Abstract
Melatonin (Mel), a classical antioxidant, has the potential to mediate ferroptosis. Cadmium (Cd) poses a substantial threat to the male reproductive system, as it can induce testicular injury by triggering ferroptosis. The study aimed to explore the protective role and mechanism of Mel in Cd-induced ferroptosis in spermatogonia (spg). Our results demonstrated that Cd disrupted the mitochondrial ultrastructure and induced more autophagosomes in spg. Exposure to Cd resulted in a reduction of the mitochondrial membrane potential of the cells. The transcriptomics analysis revealed significant differences in gene expression associated with ferroptosis and autophagy. Mel could reverse the changes caused by Cd in the genes mentioned above. Furthermore, Cd increased cellular iron content and elevated reactive oxygen species levels, which induced oxidative stress in spg. Mel pretreatment reduced iron accumulation and oxidative damage caused by Cd exposure. Additional studies demonstrated that Cd exposure activated NCOA4-mediated ferritinophagy in spg. Mel pretreatment, as anticipated, inhibited the increased the mRNA and protein expression of ATG5, LC3B, and NCOA4 caused by Cd, ameliorated Cd-caused iron overload and oxidative stress, and protected spg from ferroptosis. Our study provides a therapeutic basis for the use of Mel to treat Cd-induced testicular injury.
Similar content being viewed by others
Introduction
The decline in male fertility rates is a critical health problem. Reduced sperm count is implicated in approximately 90% of male infertility problems. The global sperm count has been declining at an accelerated pace in the recent decade1,2. A previous study on human sperm count reported that the total sperm count decreased from 335.7 million to 126.6 million among young men between 1973 and 2018, with a decrease of 1.4% per year and 62.3% overall2. The sperm concentration in European men has demonstrated a time-dependent decline, with an overall decrease of 32.5% over the past 50 years, according to a previous study3. Thus, it is imperative to investigate the causes and mechanisms of the decline in sperm count to prevent further damage to the reproductive health of men.
Environmental factors are implicated in the decrease in sperm quality4. Cadmium (Cd), a common toxic heavy metal, easily penetrates the blood-testis barrier, thereby damaging testicular cells and reducing sperm count, finally affecting spermatogenesis5. Cd can induce oxidative stress and cell death to cause reproductive toxicity through various mechanisms, including the damaging effects on Sertoli and Leydig cells. Previous studies reported that Cd induces testicular pyroptosis via m6A-methylated mitochondrial proteostasis stress or activating the AIM2 inflammasome pathway6,7. Wang T et al. reported that low-dose Cd exposure can cause Parkin translocation and mitochondrial fusion disorder, which contribute to the reduction of testicular testosterone synthesis8. Our recent study revealed that iron overload-induced ferroptosis is implicated in the declining sperm count caused by Cd9. Additionally, our previous study revealed that ferritin degradation through NCOA4-mediated ferritinophagy is associated with Cd-induced spermatogonia (spg) ferroptosis10. The studies on the mechanism provide new insights for preventing and treating Cd-induced male reproductive toxicity.
Melatonin (N-acetyl-5-methoxytryptamine, Mel) is an indoleamine neurohormone secreted primarily by the pineal gland and the retina11,12. Mel has various physiological functions, including regulating circadian rhythms, improving sleep, enhancing body immunity, and delaying aging. Mel, an endogenous antioxidant, can eliminate free radicals produced in the body and maintain the metabolic balance of intracellular ROS. Mel has powerful pharmacological effects and is used to treat cancer, treat or prevent metabolic disorders, and retard brain aging through various mechanisms13,14,15,16. Mel can exert therapeutic effects through many new molecular mechanisms, including apoptosis, autophagy, and pyroptosis17,18,19,20. Mel can modulate ferritinophagy to mitigate the acute cardiotoxicity caused by doxorubicin21. Ferroptosis is a potential therapeutic target of Mel22. Herein, we created an in vitro cellular model to investigate the protective role and mechanism of Mel in Cd-induced spg ferroptosis.
Materials and methods
Chemicals
All the reagents were listed in Supplementary Table 1.
Cell cultures
GC-1 spg (a mouse-derived spermatogonial cell line) was obtained from the BeNa Culture Collection (Beijing, China). Cells were cultured following Jia’s protocol10. The cells were cultured in DMEM containing high glucose medium supplemented with 10% FBS and kept at 37 ℃ in a humidified atmosphere with 5% carbon dioxide. At 80% confluence, the cells were used for subsequent experiments.
This study consists of four groups: Control, Mel, Cd, and Mel + Cd. The cells were treated with Phosphate Buffered Saline (PBS) or Mel (10 µM) in the control or Mel group, respectively. The cells in the Cd group were treated with CdCl2 (10 µM) for 24 h. The cells in the Mel + Cd group were pre-treated with Mel (10 µM) for 2 h before CdCl2 (10 µM).
Enhanced-cell counting Kit-8 assay
The cell activity was determined with Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, China) assay according to the manufacturer’s instructions. Briefly, GC-1 spg cells (5 × 103 cells/well) were plated in 96-well plates and treated with PBS or CdCl2 (10 µM) or Mel (10 µM) in the presence or absence of CdCl2. Thereafter, 10 µL of CCK8 working solution was added to the cells and incubated for 2 h at 37 °C. Absorbance was measured at 450 nm using a microplate reader (TECAN, Austria).
