Abstract
Staphylococcus aureus is a common bacterium that sometimes causes various pyogenic diseases. Methicillin resistant S. aureus (MRSA) infections are particularly difficult to treat. Recently, MRSA has been spreading in the community, so it is important to determine the prevalence of MRSA in the community and to conduct epidemiological studies from genetic and statistical perspectives. In this study, S. aureus/MRSA was isolated from the oral and nasal cavities of 504 dental patients (65 inpatients and 439 outpatients). Sixty-two S. aureus strains and 9 MRSA strains were isolated from the oral cavity, and 112 S. aureus strains and 21 MRSA strains were isolated from the nasal cavity. Multi-locus sequence typing (MLST) analysis showed ST8 was high (18 isolates) among 30 MRSA isolates, whereas among 144 methicillin sensitive isolates, ST15 (25 isolates) and ST8 (20 isolates) were high. Statistical analysis of the patients’ clinical status revealed a correlation between oral S. aureus and denture use. Among the 34 patients from whom S. aureus was isolated from both sites, 25 had the same ST, and 23 showed below 40 single-nucleotide polymorphisms which are considered to be identical strains. Our study revealed various properties of S. aureus/MRSA in the oral and nasal cavities as commensals.
Similar content being viewed by others
Introduction
Staphylococcus aureus is prevalent to human skin and nasal mucosa. This organism causes food poisoning and opportunistic infections such as pyogenic skin diseases, enteritis and pneumonia1,2. This diversity of pathologies caused by S. aureus is due to the wide variety of virulence factors involved3. Furthermore, the emergence of methicillin-resistant S. aureus (MRSA) has been a serious problem for chemotherapy treatment4,5. MRSA generally exhibits multidrug resistance to several antibiotics. Therefore, MRSA infectious diseases such as soft skin tissue infection, pneumonia and sepsis are frequently refractory to chemotherapy treatments and have a poor prognosis. MRSA was initially prevalent mainly in health care facilities as a major cause of nosocomial infections6, but in recent years, it has also attracted attention as a cause of community-acquired infections7,8.
S. aureus, including MRSA, can sometimes colonize the oral cavity9,10. We also reported that MRSA has been isolated from the oral cavity of elderly people in long-term care facilities (LTCFs)11. Since S. aureus is one of the causative pathogens for aspiration pneumoniae12,13, it is important to recognize the prevalence of S. aureus and MRSA colonization in the oral cavity of not only patients/residents in hospitals and facilities but also individuals in general. In this context, there is a need to understand the prevalence of MRSA in the oral cavity of home residents, who are considered to have a relatively high degree of independence, along with hospital patients and elderly care facilities.
Genetical analysis is a powerful tool for epidemiologic analysis such as infection control, public health and food microbiology. Various techniques including pulsed-field electrophoresis, random amplification of polymorphic DNA (RAPD) analysis and multi-locus sequence typing (MLST) have been used14,15. Furthermore, single-nucleotide polymorphisms (SNPs) analysis using whole genome sequence (WGS) is a more accurate method for determining identity between strains14. In investigating the transmission of S. aureus within a facility using SNP analysis, a number of SNPs between strains of less than 40 or 50 indicates the possibility of transmission11,16,17. In addition, WGS analysis is expected to provide the genetic determinants responsible for the virulence and antibiotic resistance of S. aureus.
In this study, we attempted to isolate S. aureus and MRSA from the oral and nasal cavities of patients visiting dental clinic of Hiroshima University Hospital. Also, the patient information obtained was used to examine the associations between the presence of S. aureus/MRSA and the participants. In addition, we used whole-genome sequencing data of all the S. aureus isolates to perform various genetic analysis including MLST analysis, phylogenetic tree analysis and SNP analysis for the characterization of isolates in detail.
