sircar, P. Automated emotion recognition based on higher order statistics and deep learning algorithm. Biomed. Signal Process. Control. 55, 123–135 (2020).
Sharma, R. Localization of epileptic surgical area using automated hybrid approach based on higher-order statistics with sensitivity analysis and residual wavelet transform. Biomed. Signal Process. Control. 80, 22–30 (2023).
Sipper, M. High per parameter: A large-scale study of hyperparameter tuning for machine learning algorithms. Algorithms 15 (9), 315 (2022).
Sahlaoui, H., Nayyar, A., Agoujil, S. & Jaber, M. M. Predicting and interpreting student performance using ensemble models and Shapley additive explanations. IEEE Access. 9, 128453–128467 (2021).
Maschke, C. et al. Functional outcome prediction after cardiac arrest using machine learning and network dynamics of resting-state electroencephalography. Crit. Care Med. ;51(6) (2023).
Amorim, E. et al. Predicting neurological outcome from electroencephalogram dynamics in comatose patients after cardiac arrest with deep learning. Neurocrit Care. 39 (1), 137–146 (2023).
Sung, C. W. et al. Machine learning analysis of heart rate variability for the detection of seizures in comatose cardiac arrest survivors. J. Clin. Monit. Comput. 34 (5), 917–926 (2020).
Uslenghi, F. et al. Recovery from coma after cardiac arrest: which time-window counts the most for deep learning predictions? Resuscitation 182, 40–48 (2023).
Dai, C. et al. Combining early post-resuscitation EEG and HRV features improves the prognostic performance in cardiac arrest model of rats. Am J Emerg Med. 36(10):1857.e1-1857. (2018).
Callaway, C. W. et al. Part 8: Post-cardiac arrest care: 2015 American heart association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation ;132 (2015). (18 Suppl 2).
Nolan, J. P. et al. European resuscitation Council and European society of intensive care medicine guidelines for post-resuscitation care. Intensive Care Med. 41 (12), 2039–2056 (2015).
Sandroni, C. et al. Prognostication in comatose survivors of cardiac arrest: an updated European resuscitation Council and European society of intensive care medicine consensus statement. Intensive Care Med. 47 (9), 1059–1083 (2021).
Fridman, E. A. & Schiff, N. D. The neural basis of cognitive recovery post-brain injury and the role of the frontal lobe in restoring higher-order cognitive functions. Front Neurol.
Thibaut, A. et al. Parietal lobe involvement in sensory integration and working memory as a determinant of recovery following severe brain injury. J Neurol Neurosurg Psychiatry.
Yperzeele, L. et al. Automated EEG amplitude suppression analysis for early prognosis in cardiac arrest patients: A prospective observational study. Clin Neurophysiol.
Ghassemi, M. M. et al. Quantitative electroencephalogram trends predict recovery in hypoxic-Ischemic encephalopathy. Crit Care Med.
Jeanne, M., Logier, R., De Jonckheere, J., Tavernier, B. & Vallet, B. Heart rate variability during total intravenous anesthesia: effects of nociception and analgesia. Anesth. Analgesia. 108 (2), 691–695 (2009).
Campbell, J. P. & Sleigh, J. W. The role of EEG in monitoring and management of anaesthesia and its recovery. Anesthesiology 129 (2), 398–341 (2018).
Acknowledgements
We would like to thank the I-CARE database management team for providing data support, and all the clinicians and data scientists involved in this project for their valuable contributions.
Funding
This research was financially supported by the National Key R&D Program of China (No. 2021YFC3002200).
Author information
Authors and Affiliations
Contributions
Y.N., X.C., and J.F. contributed equally to this work and were involved in data analysis and model development. C.L., M.F., and Z.L. were responsible for data preprocessing and feature engineering. X.M., Y.L., and L.L. assisted with model validation and result interpretation. H.F. conceptualized and supervised the study, provided project management, and drafted the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Niu, Y., Chen, X., Fan, J. et al. Explainable machine learning model based on EEG, ECG, and clinical features for predicting neurological outcomes in cardiac arrest patient. Sci Rep 15, 11498 (2025). https://doi.org/10.1038/s41598-025-93579-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93579-0
Keywords
This article is cited by
-
Cross-modal synchronization of EEG and ECG reveals hidden signatures of recovery in traumatic brain injury
Journal of NeuroEngineering and Rehabilitation (2026)
-
Pain Induced by Friction Based on fMRI and EEG
Journal of Bionic Engineering (2026)