Mitochondrial ultrastructure
The cells were fixed with a 2.5% glutaraldehyde solution, scraped with a cell scraper, and transferred to a centrifuge tube. After centrifugation the cells were fixed with 1% osmium tetroxide, dehydrated with graded alcohol, embedded, sectioned, and stained with 2% uranyl acetate and 2.6% lead citrate solutions before observation under transmission electron microscopy (TEM) (Thermo Scientific Talos L120C G2, America).
Detection of mitochondrial membrane potential (MMP)
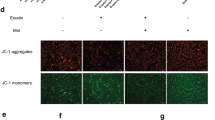
Following different treatments, the cells were gathered into flow tubes, incubated with JC-1 fluorescent probes (10 µg/mL) at 37℃ for 20 min in the dark, and washed with JC-1 assay buffer twice. Subsequently, 1 × 105 cells were resuspended with JC-1 Assay Buffer and the fluorescence intensity was measured using a flow cytometry (Beckman, USA). The ratio of the fluorescence intensity of JC-1 monomers to aggregates was calculated to assess the change in MMP.
Transcriptomics analysis
RNA was extracted using TRIzol, stored in an ultra-low-temperature freezer, and quickly sent to Personalbio Technology Co. Ltd. (Shanghai, China) for RNA library construction. The obtained results were subjected to differential gene expression analysis and visualization on the Personalbio GenesCloud. The selection criteria for differential expression genes were generally set as |FC| > 2 and P < 0.05 unless otherwise specified.
Total iron
The cells were lysed and centrifuged. The supernatant was supplemented with iron ion detection reagents in proportion (total volume of the supernatant: iron ion detection agent = 20:3), incubated in the dark at room temperature for 30 min, and 200 µL of the reaction solution was added. The OD value was detected at 550 nm using an ELISA analyzer.
Ferrous irons and ROS
Intracellular ferrous iron and ROS were performed as previously reported10. The fluorescence intensity was observed under a fluorescence microscope (Leica, Weztlar, Germany).
qRT-PCR
After different treatments, the total RNA was extracted using the TRI reagent in GC-1 spg cells. The reverse transcription kit from Sparkjade Biotechnology Co.Ltd was used to reverse transcribe the RNA into cDNA. According to the instructions provided with the kit, samples were aliquoted and amplified using a Roche LightCycler® 480 PCR sequence detection system (Roche, Basel, Switzerland). The relative gene expression levels were calculated using the 2–ΔΔCt method. All the primers were listed in Supplementary Table 2.
Western blot
The total protein of cells was extracted using RIPA buffer containing a mixture of protease inhibitors and phosphatase inhibitors, and protein quantification was performed using the BCA assay kit. The protein samples were separated using SDS-PAGE, transferred to a PVDF membrane, and blocked with 5% skimmed milk for 2 h. Subsequently, the PVDF membranes were incubated with primary antibodies. All the antibodies were listed in Supplementary Table 3. After washing, the PVDF membranes were incubated with corresponding secondary antibodies for 1 h. Protein levels were determined and imaged using a fully automated chemiluminescence image analysis system (Tanon 4600, Shanghai, China).
MDA, SOD, GSH, and H2O2
The cells were collected, lysed and centrifuged, and the precipitation and supernatant were obtained. MDA, SOD, and GSH in the precipitation and H2O2 in the supernatant were measured following the guidelines provided with the respective reagents, respectively. Protein concentrations were measured for quantification of MDA, SOD, GSH, and H2O2 accordingly.
Immunofluorescence
After different treatments, the cells were fixed for 10 min with 4% paraformaldehyde at room temperature, then permeabilized with 0.1% Triton X-100 for 10 min and blocked with BSA blocking solution for 1 h. Overnight at 4℃ with the LC3B or NCOA4 antibody (1:100 dilution). After rinsing with PBS, the accordingly secondary antibodies (1:200 diluted) were applied for 1 h at room temperature and washed with PBS again. Finally, DAPI was stained to mark the nuclei. Images were acquired by a fluorescence microscope (Leica, Wetzlar, Germany).
Statistical analysis
In the present study, all data are expressed as mean ± standard error of the mean (SEM). Differences between the groups were evaluated by one-way analysis of variance (ANOVA). A p < 0.05 was considered statistically significant.
Results
Mel alleviates Cd-induced damage and reduces autophagosomes in Spg
Firstly, we examined the effect of melatonin on the activity of spermatogonia. The results indicated that low-dose Mel had no significant effect on the cell viability, while the cell viability was slightly decreased in the 100 µM and 200 µM of Mel groups (Fig. 1A). To investigate whether melatonin could alleviate Cd-induced spermatogonial toxicity, the cells were pre-treated with Mel for 2 h before CdCl2 (10 µM). The results showed that Cd significantly decreased cell viability (Fig. 1B). Low-dose Mel partially restored the reduced viability caused by Cd, and 10 µM of Mel has the optimal protective effect (Fig. 1B). Therefore, 10 µM of Mel was chosen to explore its protective effect on cadmium toxicity.