Materials and methods
Study design
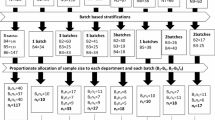
Specimens were collected between August 2021 and December 2022 to assess the prevalence of S. aureus/MRSA in the oral and nasal cavities. A total of 504 patients (65 inpatients and 439 outpatients) who visited seven departments of dentistry (Department of Endodontics and Operative Dentistry, Department of Periodontology, Department of Oral Implantology, Department of Oral Reconstructive Surgery, Department of Oral General and Oral Health Sciences and Oral Examination Center) at Hiroshima University Hospital (total 742 beds, secondary medical institution, located in Hiroshima city) were included (Supplemental Fig. 1). Informed consent was obtained from the patients, prior to specimen collection. The study was approved by the Ethics Committee of Hiroshima University Hospital (approval number: E-2525). All research procedures were conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from the study participants or their family members prior to enrollment in the study.
Sample collection
Specimens were collected from the oral and nasal cavities of patients aged 40 years and older who visited the dental clinic at Hiroshima University Hospital. Oral samples were collected from patients before mouthwash. Bacteria were isolated from the oral mucosa (buccal mucosa, tongue, palate, oral vestibule, etc.) via sterile swabs (Kawamoto Meckin Swab #104, Hydraflox Swab 25-3706-H). Nasal samples were collected by wiping the nasal mucosa. Each swab was applied directly to staphylococcal selective medium (Nissui Pharmaceuticals), and the medium was incubated under aerobic conditions at 37°C for 18–24 hours. Single yellow colonies (up to four colonies in each sample) were selected and streaked onto the same agar medium. A small part of bacterial cells was taken from each colony and suspended in 100 µL of CS buffer (100mM Tris-HCl (pH 7.5), 150mM NaCl, 10mM EDTA) containing 10 µg of lysostaphin (Sigma‒Aldrich). The bacterial suspension was incubated at 37°C for 15 min, after which the samples were heated at 95°C for 10 min. The supernatant was used as template DNA for PCR, which was performed with a specific primer for nuclease gene (nuc) (F: 5’- GTTGTAGTTTCAAGTCAAG-3’, R: 5’- TATCACCATCAATCGCT-3’) for S. aureus identification. The isolated S. aureus strains were stored in a freezer (-80 °C) before use. MRSA was defined as positive for the mecA gene based on the genomic data for each isolate. When more than two S. aureus strains were isolated from one participant, only one stain was used for further experiments.
Genetic analysis
Illumina whole-genome sequences of 174 S. aureus clinical isolates were used for genome analysis. Genomic DNA extraction, DNA library preparation and paired-end sequencing were performed11. The bacterial species were determined via KmerFinder 3.2 (Center for Genomic Epidemiology: URL: https://cge.food.dtu.dk/services/KmerFinder/); MLST analyses were performed via MLST 2.0 (Center for Genomic Epidemiology); and MLST alleles (arcC, aroE, glpF, gmk, pta, tpi and yqiL) were identified. The Clonal complexes (CC) were identified via PubMLST (https://pubmlst.org/). Antibiotic resistance genes were analyzed via ResFinder 4.1 (Center for Genomic Epidemiology), and pathogenic genes were analyzed via VirulenceFinder 2.0 (Center for Genomic Epidemiology). SCCmec typing was performed via SCCmecFinder 1.2 (Center for Genomic Epidemiology). To create the phylogenetic tree of S. aureus isolates, the assembled genome data were annotated by Prokka18, and the resulting gff files were aligned to the core genome linked by the Panaroo pipeline19. The maximum-likelihood tree was subsequently generated via RAxML-NG version 1.2.1 with 1000 bootstrap replicates20. The tree and metadata were constructed via the interactive tree of life (iTOL) version 7 software (https://itol.embl.de)21. A minimum spanning tree was also created via Grape tree version 1.5.0 software (https://achtman-lab.github.io/GrapeTree/MSTree_holder.html)22. The number of SNPs between oral and nasal isolates was analyzed using SKA version 1.0 package (split kmer analysis: https://github.com/simonrharris/SKA)23. The k-mer files were generated from paired fastq files using the fastq subcommand with default settings. The pairwise SNP distance between isolates were calculated using distance subcommand with default settings. The criterion for the intra-facility transmission of S. aureus was defined below 40 or 50 SNPs based on previous results11,16,17.