Mitochondrial damage is a typical characteristic of ferroptosis, so we evaluate the ultrastructure of mitochondria and MMP. The results of TEM revealed that Cd-induced changes in mitochondrial ultrastructure manifested as mitochondrial swelling, deformation, and cristae disruption (Fig. 1C). Perform statistical analysis on the results and generate a bar chart. There were approximately 70% of the mitochondria in the Cd-treated group exhibited swelling (Fig. 1D). Many autophagosomes were observed in spg exposed to Cd (Fig. 1C). Mel pretreatment significantly restored Cd-induced mitochondrial ultrastructural damage and reduced autophagosomes in spg (Fig. 1C–E). Cd exposure decreased MMP, while Mel pretreatment enhanced MMP reduced by Cd (Fig. 1F–G). These results indicated that Cd disrupts mitochondrial ultrastructure, causes autophagy and mitochondrial dysfunction in spg. Mel mitigated the damages caused by Cd, suggesting that Mel protects spg against Cd-induced toxicity.
Mel alleviates Cd-induced autophagy and ferroptosis, as revealed by transcriptomics analysis
The transcriptome analysis was employed to analyze the differentially expressed genes (DEGs). Compared with the control group, 2,429 genes were upregulated, and 1,080 genes were downregulated in the cells exposed to Cd. While the cells in the Mel + Cd group exhibited an increase in 116 genes and a decrease in 44 genes. Compared with the Cd group, 782 genes exhibited increased expression, while 2,240 genes displayed decreased expression in the Mel + Cd group (Fig. 2A–C).
Because Cd causes injury to the mitochondrial ultrastructure, which reveals the characteristic changes of ferroptosis and the presence of many autophagosomes in Cd-treated cells, we focused on the effects of Cd on ferroptosis and autophagy signaling pathways. The top 20 signaling pathways enriched in the DEGs included pathways related with ferroptosis and autophagy (Fig. 2D–E). The results of Heatmap reported that the expression patterns of DEGs were similar between the control and Mel + Cd groups; however, statistically differences were observed between the control and Cd groups or the Cd and Mel + Cd groups (Fig. 2F). Additionally, the analysis revealed significant differences in gene expression associated with ferroptosis and autophagy (Fig. 2G). The gene biomarkers of ferroptosis (Gpx4, Fth1, Fsp1, Tfrc, Ptgs2, xCT, and Hmox1) were significantly increased, while Sod2, Fpn1, and Dmt1 decreased in the Cd group. Similarly, the gene biomarkers of autophagy (Lc3b, Atg5, Atg7, and Ncoa4) were upregulated in the Cd group. Mel pretreatment could reverse the changes in the genes mentioned above caused by Cd (Fig. 2G). The above results indicated that Cd may activate ferroptosis and autophagy signaling pathways in spg, while Mel could protect spg against injury via antagonizing Cd-induced ferroptosis and autophagy.
Mel ameliorates Cd-induced iron metabolism disorder in Spg
Our previous study found that Cd can disrupt iron metabolism in testicular cells, and excessive iron can induce ferroptosis through the Fenton reaction23. An important feature of ferroptosis is the dysregulation of iron metabolism. Transcriptomic analysis indicated that the enriched signaling pathways in the Mel + Cd group compared to the Cd group included ferroptosis. Therefore, we assessed indicators related to iron metabolism. Figure 3A–C depict that Cd significantly increased the cellular Fe2+ and total iron levels. Ferritin is the primary storage form of intracellular iron, comprising ferritin heavy chain 1 (FTH1) and ferritin light chain (FTL). We found that Cd increased FTH1 expression (Fig. 3D and E). Heme oxygenase 1 (HO1), an enzyme that can produce free iron by degrading heme, increased mRNA and protein expression in the cells treated with Cd (Fig. 3F and G). However, the expression of FPN1 and TFR1, two critical proteins involved in iron transportation, decreased in the Cd group (Fig. 3H–J). The mRNA of Dmt1 was down-regulated in the cells exposed to Cd (Fig. 3K). Unexpectedly, the pretreatment of Mel antagonized the changes mentioned above, effectively reducing the iron accumulation induced by Cd in spg.
Mel reduced Cd-induced oxidative stress
Our results demonstrated that Cd elevated ROS levels (Fig. 4A and B). Cd stimulated excessive H2O2 production in spg, manifested as increased H2O2 content in the culture medium (Fig. 4D). Malondialdehyde (MDA) is one of the main products of lipid peroxidation and is widely used as an indicator of lipid oxidation. Glutathione (GSH) is the main antioxidant in cells and a substrate for GPX4. Superoxide dismutase (SOD) is an important antioxidant enzyme that clears superoxide anion radicals. The changes in SOD levels can indirectly reflect the degree of lipid peroxidation. The results showed that the level of MDA, was significantly elevated in the Cd group (Fig. 4C). Conversely, Cd reduced GSH content and SOD activity (Fig. 4E–F). Moreover, Cd reduced the mRNA expression of SOD2 (Fig. 4G). Additionally, Cd significantly increased mRNA and COX2 protein expression (Fig. 4H–I). Mel significantly mitigated the changes caused by Cd, indicating a reduction in Cd-induced oxidative stress and protection of spg from ferroptosis.
Mel alleviates Cd-induced ferritinophagy via inhibiting NCOA4 in Spg
NCOA4 serves as an autophagy-related cargo receptor that specifically binds ferritin and facilitates FTH degradation through autophagy, releasing a large amount of ferrous ions2. TEM and transcriptomics analysis revealed that Cd could cause autophagy in spg. Cd increased the expression of ATG5, LC3B and NCOA4 (Fig. 5A–F). However, Mel pretreatment antagonized the upregulation of ATG5, LC3B, NCOA4 mRNA, and protein induced by Cd exposure (Fig. 5A–F). Furthermore, the LC3B and NCOA4 fluorescence of the Cd group was significantly enhanced, and Mel reversed the Cd-induced LC3B and NCOA4 elevation (Fig. 5G–J). These results indicate that Mel pretreatment may ameliorate Cd-induced iron metabolism disorder via inhibiting Cd-induced ferritinophagy.
Discussion
Mel is a neurohormone primarily released from the pineal gland and has multiple physiological functions. The role of Mel in reproductive medicine has attracted increased attention24,25. Mel is essential for follicular formation, spermatogenesis, and oocyte maturation, which increases fertilization rates and promotes embryonic development. Additionally, it improves the results of in vitro fertilization and embryo transfer25,26,27. Mel works through various mechanisms, including basic circadian rhythm regulation, potent free radical scavenging ability, and anti-inflammatory effects. New molecular mechanisms are continuously being investigated. A recent study reported that Mel prevents Cd-induced fetal growth retardation by inhibiting mitophagy28. 1,2-Dichloroethane (1,2-DCE) can cause testicular damage in mice. Mel can mitigate cellular pyroptosis and reverse 1,2-DCE-caused testicular toxicity29. The studies of mechanisms are of great significance for exploring new targets of Mel in preventing and treating reproductive diseases.
In order to explore the protective mechanisms of melatonin in Cd-induced spermatogonial toxicity, the transcriptomic analysis was used to identify potential molecular pathways. The results revealed that the DEGs between the control and the Cd group were enriched in signaling pathways associated with ferroptosis and autophagy. Furthermore, the DEGs between the Mel and the Cd group were also enriched in signaling pathways associated with ferroptosis. Previous research has established ferroptosis as a form of cell death that is dependent on autophagy30. Autophagy can lead to the degradation of ferritin. The degradation of ferritin subsequently releases a significant amount of iron ions, triggering ferroptosis. The transcriptomics results suggested Cd may activate ferroptosis and autophagy signaling pathways in spg, while Mel could protect spg against injury via antagonizing Cd-induced ferroptosis and autophagy.
Ferroptosis is a promising therapeutic target and a new regulatory mechanism for cell death31,32. It is characterized by the accumulation of lipid peroxides resulting from iron overload, which disrupts the antioxidant system and induces cellular oxidative stress, leading to cell death33. Herein, we demonstrated that the mitochondria of spg in the Cd group exhibited vacuolar-like changes, including the absence of cristae and damaged outer membranes. These changes are indicative of ferroptosis, a type of mitochondrial dysfunction. Numerous autophagosomes were observed in the cells treated with Cd. Moreover, Cd significantly reduced MMP in spg. The above changes suggested that Cd could induce ferroptosis and autophagy in spg.
Iron overload is primarily implicated in ferroptosis. In a physiological state, intracellular iron balances iron input, storage, and output. Exogenous Fe3+ binds to transferrin and enters the cell through the cell surface transferrin receptor 1 (TfR1), which is acidified and converted to Fe2+34. Subsequently, Fe2+ is transported into an unstable iron pool through DMT and utilized for various cellular metabolic requirements. Excess Fe2+ binds to intracellular ferritin, producing complexes temporarily stored in the cell or transported out via the iron exporter ferroportin 1 (FPN1)35. Another source of iron is mediated by HO1, which can break down heme into ferrous ions, CO, and biliverdin.
Iron overload can be found in numerous chronic and common human diseases. When the iron concentration exceeds the cellular iron sequestration capacity, the amount of iron in the labile iron pool increases, causing oxidative damage and cell death. Age-related iron overload may be an essential factor contributing to neurodegenerative processes36,37. Excessive iron accumulation in the deep gray matter nucleus is associated with neurodegenerative changes38,39. Magnetic resonance imaging revealed changes in brain iron metabolism and increased iron accumulation, particularly in the basal ganglia in the striatum of patients with Huntington’s disease40. Iron contributes to the occurrence and growth of tumors and has become a target of therapeutic intervention41,42. Excess iron can produce excessive free radicals, leading to liver, kidney, and lung diseases. Retinal iron deposition contributes to the development of macular degeneration, or glaucoma43,44,45. Thus, there is a need to investigate the molecular mechanisms of iron homeostasis disorders in these diseases.
Mel and its metabolites can bind to iron ions, eliminate free radicals, and inhibit ROS, making them promising new drugs for reducing iron toxicity. Herein, we observed the protective effects of Mel on Cd-induced spermatogonial toxicity. As hypothesized, Mel could significantly increase MMP, inhibit ROS, reduce autophagy, and prevent spg from Cd-induced ferroptosis. The transcriptomic analysis revealed that Mel could protect spg against Cd-induced injury via antagonizing Cd-induced ferroptosis and autophagy.