Clinical data
Clinical information was obtained from 472/504 participants. Information included demographics (age, sex, inpatient/outpatient), oral status (number of remaining teeth, obvious tongue or mucosa disease, and denture wear) and history of antibiotic use within one month. Among 472 patients, information about primary diseases including heart disease, cerebrovascular disease, diabetes, respiratory disease and malignant neoplasm were taken from 397 patients.
Statistics
Correlations between clinical information and bacterial resistance were analyzed by Fisher’s exact test for categorical variables and the Mann‒Whitney U test for continuous variables; results with a p value less than 0.05 were considered significant for all the statistical analyses. All the statistical analyses were performed with JMP Pro version 17 (SAS Institute, Cary, NC, USA).
Accession number
The genomic data of the S. aureus isolates used in this study have been deposited in NCBI database (accession number: PRJDB18935).
Results
Participant characteristics
In this study, specimens were collected from 504 patients (outpatients: 439, inpatients: 65) who visited the dental outpatient department of Hiroshima University Hospital. Of these, 472 (outpatients: 409, inpatients: 63) were selected and included in the statistical analysis, with a mean age of 69.6 ± 11.1 years (40–95 years). In terms of sex, 225 (47.7%) were male, with a mean age of 70.0 ± 10.9 years (40–93 years), and 247 (52.3%) were female, with a mean age of 69.2 ± 11.2 years (40–95 years).
The specimens were collected by the following seven departments: 73 from the Department of Endodontics and Operative Dentistry; 98 from the Department of Periodontology; 57 from the Department of Oral Implantology; 72 from the Department of Oral and Maxillofacial Reconstructive Surgery; 98 from the Department of Oral General Medicine; 33 from the Department of Oral Health; and 73 from the Oral Examination Center, of which 70, 97, 57, 55, 97, 24 and 72, respectively, were selected for statistical analysis.
Isolation of S. aureus/MRSA from the oral and nasal cavities
S. aureus was isolated from the oral or nasal cavities of 140 (27.8%) of 504 patients, of whom MRSA was isolated in 24 patients (4.8%) (Table 1). S. aureus isolates including MRSA isolates were found in 28 patients (5.6%) oral-only samples, 78 patients (15.5%) nasal-only samples and 34 patients (6.7%) both samples. For MRSA alone (24/504 patients), MRSA isolates were found in 3 patients (0.6%) oral-only samples, 15 patients (3.0%) from nasal-only samples and 6 patients (1.2%) both samples. The isolation rates of S. aureus and MRSA in the oral cavity were 12.3% (62/504 patients) and 1.8% (9/504 patients), respectively. The isolation rates of S. aureus and MRSA in the nasal cavity were 22.2% (112/504 patients) and 4.2% (21/504 patients), respectively. There were no significant differences in the isolation rates of S. aureus and MRSA from the oral and nasal cavities among the seven dental departments. S. aureus was isolated from 122 (27.8%) of 439 outpatients, of whom 21 (4.8%) had MRSA; S. aureus was isolated from 18 (27.7%) of 65 inpatients, of whom 3 (4.6%) had MRSA (Table 1).
Genomic analysis of S. aureus isolates
Among the 174 S. aureus (144 MSSA and 30 MRSA) isolates from 140 patients, 26 sequence types (STs) were identified, whereas 19 isolates did not match the deposited STs (Figs. 1 and 2, Supplemental Tables 1 and 2). The major STs of isolates in the oral and nasal cavities were ST8 (38 isolates: 21.8%) and ST15 (22 isolates: 12.6%). Among 30 MRSA isolates (9 isolates from the oral cavity and 21 isolates from the nasal cavity), ST8 was high (6 oral isolates: 66.7%, 12 nasal isolates: 57.1%), whereas among 144 MSSA isolates (53 isolates from the oral cavity and 91 isolates from the nasal cavity), ST15 (8 isolates: 15.1%) and ST97 (6 isolates: 11.3%) were high in the oral cavity, and ST8 (17 isolates: 18.7%) and ST15 (14 isolates: 15.4%) were high in the nasal cavity.
We subsequently analyzed SCCmec types among the isolates. Four SCCmec types were identified among the 30 MRSA isolates from 24 patients (Supplemental Table 3). In terms of distribution in the outpatient/inpatient and oral/nasal cavities, SCCmec-IV was the most common type, with 18/24 (66.7%) isolates from the outpatient group and 6/6 (100%) isolates from the inpatient group and 7/9 (77.8%) oral isolates and 17/21 (80.9%) nasal isolates. Regarding the untypable strain (HSA344), the SCCmecfinder identified that HSA344 had a class A mec gene complex, and the cassette showed highest homology with SCCmec type VIII, but since the ccrA and ccrB genes were absent, the exact SCCmec type could not be identified.
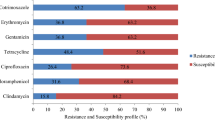
We identified antibiotic resistance and virulence genes in each isolate (Supplemental Table 4) and compared the distribution of antibiotic resistance genes between MSSA and MRSA isolates (Fig. 2, Supplemental Table 5). Among antibiotic resistance genes, the proportions of aminoglycoside resistance genes (ant(9)-Ia: 70%, aac(6’)-aph(2’’): 40%, aadD: 36.7%, bleO: 36.7%), macrolide resistance genes (erm(A): 70%), tetracycline resistance genes (tet(K, L, M): 33.3%) and quinolone resistance genes (gyrA: 66.7%, grlA: 66.7%) in MRSA isolates were greater than those of MSSA isolates with aminoglycoside resistance genes (ant(9)-Ia: 20.1%, aac(6’)-aph(2’’): 20.8%, aadD: 0.7%, bleO: 0.7%), macrolide resistance genes (erm(A): 20.1%), tetracycline resistance genes (tet(K, L, M): 3.5%), the mutation of gyrA (9.7%) and grlA (18.8%) for quinolone resistance. Regarding virulence genes, although the proportion of some genes was different between MSSA and MRSA isolates, no significant difference was detected (Supplemental Table 5).
Associations between clinical characteristics and S. aureus/MRSA infection status
For statistical analysis, the correlation between S. aureus/MRSA in the oral and nasal cavities and the clinical characteristics of the 472 patients was analyzed (Table 2, Supplemental Table 6). The results of the statistical analysis revealed a statistically significant difference in oral isolation rates for S. aureus between denture wearers and non-denture wearing patients. Further logistic regression analysis revealed that denture use was predominantly associated with the presence of S. aureus in the oral cavity after controlling for the influence of confounding factors (Table 3). On the other hand, no significant differences (p < 0.05) were found for MRSA (Supplemental Table 6).
In addition, we analyzed the relation of S. aureus/MRSA with primary diseases including heart disease, cerebrovascular disease, diabetes, respiratory disease and malignant neoplasm, and we found no correlation with them among 397 patients (Supplemental Table 7).
Comparison of S. aureus isolates from the oral and nasal cavities in the genetically identical participant
Among the 504 participants, 34 had S. aureus isolated from both the oral cavity and nasal cavity. Among 34 participants, 25 (73.5%) had the same ST of isolates from both sites. The most common ST was ST8 (6 participants: 24.0%), followed by ST15 (3 participants: 12.0%). MLST of S. aureus isolates from three participants revealed “untypable” STs, but the MLST alleles were all the same between the oral and nasal isolates. Therefore, three sets were included in the same ST.