The following results confirmed that Cd exposure significantly increased cellular Fe2+ and total iron levels. Additionally, Cd increased FTH1 and HO1 expression, while DMT1, FPN1, and TFR1 expression decreased in Cd-induced spg. The above changes suggested that Cd could disrupt iron homeostasis in spg. Ferroptosis can be mitigated by alleviating iron overload. Mel can directly form complexes with metal ions, serving as a metal chelating agent to alleviate the negative impacts of free metal ions by decreasing their reactivity46,47,48. Mel exhibited a potential chelating effect on ferrous ions, and the percentage of the metal scavenging capacity of Mel was significantly higher than that of α-Tocopherol46,47,48. Galano et al. reported that Mel could completely inhibit oxidative stress via chelating copper ions48. Our study demonstrated that Mel pretreatment could significantly decrease FTH1 and HO1 expression and alleviate the cellular Fe2+ and total iron levels induced by Cd. Furthermore, Mel pretreatment antagonized DMT1, FPN1, and TFR1 downregulation caused by Cd, indicating that Mel relieved Cd-induced iron overload in spg.
A previous study reported that Cd induces ferroptosis in renal tubular epithelial cells through excessive activation of autophagy49. However, other studies have reported that autophagy plays a protective role in Cd-induced testicular Sertoli cells, and inhibition of autophagy exacerbates Cd-induced germ cell apoptosis50. The discrepancy may be attributed to the different concentrations of Cd, which result in varying levels of autophagy. Mild levels of autophagy can protect cells, while excessive activation of autophagy can induce cell death51,52. NCOA4-mediated ferritinophagy is an essential factor contributing to ferroptosis53. Ferritin can interact with NCOA4 and be transported to lysosomes for degradation through autophagy, maintaining cellular iron homeostasis54. Excessive NCOA4 activation can increase ferritin degradation, releasing excess ferrous ions and aggravated ferroptosis. Our recent study found that ferritinophagy participates in Cd-triggered ferroptosis10. Herein, we found that NCOA4, LC3B, ATG5 expression, and ferritinophagy-related genes increased, indicating ferritinophagy activation by Cd treatment.
Increasing evidence suggests that the regulatory role of Mel on autophagy varies in different stress conditions and cell lines. Mel plays a dual role by activating or inhibiting mitochondrial autophagy55. Mel can protect cardiomyocytes from simulated ischemia/reperfusion injury by inhibiting mitochondrial autophagy through the MT2/SIRT3/FoxO3a axis in H9c2 cells56. Inhibiting macrophage ferroptosis can effectively alleviate pneumonia-induced injury. Mel reduces the expression of NCOA4 and FTH1 proteins in alveolar macrophages of septic mice and inhibits ferroptosis, thereby ameliorating septic acute respiratory distress syndrome57. Yu et al. reported that Mel regulates the interaction between FTH1 and NCOA4 through SIRT6, affecting ferritinophagy, effectively inhibiting intracellular Fe2+ levels, inhibiting ferroptosis, and alleviating brain injury58. We demonstrated that Mel could alleviate ferritinophagy by inhibiting NCOA4 and FTH1 expression and protecting spg against Cd-induced autophagy and ferroptosis.
Conclusion
We demonstrated that Mel improves iron metabolism and relieves Cd-induced ferroptosis by regulating the storage and output of iron ions, inhibiting HO1 activity, weakening the Fenton reaction, and reducing ROS production in spg. Furthermore, Mel could alleviate ferroptosis and protect spg against Cd toxicity by inhibiting NCOA4/FTH1-mediated ferritinophagy, suggesting that Mel is an effective approach for preventing and treating Cd-induced male reproductive system injury.
Mel alleviates Cd-induced mitochondrial ultrastructure damage and reduces autophagosomes in spg. (A–B) Cell viability. (C) The representative images observed under TEM. Green arrows: intact mitochondria. Red arrows: swollen mitochondria with absent cristae. Yellow arrows: autophagosomes and autolysosomes. (D) The proportion of intact mitochondria. (E) The number of autophagosomes. (F) The JC-1 red/green ratio. (G) The representative images of MMP, which were detected by flow cytometry using JC-1 as a fluorescent probe. The data were analyzed using One-way ANOVA. Data are presented as mean ± SEM (n = 3). * P < 0.05, ** P < 0.01 compared to the control group. # P < 0.05, ## P < 0.01 compared to the Cd group.
Melatonin alleviates Cd-induced iron metabolism disorder in spg. (A and B) ferrous iron. Scale bar = 100 μm. (C) Total iron. (D) Fth1 mRNA. (E) FTH1 protein. (F) Hmox1 mRNA. (G) HO-1 protein. (H) TFR1 protein. (I) FPN1 mRNA. (J) FPN1 protein. (K) Dmt1 mRNA. The data were analyzed using One-way ANOVA. Data were presented as mean ± SEM (n = 3). * P < 0.05, ** P < 0.01 compared to the control group. # P < 0.05, ## P < 0.01 compared to the Cd group.
Mel alleviates Cd-induced oxidative stress in spg. (A and B) ROS. Scale bar = 100 μm. (C) MDA. (D) H2O2. (E) GSH. (F) SOD. (G) Sod2 mRNA. (H) Cox2 mRNA. (I) COX2 protein. The data were analyzed using One-way ANOVA. Data are presented as mean ± SEM (n = 3). * P < 0.05, ** P < 0.01 compared to the control group. # P < 0.05, ## P < 0.01 compared to the Cd group.