Among the 25 participants whose isolates from both sites had the same ST, we investigated pairwise distance analysis for S. aureus strains of the same STs (Table 4). The pairwise distances were varied, with 0 to 70 SNPs. Based on the criterion for the intra-facility transmission of S. aureus (below 40 or 50 SNPs11,16,17, 23/25 participants showed SNPs below 40. In addition, the profile of antibiotic resistance genes was investigated, and 22 (88.0%) participants had the same pattern (Table 4). The qacA gene in patient F1, blaZ gene in patient C34 and aac(6’)-aph(2’’) gene in patient E57 were found in the oral isolate but not in the nasal isolate. In terms of virulence genes, 22/25 (88.0%) of the participants whose isolates from both sites had the same ST presented the same distribution of virulence genes (Table 4 and Supplemental Table 4). The sen gene in the patient C34 was detected only in the nasal isolate, but the seu gene was detected only in the oral isolate. The seu gene in the patient G52 and scn gene in the patient E65 were found only in the nasal isolates. Therefore, 20/25 participants with the same ST of S. aureus in the oral and nasal cavities presented identical patterns of AMR and virulence gene carriage.
Among the 9 participants whose isolates from both sites had different STs, 6 presented the same pattern of AMR genes, although AMR genes were not found in 5 of the isolates, whereas 7/9 participants presented different patterns of virulence genes (Supplemental Table 4).
Discussion
This study investigated the isolation of S. aureus/MRSA from dental patients aged 40 to 95 years (mean age of 69.6 years). The isolation rates of S. aureus/MRSA in the oral and nasal cavities were 12.3%/1.8% and 22.2%/4.2%, respectively. Previous reports regarding the prevalence of S. aureus/MRSA in the oral cavity demonstrated variable isolation rates (Supplemental Table 8); Campos, J. et al. reported that the prevalence of S. aureus in the oral and nasal cavities was 12.0% in only the oral cavity, 13.9% in only the nasal cavity and 9.9% in both cavities among 101 healthy adults (mean age of 21.8 ± 3.5 years, 18–45 years)9. Petti, S. et al. reported that the S. aureus and MRSA carriage rates in the oral cavity were 8.9% and 1.9%, respectively, whereas those in the nasal cavity were 8.9% and 1.3%, respectively, among 157 dental students24. Patil, A. K. et al. also reported isolation rates of S. aureus/MRSA in the oral and nasal cavities (healthy children aged 4–13 years living in rural areas), which were 36%/16% and 45%/30% from the oral and nasal cavities, respectively25. Kusaka, S. et al. reported isolation rates of S. aureus (34.3%)/MRSA (15.7%) from the oral cavity in geriatric health facility (GHSF) participants (67 participants: mean age of 85.7 years) and in long-term care welfare facility (WF) participants (111 participants: mean age of 87.8 years)11. Silva, L. P. et al. reported the prevalence of S. aureus and MRSA in the nasal, oral, and rectal cavities of 150 LTCF residents and 76 bedridden patients26. The prevalence rates of total S. aureus and MRSA were 33.6% (n = 76) and 8% (n = 18), respectively. Considering these previous findings and our results, the prevalence of S. aureus and MRSA may vary depending on various factors, such as age, lifestyle including denture wearing and the environment. Compared with the isolation rates reported by Kusaka, S. et al. and Silva, L. P. et al.11,26 in elderly care facilities, the S. aureus isolation rates recorded in this study were lower. Most dental patients in this study lived their daily life at home, whereas participants in the facilities had problems living at home because of various health conditions. Therefore, this difference may affect the isolation rate between dental patients who live at home and participants in facilities. Moreover, horizontal transmission may occur relatively easily in institutions, where people are forced to live in a limited community and are exposed to many shared environments. Kusaka, S. et al. reported the possibility of horizontal transfer among some participants in the same facility11. However, in this study, most participants (87.1%) were outpatients, so the occurrence of horizontal transfer among participants was considered quite low.
In this study, 30 MRSA isolates were isolated from the oral and nasal cavities of participants; ST8 was the most common ST, accounting for 18 (60.0%) isolates; and the SCCmec types of the MRSA isolates were types I, II and IV. This tendency was observed in the MRSA isolates from the oral and nasal cavities. Most of the MRSA strains detected in clinical specimens (bacteremia) in Japan thus far are type I, II or IV27,28. It is important to note that SCCmec types of MRSA isolated from the oral and nasal cavities showed a similar trend to MRSA isolated from bacteremia. Until roughly 2010, SCCmec type II, which is the predominant type of HA-MRSA, accounted for approximately 70–80% of MRSA cases29, and SCCmec type IV has recently become common in Japan30,31. Among the participants in this study, type IV was the most prevalent type (Supplemental Table 3), with a similar trend as reported in recent years. Furthermore, among the 30 MRSA strains, ST8-SCCmec IV accounted for the greatest proportion of type IV isolates (66.7%), and in recent years30, a marked increasing trend was also observed in the present study.
We found a significant difference in S. aureus isolation between patients with and without dentures. Therefore, we further investigated S. aureus isolation rates according to the denture wearing site (maxillary only, mandibular only and both upper and lower jaws) (Supplemental Table 9). The results revealed that S. aureus was more prevalent in maxillary dentures only (16.7%) than in mandibular dentures only (15.8%) and in bilateral dentures (19.2%) than in unilateral dentures only (16.4%) although significant difference was not observed among denture wearing sites. Since the maxillary denture area is larger than the mandible denture area is, the denture area in the oral cavity is considered to be one of the factors associated with S. aureus colonization. A previous report32 noted a correlation between denture use and S. aureus retention in the mouth. Considering the results of this study, it is possible that the patient background of denture use may affect S. aureus colonization in the oral cavity.
Among the 140 participants whose S. aureus was isolated, including MRSA from the oral or nasal cavity, 34 (24.1%) had S. aureus isolated from both cavities (Table 1). We compared STs between nasal and oral isolates to determine whether different types of S. aureus colonize both sites. We found that 25/34 (73.5%) of the isolates from the oral and nasal cavities of the genetically identical participant were of the same ST (Supplemental Table 4). We further investigated the pairwise distance analysis of S. aureus strains of the same STs was performed (Table 4). Based on the criteria for intra-facility transmission11,16,17, the oral and nasal S. aureus isolates from 23/25 participants were considered the same clone. Since 23/34 (67.6%) participants had the same clones from the oral and nasal cavities, there may be a possibility of S. aureus trafficking between the oral and nasal cavities. However, other participants had different clones from both cavities. This finding provides important information that two strains isolated from the oral and nasal cavities in one individual may not be identical, even if they exhibit the same ST. In addition, S. aureus isolates were found in 28 (5.6%) oral-only samples and 78 (15.5%) nasal-only samples among 140 participants with S. aureus isolation (Table 1). Other reports also indicated S. aureus colonization in the oral cavity without being present in the nasal cavity of the same individual9,33. These findings suggest that the oral cavity is an independent reservoir for S. aureus colonization. Therefore, it is considered the necessary to screen the oral cavity for infection prevention and transmission within hospitals and facilities.
There are some limitations in this study. We suppose S. aureus isolation rates in this study reflect the isolation rates in the general population because most dental patients live at home. However, dental patients are undergoing oral treatment, so their situation may not necessarily be the same as that of the general population. It is expected that further studies with dental patients at other facilities and the studies from general volunteers will help to determine if the results of this study show a general trend. In addition, to investigate the S. aureus clonal association between the oral and nasal cavities, we utilized the criteria for an intra-facility transmission study because there are no obvious criteria regarding oral-nasal transmission. Further study will be required to clarify this association.