Mel alleviates Cd-induced ferritinophagy in spg. (A) Lc3b mRNA. (B) Atg5 mRNA. (C)Ncoa4 mRNA. (D) LC3B protein. (E) ATG5 protein. (F) NCOA4 protein. (G–J) The representative images and relative fluorescence intensity of LC3B and NCOA4. Scale bar = 100 μm. The data were analyzed using One-way ANOVA. Data are presented as mean ± SEM (n = 3). * P < 0.05, ** P < 0.01 compared to the control group. # P < 0.05, ## P < 0.01 compared to the Cd group.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author (ylji@ahmu.edu.cn) on reasonable request.
References
Levine, H. et al. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum. Reprod. Update 23, 646–659. https://doi.org/10.1093/humupd/dmx022 (2017).
Levine, H. et al. Temporal trends in sperm count: A systematic review and meta-regression analysis of samples collected globally in the 20th and 21st centuries. Hum. Reprod. Update 29, 157–176. https://doi.org/10.1093/humupd/dmac035 (2023).
Sengupta, P., Borges, E. Jr., Dutta, S. & Krajewska-Kulak, E. Decline in sperm count in European men during the past 50 years. Hum. Exp. Toxicol. 37, 247–255. https://doi.org/10.1177/0960327117703690 (2018).
Mirnamniha, M., Faroughi, F., Tahmasbpour, E., Ebrahimi, P. & Beigi Harchegani, A. An overview on role of some trace elements in human reproductive health, sperm function and fertilization process. Rev. Environ. Health. 34, 339–348. https://doi.org/10.1515/reveh-2019-0008 (2019).
Pappalardo, C. et al. Seminal cadmium affects human sperm motility through stable binding to the cell membrane. Front. Cell. Dev. Biol. 11, 1134304. https://doi.org/10.3389/fcell.2023.1134304 (2023).
Kolb, P. S. et al. The therapeutic effects of 4-phenylbutyric acid in maintaining proteostasis. Int. J. Biochem. Cell Biol. 61, 45–52. https://doi.org/10.1016/j.biocel.2015.01.015 (2015).
Zhou, J. et al. Cadmium exposure induces pyroptosis in testicular tissue by increasing oxidative stress and activating the AIM2 inflammasome pathway. Sci. Total Environ. 847, 157500. https://doi.org/10.1016/j.scitotenv.2022.157500 (2022).
Wang, R. et al. PBA alleviates cadmium-induced mouse spermatogonia apoptosis by suppressing endoplasmic reticulum stress. Toxicology 96 https://doi.org/10.1016/j.tiv.2024.105784 (2024).
Wang, Y. et al. Cadmium exposure during puberty damages testicular development and spermatogenesis via ferroptosis caused by intracellular iron overload and oxidative stress in mice. Environ. Pollut. (Barking, Essex) 325, 121434. https://doi.org/10.1016/j.envpol.2023.121434 (2023).
Jia, D. et al. NCOA4-mediated ferritinophagy participates in cadmium-triggered ferroptosis in spermatogonia. Toxicology 505, 153831. https://doi.org/10.1016/j.tox.2024.153831 (2024).
Challet, E. & Pévet, P. Melatonin in energy control: Circadian time-giver and homeostatic monitor. J. Pineal Res. 76, e12961. https://doi.org/10.1111/jpi.12961 (2024).
Felder-Schmittbuhl, M. P., Hicks, D., Ribelayga, C. P. & Tosini, G. Melatonin in the mammalian retina: Synthesis, mechanisms of action and neuroprotection. J. Pineal Res. 76, e12951. https://doi.org/10.1111/jpi.12951 (2024).
Moloudizargari, M. et al. Therapeutic targets of cancer drugs: Modulation by melatonin. Life Sci. 267, 118934. https://doi.org/10.1016/j.lfs.2020.118934 (2021).
Yi, Y. J. et al. Melatonin in cancer biology: Pathways, derivatives, and the promise of targeted delivery. Drug Metab. Rev. 56, 62–79. https://doi.org/10.1080/03602532.2024.2305764 (2024).
Bocheva, G., Bakalov, D., Iliev, P. & Tafradjiiska-Hadjiolova, R. The vital role of melatonin and its metabolites in the neuroprotection and retardation of brain aging. Int. J. Mol. Sci. 25 https://doi.org/10.3390/ijms25105122 (2024).
Davoodvandi, A., Nikfar, B., Reiter, R. J. & Asemi, Z. Melatonin and cancer suppression: Insights into its effects on DNA methylation. Cell. Mol. Biol. Lett. 27, 73. https://doi.org/10.1186/s11658-022-00375-z (2022).
Zhu, H. L. et al. Gestational exposure to environmental cadmium induces placental apoptosis and fetal growth restriction via Parkin-modulated MCL-1 degradation. J. Hazard. Mater. 424, 127268. https://doi.org/10.1016/j.jhazmat.2021.127268 (2022).
Zhu, H. L. et al. Environmental cadmium exposure induces fetal growth restriction via triggering PERK-regulated mitophagy in placental trophoblasts. Environ. Int. 147, 106319. https://doi.org/10.1016/j.envint.2020.106319 (2021).
Liu, L. et al. Melatonin inhibits atrazine-induced mitochondrial impairment in cerebellum of mice: modulation of cGAS-STING-NLRP3 axis-dependent cell pyroptosis. Sci. Total Environ. 912, 168924. https://doi.org/10.1016/j.scitotenv.2023.168924 (2024).
Cong, L. et al. Melatonin alleviates pyroptosis by regulating the SIRT3/FOXO3α/ROS axis and interacting with apoptosis in atherosclerosis progression. Biol. Res. 56, 62. https://doi.org/10.1186/s40659-023-00479-6 (2023).
Hanna, M. et al. Synergistic cardioprotective effects of melatonin and deferoxamine through the improvement of ferritinophagy in doxorubicin-induced acute cardiotoxicity. Front. Physiol. 13 https://doi.org/10.3389/fphys.2022.1050598 (2022).
Zhang, D., Jia, X., Lin, D. & Ma, J. Melatonin and ferroptosis: Mechanisms and therapeutic implications. Biochem. Pharmacol. 218, 115909. https://doi.org/10.1016/j.bcp.2023.115909 (2023).
Wang, Y. et al. Cadmium exposure during puberty damages testicular development and spermatogenesis via ferroptosis caused by intracellular iron overload and oxidative stress in mice. Environ. Pollut. 325 https://doi.org/10.1016/j.envpol.2023.121434 (2023).
Shao, R., Wang, Y., He, C. & Chen, L. Melatonin and its emerging physiological role in reproduction: A review and update. Curr. Mol. Med. 24, 449–456. https://doi.org/10.2174/1566524023666230417103201 (2024).
Srinivasan, V. et al. Melatonin and human reproduction: Shedding light on the darkness hormone. Gynecol. Endocrinology: Official J. Int. Soc. Gynecol. Endocrinol. 25, 779–785. https://doi.org/10.3109/09513590903159649 (2009).
Tamura, H. et al. Importance of melatonin in assisted reproductive technology and ovarian aging. Int. J. Mol. Sci. 21 https://doi.org/10.3390/ijms21031135 (2020).
Yong, W. et al. Roles of melatonin in the field of reproductive medicine. Biomed. Pharmacother. Biomed. Pharmacother. 144, 112001. https://doi.org/10.1016/j.biopha.2021.112001 (2021).
Zhu, H. L. et al. Melatonin protects against environmental stress-induced fetal growth restriction via suppressing ROS-mediated GCN2/ATF4/BNIP3-dependent mitophagy in placental trophoblasts. Redox Biol. 40, 101854. https://doi.org/10.1016/j.redox.2021.101854 (2021).
Yang, Y. et al. Spatial - temporal mapping of urine cadmium levels in China during 1980–2040: dietary improvements lower exposure amid rising pollution. J. Hazard. Mater. 473, 134693. https://doi.org/10.1016/j.jhazmat.2024.134693 (2024).
Gao, M. et al. Ferroptosis is an autophagic cell death process. Cell. Res. 26, 1021–1032. https://doi.org/10.1038/cr.2016.95 (2016).
Stockwell, B. R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 185, 2401–2421. https://doi.org/10.1016/j.cell.2022.06.003 (2022).
Bayır, H., Dixon, S. J., Tyurina, Y. Y., Kellum, J. A. & Kagan, V. E. Ferroptotic mechanisms and therapeutic targeting of iron metabolism and lipid peroxidation in the kidney. Nat. Rev. Nephrol. 19, 315–336. https://doi.org/10.1038/s41581-023-00689-x (2023).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. https://doi.org/10.1016/j.cell.2012.03.042 (2012).
Feng, H. et al. Transferrin receptor is a specific ferroptosis marker. Cell. Rep. 30, 3411–3423e3417. https://doi.org/10.1016/j.celrep.2020.02.049 (2020).
Plays, M., Müller, S. & Rodriguez, R. Chemistry and biology of ferritin. Metallomics Integr. Biometal Sci. 13 https://doi.org/10.1093/mtomcs/mfab021 (2021).
Ward, R. J., Zucca, F. A., Duyn, J. H., Crichton, R. R. & Zecca, L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13, 1045–1060. https://doi.org/10.1016/s1474-4422(14)70117-6 (2014).
Cerri, S., Milanese, C. & Mastroberardino, P. G. Endocytic iron trafficking and mitochondria in Parkinson’s disease. Int. J. Biochem. Cell Biol. 110, 70–74. https://doi.org/10.1016/j.biocel.2019.02.009 (2019).
Cheng, R., Dhorajia, V. V., Kim, J. & Kim, Y. Mitochondrial iron metabolism and neurodegenerative diseases. Neurotoxicology 88, 88–101. https://doi.org/10.1016/j.neuro.2021.11.003 (2022).
Reichert, C. O. et al. Ferroptosis mechanisms involved in neurodegenerative diseases. Int. J. Mol. Sci. 21 https://doi.org/10.3390/ijms21228765 (2020).
Chen, L. et al. Altered brain iron content and deposition rate in Huntington’s disease as indicated by quantitative susceptibility MRI. J. Neurosci. Res. 97, 467–479. https://doi.org/10.1002/jnr.24358 (2019).
Hassannia, B., Vandenabeele, P. & Vanden Berghe, T. Targeting ferroptosis to iron out cancer. Cancer Cell. 35, 830–849. https://doi.org/10.1016/j.ccell.2019.04.002 (2019).