In conclusion, we isolated S. aureus and MRSA from the oral and nasal cavities of 504 dental patients and isolated 62 S. aureus and nine MRSA strains from the oral cavity and 112 S. aureus and 21 MRSA strains from the nasal cavity. Statistical analysis of the patients’ clinical information revealed a correlation between S. aureus isolation in the oral cavity and denture wear. Since a certain percentage of MRSA is present in the oral cavity of dental patients, attention should be given to the spread of infection from patients to health care workers or other patients during dental treatment and other medical procedures.
Data availability
The genomic data of the S. aureus isolates used in this study have been deposited in NCBI database (accession number: PRJDB18935).
References
Lindsay, J. A. & Holden, M. T. G. Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 12, 378–385 (2004).
Lowy, F. D. Staphylococcus aureus infections. N Engl. J. Med. 339, 520–532 (1998).
Cheung, G. Y. C., Bae, J. S. & Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 12, 547–569 (2021).
Chambers, H. F. & Deleo, F. R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641 (2009).
Lee, A. S. et al. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers. 4, 18033 (2018).
Molton, J. S., Tambyah, P. A., Ang, B. S. P., Ling, M. L. & Fisher, D. A. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin. Infect. Dis. 56, 1310–1318 (2013).
Turner, N. A. et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218 (2019).
Witte, W. Community-acquired methicillin-resistant Staphylococcus aureus: what do we need to know? Clin. Microbiol. Infect. 15 (Suppl 7), 17–25 (2009).
Campos, J. et al. Unveiling the Relevance of the Oral Cavity as a Staphylococcus aureus Colonization Site and Potential Source of Antimicrobial Resistance. Pathogens 12, (2023).
Smith, A. J., Jackson, M. S. & Bagg, J. The ecology of Staphylococcus species in the oral cavity. J. Med. Microbiol. 50, 940–946 (2001).
Kusaka, S. et al. Oral and rectal colonization of methicillin-resistant Staphylococcus aureus in long-term care facility residents and their association with clinical status. Microbiol. Immunol. 68, 75–89 (2024).
Rathbun, K. P., Bourgault, A. M. & Sole, M. Lou. Oral microbes in Hospital-Acquired pneumonia: practice and research implications. Crit. Care Nurse. 42, 47–54 (2022).
Khadka, S. et al. Poor oral hygiene, oral microorganisms and aspiration pneumonia risk in older people in residential aged care: a systematic review. Age Ageing. 50, 81–87 (2021).
Dendani Chadi, Z., Dib, L., Zeroual, F. & Benakhla, A. Usefulness of molecular typing methods for epidemiological and evolutionary studies of Staphylococcus aureus isolated from bovine intramammary infections. Saudi J. Biol. Sci. 29, 103338 (2022).
Enright, M. C. & Spratt, B. G. Multilocus sequence typing. Trends Microbiol. 7, 482–487 (1999).
Tsujiwaki, A. et al. Epidemiology of methicillin-resistant Staphylococcus aureus in a Japanese neonatal intensive care unit. Pediatr. Int. 62, 911–919 (2020).
Harris, S. R. et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 13, 130–136 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Tonkin-Hill, G. et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21, 180 (2020).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M. & Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 9, e104984 (2014).
Zhou, Z. et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 28, 1395–1404 (2018).
Harris, S. R. S. K. A. Split Kmer Analysis Toolkit for Bacterial Genomic Epidemiology. Preprint at (2018). https://doi.org/10.1101/453142
Petti, S. et al. Low methicillin-resistant Staphylococcus aureus carriage rate among Italian dental students. Am. J. Infect. Control. 43, e89–91 (2015).
Patil, A. K. et al. Prevalence of Community-Associated Methicillin-Resistant Staphylococcus aureus in oral and nasal cavities of 4 to 13-year-old rural school children: A Cross-sectional study. Contemp. Clin. Dent. 10, 99–104 (2019).