Rockfield, S., Raffel, J., Mehta, R., Rehman, N. & Nanjundan, M. Iron overload and altered iron metabolism in ovarian cancer. Biol. Chem. 398, 995–1007. https://doi.org/10.1515/hsz-2016-0336 (2017).
Zhao, T., Guo, X. & Sun, Y. Iron accumulation and lipid peroxidation in the aging retina: Implication of ferroptosis in Age-Related macular degeneration. Aging Dis.12, 529–551. https://doi.org/10.14336/ad.2020.0912 (2021).
Yao, F. et al. Pathologically high intraocular pressure disturbs normal iron homeostasis and leads to retinal ganglion cell ferroptosis in glaucoma. Cell Death Differ. 30, 69–81. https://doi.org/10.1038/s41418-022-01046-4 (2023).
Tang, Z. et al. HO-1-mediated ferroptosis as a target for protection against retinal pigment epithelium degeneration. Redox Biol. 43, 101971. https://doi.org/10.1016/j.redox.2021.101971 (2021).
Gulcin, I., Buyukokuroglu, M. E. & Kufrevioglu, O. I. Metal chelating and hydrogen peroxide scavenging effects of melatonin. J. Pineal Res. 34, 278–281. https://doi.org/10.1034/j.1600-079x.2003.00042.x (2003).
Chakrabarty, S., DiTucci, M. J., Berden, G., Oomens, J. & Williams, E. R. Structural investigation of the hormone melatonin and its alkali and alkaline Earth metal complexes in the gas phase. J. Am. Soc. Mass. Spectrom. 29, 1835–1847. https://doi.org/10.1007/s13361-018-2020-0 (2018).
Galano, A., Medina, M. E., Tan, D. X. & Reiter, R. J. Melatonin and its metabolites as copper chelating agents and their role in inhibiting oxidative stress: A physicochemical analysis. J. Pineal Res. 58, 107–116. https://doi.org/10.1111/jpi.12196 (2015).
Zhao, C. et al. Endoplasmic reticulum stress-mediated autophagy activation is involved in cadmium-induced ferroptosis of renal tubular epithelial cells. Free Radic Biol. Med. 175, 236–248. https://doi.org/10.1016/j.freeradbiomed.2021.09.008 (2021).
Zhou, G. X. et al. Autophagy in Sertoli cell protects against environmental cadmium-induced germ cell apoptosis in mouse testes. Environmental pollution (Barking, Essex) (1987) 270, 116241. https://doi.org/10.1016/j.envpol.2020.116241 (2021).
Parzych, K. R. & Klionsky, D. J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox. Signal. 20, 460–473. https://doi.org/10.1089/ars.2013.5371 (2014).
Liu, Y. & Levine, B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 22, 367–376. https://doi.org/10.1038/cdd.2014.143 (2015).
Zhou, H. et al. NCOA4-mediated ferritinophagy is involved in ionizing radiation-induced ferroptosis of intestinal epithelial cells. Redox Biol. 55, 102413. https://doi.org/10.1016/j.redox.2022.102413 (2022).
Santana-Codina, N., Gikandi, A. & Mancias, J. D. The role of NCOA4-Mediated ferritinophagy in ferroptosis. Adv. Exp. Med. Biol. 1301, 41–57. https://doi.org/10.1007/978-3-030-62026-4_4 (2021).
Wu, J., Bai, Y., Wang, Y. & Ma, J. Melatonin and regulation of autophagy: mechanisms and therapeutic implications. Pharmacol. Res. 163, 105279. https://doi.org/10.1016/j.phrs.2020.105279 (2021).
Zhou, H. et al. Melatonin protects cardiac microvasculature against ischemia/reperfusion injury via suppression of mitochondrial fission-VDAC1-HK2-mPTP-mitophagy axis. J. Pineal Res. 63 https://doi.org/10.1111/jpi.12413 (2017).
Xu, W. et al. Melatonin alleviates septic ARDS by inhibiting NCOA4-mediated ferritinophagy in alveolar macrophages. Cell. Death Discov. 10 https://doi.org/10.1038/s41420-024-01991-8 (2024).
Yu, X., Wang, S., Wang, X., Li, Y. & Dai, Z. Melatonin improves stroke by inhibiting autophagy-dependent ferroptosis mediated by NCOA4 binding to FTH1. Exp. Neurol. 114868. https://doi.org/10.1016/j.expneurol.2024.114868 (2024).
Author information
Authors and Affiliations
Contributions
D.J., W.H. and Q.Y.: Performed the experiments and analyzed the data. H.W. and Z.W.: Validation and data curation. M.Z. and W.G.: Formal analysis and project administration. R.W. and Y.Z.: Writing—review & editing. Y.J.: Conceived and designed the experiments; Wrote and revised the manuscript and Project administration; Funding support.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jia, D., Huang, W., Yin, Q. et al. Melatonin alleviates ferroptosis triggered by cadmium via regulating ferritinophagy and iron metabolism in spermatogonia. Sci Rep 15, 8910 (2025). https://doi.org/10.1038/s41598-025-93822-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93822-8
Keywords
This article is cited by
-
Overview of Ferroptosis in Cadmium Toxicity
Biological Trace Element Research (2025)