Silva, L. P. et al. Molecular epidemiology of Staphylococcus aureus and MRSA in bedridden patients and residents of Long-Term care facilities. Antibiot. (Basel) 11, 1526 (2022).
Kaku, N. et al. Influence of antimicrobial regimen on decreased in-hospital mortality of patients with MRSA bacteremia. J. Infect. Chemother. 20, 350–355 (2014).
Yamada, K. et al. Clinical features of bacteremia caused by methicillin-resistant Staphylococcus aureus in a tertiary hospital. Tohoku J. Exp. Med. 224, 61–67 (2011).
Yanagihara, K. et al. Antimicrobial susceptibility and molecular characteristics of 857 methicillin-resistant Staphylococcus aureus isolates from 16 medical centers in Japan (2008–2009): nationwide survey of community-acquired and nosocomial MRSA. Diagn. Microbiol. Infect. Dis. 72, 253–257 (2012).
Yamaguchi, T. et al. Changes in the genotypic characteristics of Community-Acquired Methicillin-Resistant Staphylococcus aureus collected in 244 medical facilities in Japan between 2010 and 2018: a nationwide surveillance. Microbiol. Spectr. 10, e0227221 (2022).
Harada, D. et al. Change in genotype of methicillin-resistant Staphylococcus aureus (MRSA) affects the antibiogram of hospital-acquired MRSA. J. Infect. Chemother. 24, 563–569 (2018).
Pereira, C. A. et al. Opportunistic microorganisms in individuals with lesions of denture stomatitis. Diagn. Microbiol. Infect. Dis. 76, 419–424 (2013).
Lam, O. L. T., McGrath, C., Bandara, H. M. H. N. & Li, L. S. W. Samaranayake, L. P. Oral health promotion interventions on oral reservoirs of Staphylococcus aureus: a systematic review. Oral Dis. 18, 244–254 (2012).
Acknowledgements
We would like to thank all of the institutions and subjects for their cooperation in this research and the colleagues who supported us with the survey. Regarding the collection of samples, we gratefully acknowledge the contributions of Katsuhiro Takeda, Jun Nakanishi, Naoki Sadaoka, Kazuhiro Tsuga, Mineka Yoshikawa, Maho Takeuchi, Azusa Haruta, Tomonao Aikawa, Kuniko Mizuta, Takako Naruse, Mikihito Kajiya, Tomoaki Shintani, Kouji Ohta, Yoshino Kaneyasu at Hiroshima University Hospital.
Funding
This study was supported by The Japan Agency for Medical Research and Development (AMED) under Grant Number JP23fk0108606.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.K.-M., H.S., R.N., T.T., M.S. and H.K. Data curation: T.K. and M.N.-T.L. Formal analysis: M.N.-T.L., T.K., T.T. and M.K.-M. Funding acquisition: M.K.-M. and H.K. Investigation: T.K., M.K.-M., Sa.K.(Satoru Kusaka), Y.Y., M.N.-T.L., Yo.S.(Yo Sugawara), J.H., Sh.K.(Shoko Kutsuno), M.A., T.T., M.K.-M. and H.K Methodology: M.K.-M., M.S. and H.K. Project administration: M.K.-M. and H.K. Resources: M.N.-T.L., M.K.-M., H.S., M.S. and H.K. Software: M.N.-T.L., Yu.S.(Yujin Suzuki) and S.N. Supervision: H.S., R.N., T.T., M.S. and H.K. Validation: M.N.-T.L., M.K.-M. and H.K. Visualization: Yu.S., S.N., T.K. and T.T. Writing - original draft: T.K., Yu.S., S.N., M.K.-M. and H.K. All authors read, sub-edited, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kawayanagi, T., Kawada-Matsuo, M., Kusaka, S. et al. Clinical and genetic analysis of oral and nasal staphylococcus aureus isolates in dental patients. Sci Rep 15, 13149 (2025). https://doi.org/10.1038/s41598-025-93773-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93773-0